Abstract
Many plant molecules interact with and modulate key regulators of mammalian physiology in ways that are beneficial to health, but why? We propose that heterotrophs (animals and fungi) are able to sense chemical cues synthesized by plants and other autotrophs in response to stress. These cues provide advance warning about deteriorating environmental conditions, allowing the heterotrophs to prepare for adversity while conditions are still favorable.
That plants make substances of benefit to human health has been known for millennia. One-third of the current top 20 drugs on the market are plant derived and every week, it seems, another plant molecule is found to be good for our health. But why should the plant kingdom be such a pharmacological cornucopia? The question is more than of academic interest. The global economy and human health depend largely on our ability to find new and effective medicines. Yet, surprisingly little attention has been given to understanding why plants synthesize so many compounds that provide health benefits to other organisms.
Despite the success of plant molecules as drugs, they are losing favor among drug developers. One reason is the easier patentability of new chemical entities (NCEs), and the other is the perceived “dirtiness” of plant molecules. A molecule is considered “dirty” if it interacts with numerous endogenous proteins. Such compounds presumably are more likely to have negative “off-target” effects than a molecule that specifically targets one protein.
Flying in the face of this dogma are examples of plant molecules that, despite interacting with multiple human enzymes and receptors, are surprisingly safe (Corson and Crews, 2007). These molecules can impart a spectrum of reinforcing health benefits. Take for example salicylic acid (SA), which the Greek physician Hippocrates wrote about in the 5th century: a bitter powder extracted from willow bark that eases aches and pains and reduces fevers. In 1763, Reverend Edward Stone tested the bark of the white willow (Salix alba) for treating fever and concluded that it is a “very efficacious” remedy. Then, a variety of salicylates were isolated from different plants and found to help in the treatment of gout, rheumatic fever, pain, swelling, and arthritis. Today, 45,000 metric tons of the acetylated derivative of SA, also known as “aspirin,” are consumed worldwide each year to treat a variety of human ailments.
Salicylates are just one example of dozens of known plant bioactives that produce wide-ranging health benefits in humans by interacting with more than one endogenous protein. Another interesting bioactive is resveratrol, a small polyphenol produced by numerous plant species in response to stress, which famously is present in red wine. In mammals, resveratrol directly modulates over two dozen enzymes and receptors, is surprisingly nontoxic, and protects rodent models against cancer, atherosclerosis, and diabetes while boosting endurance (Baur and Sinclair, 2006; Westphal et al., 2007). Some proteins are inhibited by resveratrol whereas others are activated in a manner conducive to health (Figure 1). Similarly, green tea polyphenols, including epigallocatechin-3-gallate (EGCG), inhibit the enzyme cyclooxygenase 2 (COX-2), promote cell-cycle arrest, increase apoptosis, disable multidrug resistance pumps, and reportedly provide a large number of health benefits in animal models and in some human epidemiological studies (Khan et al., 2006). Even the common molecule curcumin from turmeric has been found to influence over 60 molecular targets implicated primarily in tumorigenesis (Goel et al., 2008).
Figure 1. Direct Modulation of Key Mammalian Enzymes by Plant Metabolites.
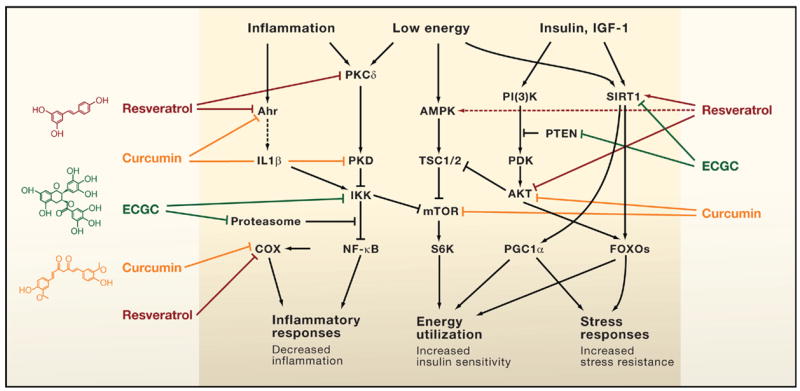
A surprising number of plant molecules in our diet interact with key regulators of mammalian physiology to provide health benefits. Shown are three examples: resveratrol found in numerous plants and concentrated in red wine; curcumin from turmeric; and epigallocatechin-3-gallate (EGCG) in green tea. These compounds modulate key pathways that control inflammation, the energy status of cells, and cellular stress responses in a way that is predicted to increase health and survival of the organism. Such observations raise the question, are these biochemical interactions merely a remnant of what existed in the common ancestor of plants and animals, or is selection maintaining interactions between the molecules of plants and animals? Some interactions activate signaling pathways (arrows) whereas others inhibit them (bars). Solid arrows or bars indicate instances where there is some evidence of a direct interaction of the plant metabolite with a mammalian protein.
Inheritance, Coincidence, or Selection?
Given this wealth of data and the importance of finding new medicines, it is surprising that we understand so little about why many plant molecules are safe and beneficial to health. The main theory is that the biosynthetic pathways for signaling compounds originated in a common ancestor of plants and animals (Kushiro et al., 2003). After the divergence of these two kingdoms, signaling molecules are thought to have been conserved due to structural constraints, such as having the same biosynthetic precursor molecules and needing to interact with protein receptor-binding pockets whose origins also predate the divergence (Kushiro et al., 2003). A particularly notable example of signaling conservation is the role played by fatty acid oxidation products in the wound responses of both plants and animals (Schultz, 2002). These signaling molecules are produced by similar synthetic pathways in plants and animals (e.g., jasmonic acid in plants, prostaglandins in animals), resulting in analogous downstream effects (herbivore resistance in plants, inflammation and immune responses in animals). Interestingly, the biological role of SA in plants as an endogenous negative regulator of jasmonic acid synthesis parallels its ability to inhibit prostaglandin synthesis in mammals by binding to COX-1 and COX-2 (Schultz, 2002). Schultz has suggested that the shared signaling heritage or convergence of plants and animals enables plants to, for example, mount signaling-based chemical defenses against herbivores while allowing herbivores, in turn, to sabotage those defensive measures. This evolutionary back and forth has been termed “phylogenetic espionage” (Schultz, 2002).
Although there is considerable validity to this theory, there is accumulating evidence that this may not be the entire story. There are abundant examples of interactions between plant and animal molecules that cannot be readily explained by the “common origin” hypothesis. For example, why do some plant signaling molecules interact directly with animal enzymes and promote health despite having no apparent homolog or chemical relative in animals? One could argue that these molecular interactions are simply a fortuitous coincidence, with the vast majority of plant molecules being either toxic or producing no benefit to animals. Indeed, given the immensity of the chemical space occupied by plant secondary metabolites, such a view seems plausible. However, several factors suggest that selection, rather than mere coincidence, may be at work. Let us examine, as an example, members of one broad chemical class in plant foodstuffs that confer human health benefits: the polyphenols. The synthesis of polyphenols (and many other phytochemicals) is induced in plants by a variety of environmental stresses. Polyphenol content provides a chemical signature of the state of the environment. This chemical cocktail, when ingested, comes into intimate contact with the receptors and enzymes within the consumer. The fact that stress-induced plant compounds tend to upregulate pathways that provide stress resistance in animals suggests that plant consumers may have mechanisms to perceive these chemical cues and react to them in ways that are beneficial. We have coined the term xenohormesis to explain this phenomenon (from xenos, the Greek word for stranger, and hormesis, the term for health benefits provided by mild biological stress, such as cellular damage or a lack of nutrition).
The Xenohormesis Hypothesis
Our xenohormesis hypothesis proposes that animals and fungi (heterotrophs) have evolved the ability to sense signaling and stress-induced molecules from other species, and that they are under selective pressure to do so (Figure 2). In essence, xenohormesis refers to inter-species hormesis, such that an animal or fungal species uses chemical cues from other species about the status of its environment or food supply to mount a preemptive defense response that increases its chances of survival. But why do we call it xenohormesis instead of hormesis? The reason is that the stress occurs in one organism and the beneficiaries include other organisms that evolved to sense those chemical cues.
Figure 2. The Xenohormesis Hypothesis.
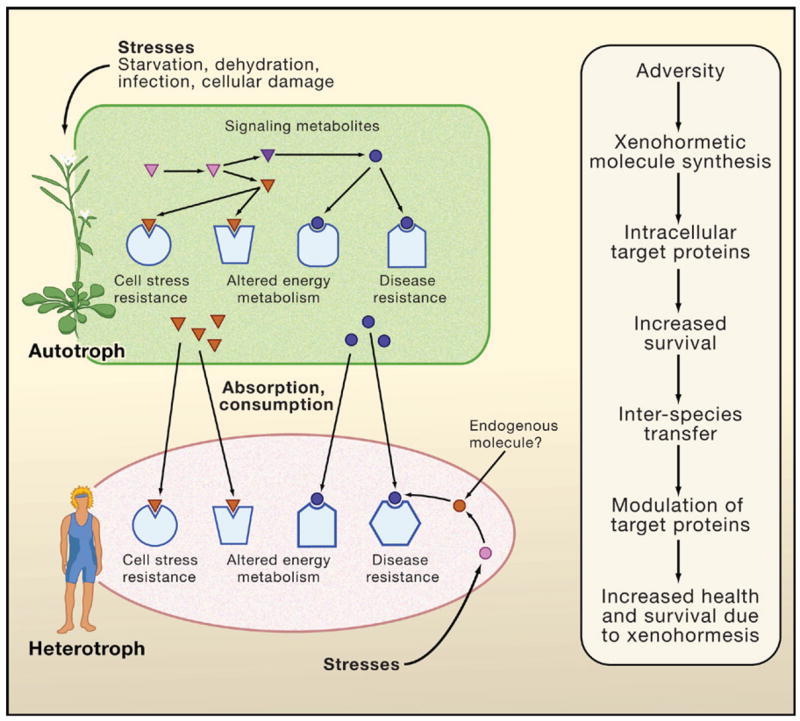
We propose that the common ancestor of plants and animals synthesized polyphenols. Since the divergence of phyla, there has been selection such that heterotrophs (animals and fungi) detect chemical cues about their environment from plants and other autotrophs (that is, organisms that derive energy from light or inorganic chemical reactions). These chemical cues would give the heterotroph advance warning about the deterioration of the environment, allowing them to prepare while conditions are still relatively favorable. The theory predicts that many key mammalian enzymes and receptors will have evolved binding pockets that allow modulation by molecules produced by other species.
When it comes to phytochemical consumption, it is important to distinguish xenohormetic effects from straightforward hormetic effects. The latter may result from low doses of “toxins” that cause a moderate biological stress, presumably cellular damage, thereby inducing a beneficial stress response (Mattson and Cheng, 2006). At least for mammals and their responses to the more common dietary phytochemicals, we consider this unlikely as the primary mode of action. Compounds such as quercetin, a flavonoid found in apples, tea, and onions, and resveratrol, found in wine and peanuts, are abundant in decidedly nontoxic foodstuffs. Even when administered in pure form, resveratrol and quercetin have remarkably low toxicity (Baur and Sinclair, 2006).
We suggest that the majority of health benefits from phytochemical consumption result not from responses to mild cellular damage or from their antioxidant properties, but rather from the evolutionarily adaptive modulation of the enzymes and receptors of stress-response pathways in mammals (Figure 1). Concurrent with the evolution of highly efficient detoxifying mechanisms or toxic food avoidance behaviors, we propose that there has been selective pressure on animals and fungi to use the “information” content of phytochemicals about the status of the environment. Mechanisms that minimize the dangers of toxins could thus have evolved concurrently without loss of the advantageous ability to use phytochemicals as molecular signals. In other words, the xenohormetic and hormetic modes of action are not mutually exclusive, even for the same compound, and may well function synergistically in response to the complex mixtures of phytochemicals in food.
Current evolutionary thinking suggests that when an animal faces adversity, such as reduced food availability or other biological stresses, there is a selective advantage to diverting limited resources away from reproduction and growth into maintenance and defense until the offspring have a better chance of survival. For example, the phenomenon of life-span extension through caloric restriction is thought to be a hormetic response, more specifically a consequence of mechanisms that evolved to promote survival in an environment with poor prospects for reproduction.
But what about the advantages of detecting possible future stresses and increasing one’s biological defenses while resources remain plentiful? As illustrated by the tale of “the ant and the grasshopper,” there is undoubted survival value in preparing for adversity while conditions are still favorable. Take fasting on alternate days, which can impart many of the same health benefits as caloric restriction (Varady and Hellerstein, 2007). One possible interpretation of this result is that this type of fasting replicates a natural circumstance in which increasing food uncertainty “foretells” of impending starvation. A selective advantage lies in the improved survival resulting from getting a head start on defensive preparations, well before starvation occurs.
Stress-induced plant molecules such as resveratrol, butein, and fisetin can induce defense responses in fungi, nematodes, flies, fish, and mice, leading to an extended life span (Westphal et al., 2007). It has been suggested that such molecules are “caloric restriction mimetics” (Howitz et al., 2003). As interpreted in the xenohormesis theory, the molecules send a chemical cue analogous to alternate-day fasting. This cue is an early warning gleaned from the environmental stresses felt by the food supply while that food supply is still available. Indeed, levels of these polyphenols required to extend life span in the laboratory (~10 μM) are detectable in the leaves and fruits of stressed plants.
Secondary Metabolites as Interspecies Signals
To understand the xenohormesis theory, it is helpful to consider why plants synthesize secondary metabolites in the first place. The synthesis of most plant secondary metabolites generally coincides with environmental stresses—UV light, lack of nutrients, disease, and predation—but with rare exceptions, their functions are unknown. SA is one phytochemical whose role in stress responses is well understood. SA serves as an endogenous signal that mediates plant defenses against pathogens and stress by inducing the production of “pathogenesis-related” proteins (Nandi et al., 2003). As noted earlier, SA also confers health benefits on animals, consistent with the xenohormesis hypothesis.
Polyphenols are a major group of plant secondary metabolites that encompass a number of chemical classes, including chalcones, stilbenes, flavones, isoflavones, catechins, and anthocyanidins. Interestingly, the majority of these molecules are synthesized by plants in response to stress. Some of the endogenous roles ascribed to polyphenols—such as UV filters, herbivory deterrents, antioxidants, antibiotics, and fungicides (that is, phytoalexin activity)—are simple chemical activities rather than signaling functions. However, members of the flavonoid class of polyphenols are also known to act as signaling molecules in pollen and seed development, auxin transport, transcription, the cell cycle, and interaction with bacterial symbionts (Taylor and Grotewold, 2005).
Stafford proposed that flavonoids originated as “[plant] physiological regulators or chemical messengers” and that functions such as UV filtration evolved later (Stafford, 1991). In light of emerging evidence for flavonoid signaling (Taylor and Grotewold, 2005), Stafford’s idea now seems prescient. Of particular interest is the finding that a yet-to-be-identified flavonoid-like molecule delays leaf senescence in the model plant Arabidopsis thaliana (Woo et al., 2005). This is reminiscent of life-span extension in fruit flies and budding yeast fed the flavonoid fisetin (Howitz et al., 2003; Wood et al., 2004). Just as some animal lineages have lost the ability to synthesize vitamin C or certain amino acids, perhaps animal phyla in general have lost an ancestral capacity to synthesize flavonoid-like “physiological regulators” while retaining the capacity to respond to them.
Polyphenols: Antioxidants and Signaling Molecules?
Scan the scientific literature and you will likely read that plant polyphenols, including flavonoids, provide health benefits because of their antioxidant activity. Yet many view the data as unpersuasive: Halliwell and colleagues, for example, have described the evidence as “confusing and equivocal” (Halliwell et al., 2005). Most damaging for the antioxidant theory is that the antioxidant capacity of polyphenols does not correlate with their efficacy (Halliwell et al., 2005). Moreover, anti-oxidants as a class of molecules do not provide the life-span extending effects across diverse species that polyphenols do. Similarly damaging is the observation that maximal plasma concentrations of polyphenols, such as resveratrol and quercetin, when provided in the diet are often far lower than the levels required for protection against oxidation (Soleas et al., 1997; Yu et al., 2002). While it is true that the metabolites of polyphenols can reach ten times higher concentrations in the bloodstream, these compounds tend to have decreased antioxidant activity compared to the parent compound (Halliwell et al., 2005). There is increasing evidence that the observed decreases in reactive oxygen species due to polyphenols are an indirect effect, the result of the induction of defense enzymes such as heme oxygenase (Dore, 2005).
Testing the Hypothesis
Unless they generate their own energy, the majority of life forms in the biosphere feed on or live in close proximity to photoautotrophs, that is, plants or other organisms that perform photosynthesis with carbon dioxide as the sole carbon source. In response to changes in their own health and changes in the environment, photoautotrophs generate a variety of secondary metabolites. These molecules could be used by other organisms in the same environment to detect and prepare for environmental changes. In discussing xenohormesis, we have presented evidence from plants and animals, but the concept is by no means limited to these kingdoms. It could equally well apply to chemical stress signals from plants, algae, photosynthetic bacteria, or chemosynthetic bacteria that could be sensed by animals, fungi, some bacteria, some protists, or even parasitic plants.
A long-term goal of such research should be to characterize the stress-induced secondary metabolites of those autotrophs that have diverged widely from land plants and to assess their interactions with the enzymes of the heterotrophs (animals and fungi) that feed on them. For example, mycosporines and mycosporine-like amino acids (MAAs) are secondary metabolites produced by cyanobacteria, among other aquatic microorganisms, that accumulate in the tissues of many marine animals. Synthesis of MAAs in cyanobacteria can be induced by a variety of stresses. The sunscreen and antioxidant functions attributed to MAAs parallel, in many ways, functions attributed to the polyphenols of land plants (Oren and Gunde-Cimerman, 2007). Are the signaling pathways of MAA-consuming organisms modulated by direct protein interactions with MAAs? Are they more receptive to marine MAAs than to “exotic” substances such as polyphenols from land plants? Answering such questions would test the general applicability of the xenohormesis idea as well as, assuming its validity, providing insight into the evolutionary mechanisms that underlie it.
We propose that xenohormetic selective pressure could have been around ever since the various phyla diverged and influenced the structure of animal enzymes and receptors. We do not mean to suggest that autotrophs produce metabolites to benefit the heterotrophs, although there are clearly examples where it is advantageous to fool the consumer (Schultz, 2002). The xenohormesis hypothesis predicts that we will find conserved domains in enzymes and receptors that do not interact with any endogenous molecule. On the other hand, given evolutionary considerations and the fact that there are likely to be a limited number of regulatory mechanisms for any given receptor or enzyme, xenohormetic molecules are predicted to bind to many of the same receptor sites as endogenous regulators do. Xenohormetic molecules could act as an antagonist against one enzyme or receptor and be an agonist against another. As we propose that these interactions result from selective pressures for particular phenotypic outcomes, we would predict that their effects on the proteins of a given signaling pathway would tend to reinforce one another (see Figure 1).
A limited number of plant polyphenol-binding sites on mammalian proteins have been characterized. One theme that emerges is the affinity of flavonoids (such as quercetin) and stilbenes (such as resveratrol) for the nucleotide-binding sites of protein kinases. There are notable examples for proteins that have nucleotide substrate-binding sites, where structural (Gledhill et al., 2007) or kinetic (Howitz et al., 2003) evidence indicates that polyphenols do not compete with the enzyme’s nucleotide substrates and instead bind elsewhere on the protein. We suggest that this is at least consistent with these interactions being driven by selective pressures rather than simply by a coincidental structural resemblance to nucleotides. In considering the 14 solved structures of resveratrol-protein or quercetin-protein complexes, Gledhill et al. (2007) did not identify a conserved polyphenol interaction domain but concluded simply that in all cases the polyphenols were bound in hydrophobic pockets “predominantly by means of van der Waals contacts and H-bonds involving the hydroxyl groups.” Clearly, identification of polyphenol interaction domains, if they exist, must await further structural work.
It should be possible to test the xenohormesis hypothesis at the level of whole organisms and their ecology. For example, a testable prediction of xenohormesis is that stressing a plant such as Arabidopsis with heat or light would lengthen the life span of insects, such as aphids, that feed on it. The life span of aphids feeding on the leaves of the herb Shepherd’s Purse (Capsella bursapastoris) is known to be extended when the plant is water-stressed due to root predation by beetle larvae (Gange and Brown, 1989). The greatest difficulty with such experiments may lie in designing sufficient controls to allow for an unambiguous interpretation. For example, one must be sure to distinguish xenohormetic lifespan effects from those that might arise from simple caloric restriction or lower levels of a particular nutrient or toxin. The genetic manipulations that are possible in Arabidopsis in the realm of polyphenol biogenesis may prove especially useful in this regard.
We propose that animals and fungi may have retained the ability to sense stress signaling molecules that their distant ancestors once synthesized to modulate their enzymes and receptors during adversity. These stress-induced molecules have been consistently encountered by animals and fungi in the context of particular environmental stresses, such that evolution may have favored the preservation of enzyme binding and other types of molecule-sensing capacities in heterotrophs. As we achieve a more detailed understanding of the numerous interactions of plant molecules with mammalian proteins, there may come a day when calling a plant molecule “dirty” will be interpreted as a compliment.
References
- Baur JA, Sinclair DA. Nat Rev Drug Discov. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- Corson TW, Crews CM. Cell. 2007;130:769–774. doi: 10.1016/j.cell.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore S. Neurosignals. 2005;14:61–70. [Google Scholar]
- Gange AC, Brown VK. Oecologia. 1989;81:38–42. doi: 10.1007/BF00377007. [DOI] [PubMed] [Google Scholar]
- Gledhill JR, Montgomery MG, Leslie AGW, Walker JE. Proc Natl Acad Sci USA. 2007;104:13632–13637. doi: 10.1073/pnas.0706290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel A, Kunnumakkara AB, Aggarwal BB. Biochem Pharmacol. 2008;75:787–809. doi: 10.1016/j.bcp.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Halliwell B, Rafter J, Jenner A. Am J Clin Nutr. 2005;81:268S–276S. doi: 10.1093/ajcn/81.1.268S. [DOI] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, et al. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- Khan N, Afaq F, Saleem M, Ahmad N, Mukhtar H. Cancer Res. 2006;66:2500–2505. doi: 10.1158/0008-5472.CAN-05-3636. [DOI] [PubMed] [Google Scholar]
- Kushiro T, Nambara E, McCourt P. Nature. 2003;422:122. doi: 10.1038/422122a. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Cheng A. Trends Neurosci. 2006;29:632–639. doi: 10.1016/j.tins.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Nandi A, Krothapalli K, Buseman CM, Li M, Welti R, Enyedi A, Shah J. Plant Cell. 2003;15:2383–2398. doi: 10.1105/tpc.015529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A, Gunde-Cimerman N. FEMS Microbiol Lett. 2007;269:1–10. doi: 10.1111/j.1574-6968.2007.00650.x. [DOI] [PubMed] [Google Scholar]
- Schultz JC. Integr Comp Biol. 2002;42:454–462. doi: 10.1093/icb/42.3.454. [DOI] [PubMed] [Google Scholar]
- Soleas GJ, Diamandis EP, Goldberg DM. Clin Biochem. 1997;30:91–113. doi: 10.1016/s0009-9120(96)00155-5. [DOI] [PubMed] [Google Scholar]
- Stafford HA. Plant Physiol. 1991;96:680–685. doi: 10.1104/pp.96.3.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor LP, Grotewold E. Curr Opin Plant Biol. 2005;8:317–323. doi: 10.1016/j.pbi.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Varady KA, Hellerstein MK. Am J Clin Nutr. 2007;86:7–13. doi: 10.1093/ajcn/86.1.7. [DOI] [PubMed] [Google Scholar]
- Westphal CH, Dipp MA, Guarente L. Trends Biochem Sci. 2007;32:555–560. doi: 10.1016/j.tibs.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Woo HH, Jeong BR, Hawes MC. Biotechnol Lett. 2005;27:365–374. doi: 10.1007/s10529-005-1521-7. [DOI] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Yu C, Shin YG, Chow A, Li Y, Kosmeder JW, Lee YS, Hirschelman WH, Pezzuto JM, Mehta RG, van Breemen RB. Pharm Res. 2002;19:1907–1914. doi: 10.1023/a:1021414129280. [DOI] [PubMed] [Google Scholar]


