Abstract
Trim33 (Tif1γ, ectodermin, moonshine), a member of the TIF1 family of transcriptional coactivators and corepressors, is a large nuclear protein that contains an N-terminal tripartite (Trim) domain composed of a RING domain, two B-box domains and a coiled coil domain. It has been suggested that Trim33 (Ectodermin) mediates ectodermal induction in the Xenopus by functioning as a Smad4 ubiquitin ligase, while in the zebrafish Trim33 (moonshine) has been reported to act as a R-Smad binding protein in induction of erythroid differentiation. Since the developmental role of Trim33 in mammals is currently unknown, we generated mice carrying the conditional Trim33 (Trim33FX) allele by flanking exons 2–4 encoding most of the functionally critical N-terminal tripartite domain by loxP sites. We confirmed the null genotype by using the EIIa-Cre transgenic approach to create mice that lack exons 2–4. Embryos deficient in Trim33 die during early somitogenesis, demonstrating that Trim33 plays an important non-redundant role in mammalian embryonic development.
Trim33 (Tif1γ) is one of 70 tripartite motif-containing Trim proteins(Yan et al., 2004). Together with Trim24 (Tif1α) and Trim28 (Tif1β), it forms a transcription intermediary factor-1 (Tif-1) subfamily of transcriptional regulators(Venturini et al., 1999). While Tif-1 proteins all share several characteristic functional domains, e.g., amino terminal Trim domains, carboxy terminal PHD finger and Bromo domains, it is thought that their biological functions are quite divergent. For instance, Trim28, but neither Trim24 nor Trim33, has been shown to interact with members of the KRAB zinc finger proteins(Friedman et al., 1996; Kim et al., 1996). Moreover, Trim28 is an intrinsic component of the histone deacetylase N-CoR1/HDAC3 complex(Underhill et al., 2000), while Trim24 does not bind nuclear hormone receptors, but it has been shown to function as a coactivator of the retinoid acid receptor(Fraser et al., 1998). Whereas mouse embryos deficient in Trim28 die soon after implantation, Trim24 is not essential for embryogenesis, but was found to be a potent liver-specific tumor suppressor(Cammas et al., 2000; Khetchoumian et al., 2007). Much less is known about the biological role of Trim33, particularly in mammals. This is largely due to the fact that knockout mice have not yet been developed. However, it was recently shown that in Xenopus Trim33 functions as an E3 ubiquitin-protein ligase promoting Smad4 degradation via the ubiquitin proteasome pathway(Dupont et al., 2005). In the zebrafish, Trim33 was shown to be required for erythroid lineage-specific control of hematopoietic gene expression(Ransom et al., 2004). Subsequently, it was shown that R-Smad/Trim33 interaction is required for Tgf-β-dependent erythroid differentiation. Interestingly, it seems that in this context Trim33 does not target Smad4 for degradation, but rather competes with it by binding activated R-Smads(He et al., 2006).
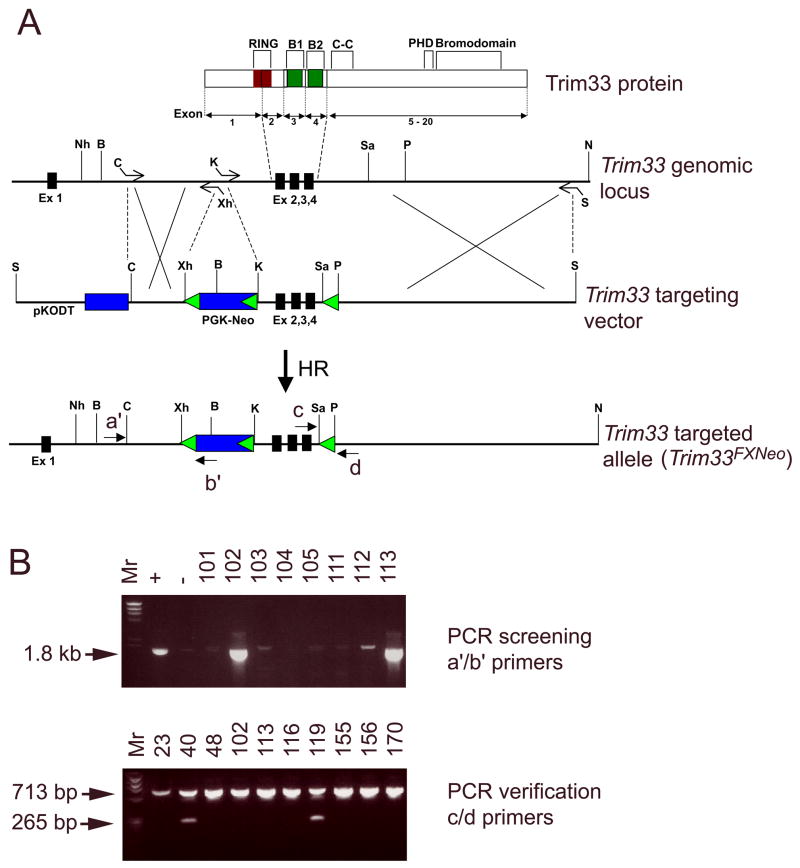
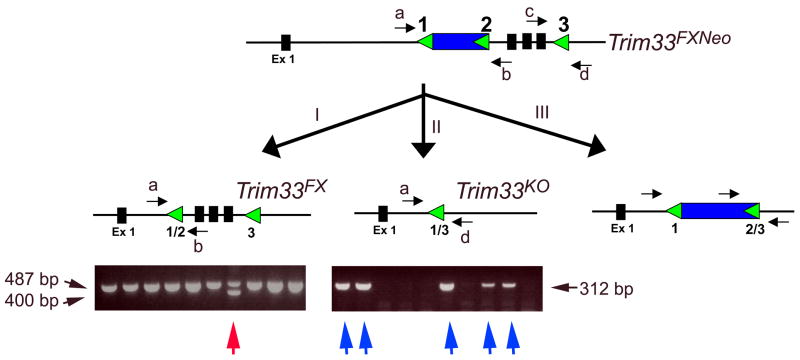
In order to address the role of Trim33 in mammalian embryogenesis, we generated mice harboring the conditional knockout allele for Trim33. Our strategy was to flank exons 2–4 encoding the functionally critical RING and B1 and B2 Box domains by loxP sites. The short (1.6 kb) and long (8.2 kb) arms of the targeting vector were PCR amplified from a Bac genomic DNA. The loxPNeoloxP cassette was inserted into intron 1 and a single loxP site was inserted into intron 4 (Fig. 1). As a negative selection marker we used the diphteria toxin (DT) gene. The linearized targeting vector was electroporated into TVB2 embryonic stem cells as described(Yang and Kaartinen, 2007). Fifteen out of 261 G418-resistant colonies were targeted to the correct locus, however, only 3 of them contained the 3′ loxP site. All three correctly targeted ES clones were able to produce highly chimeric male mice, which in turn were potent germ line transmitters. To remove the Neo selection marker and to generate a presumed knockout allele for Trim33, we crossed the Trim33FXNeo/FXNeo mice (homozygotes for the targeting vector) with EIIa-Cre transgenic mice(Xu et al., 2001). In these mice, Cre is expressed under the control of the adenoviral EIIa promoter that targets expression of the Cre recombinase to the early mouse embryo (Lakso et al., 1996). The EIIa-Cre transgene creates both partial and complete recombinations and therefore EIIa-Cre transgenic mice can be used both as a deleter mouse to generate knockout alleles and to remove selection markers (e.g., loxP-pGKNeo-loxP) when a triple loxP-strategy is used (Holzenberger et al., 2000; Xu et al., 2001). The obtained mosaic male mice progeny were subsequently crossed with wild-type female mice to obtain the floxed (Trim33FX) and null (Trim33KO) alleles (Fig. 2). Homozygote Trim33FXFX mice were viable and fertile, and they did not display any recognizable phenotypes.
Figure 1. Trim33 targeting vector and screening of ES colonies.
A, a schematic presentation of the Trim33 protein domains, Trim33 genomic locus depicting a segment from exon1 to intron 4, the Trim33 targeting vector and the Trim33 targeted allele. B, An example of a PCR screen (upper panel); positive clones #102 and #113 can be easily identified. PCR analysis using c/d primers and template DNA from clones that suggested the correct targeting demonstrated that only clones number #40, #119 and #259 (not shown) had retained the 3′ loxP site (lower panel). The wild type allele produces a 713-bp amplification product. Since the strategy to insert the 3′ loxP site into intron 4 involved the replacement of a 553-bp SacI-PstI fragment with a loxP site, the mutant allele gives rise only to a 265-bp amplification product.
Figure 2. Generation of mice carrying the floxed (Trim33FX) and knockout (Trim33KO) Trim33 alleles.
Transgenic EIIa-Cre mice were crossed with mice homozygous for the targeting vector. The obtained mosaic males were further crossed with wild-type females to obtain Trim33FX/WT mice (type I recombination; red arrow) and Trim33KO/WT mice (type II recombination; blue arrows).
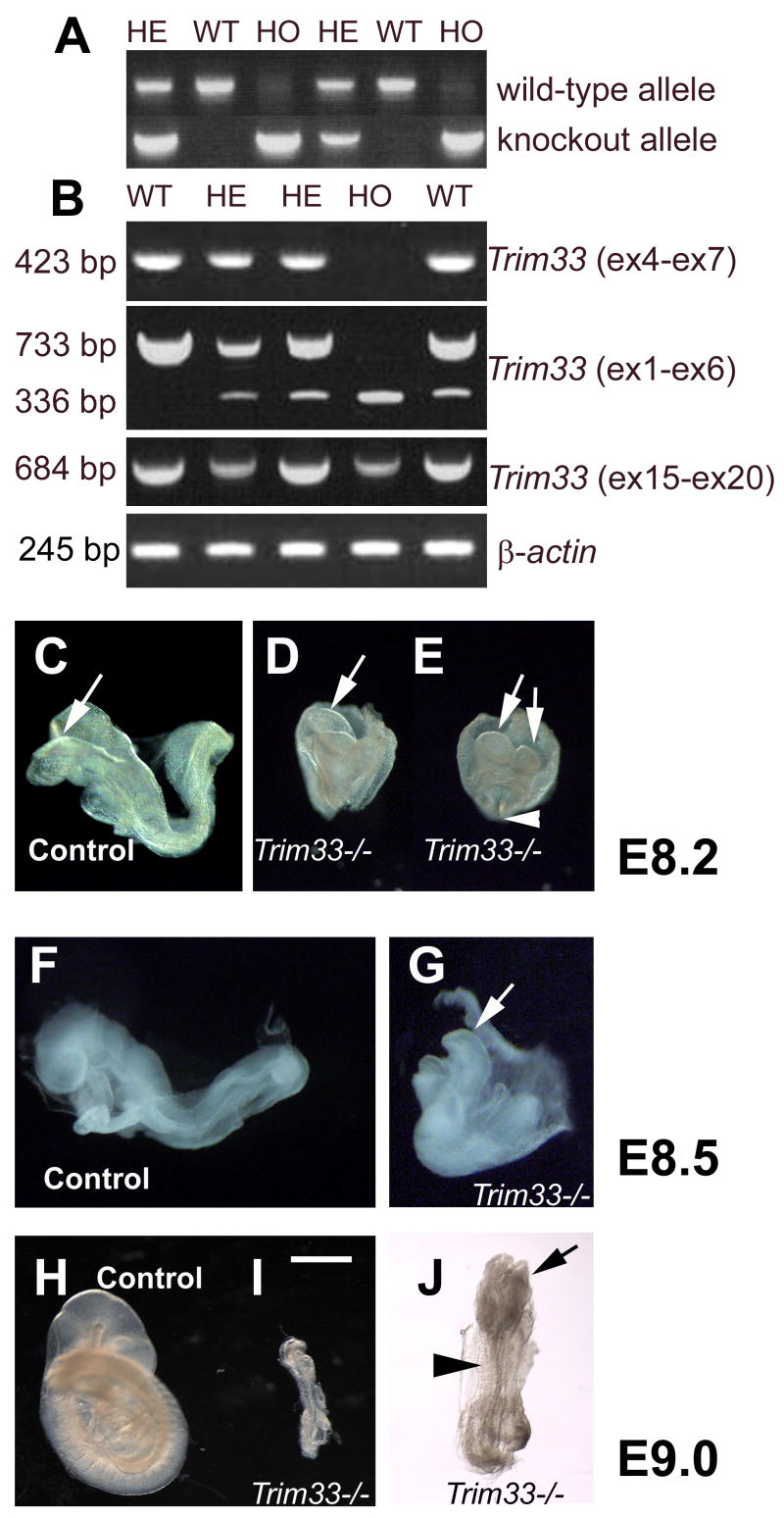
To confirm that the Trim33KO allele encoded the true null allele and to provide initial information about the biological role of Trim33 during embryogenesis, we inter-crossed the heterozygote Trim33KO/WT mice to obtain homozygote Trim33KO/KO mice (Fig 3A). Genotype analyses of newborn offspring revealed that all the homozygote mutant pups died during gestation (Table 1). To examine the time frame during which embryonic lethality occurred, we harvested embryos at different time points and discovered that at E9 the mutant embryos displayed a dramatic developmental delay when compared to controls (Table 1 and Fig. 3H–J). Nevertheless, they had formed a body axis, displayed head folds and the neural tube and showed 5–6 somite pairs. We were unable to discover any living mutant embryos after E9.5. At E8.0–8.5 (3–6 somite pairs in controls), the Trim33 mutant embryos were aligned at the base of the yolk sac, and while they had formed the anterior-posterior body axis and identifiable anterior structures, e.g., head folds (arrows in Fig. 3D, E and G), it was difficult to identify any other embryonal structures. RT-PCR using primers with target sequences in exons 4 and 7 did not produce any detectable amplification product, while the heterozygote and wild-type samples showed the expected 423-bp fragment (Fig. 3B). This is concordant with the lack of sequences encoded by exon 4. To further validate the Trim33KO/KO allele, we used exon 1- and exon 6-specific primers. The wild-type and homozygote Trim33KO/KO alleles produced the expected 733-bp and 366-bp products, respectively, while the heterozygote samples gave rise to both PCR fragments (Fig. 3B). RT-PCR analysis using primers specific for 3′ exons 15 and 20 produced the expected 684-bp fragment from wildtype, heterozygote (Trim33KO/WT) and homozygote (Trim33KO/KO) samples suggesting that the mutated allele lacking exons 2–4 is able to produce a stable mRNA (Fig. 3B). To conclude, our RT-PCR analyses demonstrated that the mRNA encoded by Trim33KO/KO allele lacks sequences encoded by exons 2–4, and that it is highly likely that the phenotype observed in homozygote samples results from a loss of Trim33 function, particularly since most of the possible splicings, e.g., exon 1 to 4, will lead to a frameshift and premature translational stop. These mutant mRNAs would produce only the very N-terminal peptide encoded by Trim33 exon 1. Only the splicing from exon 1 to 9 or exon 1 to 13 would maintain the open reading frame. In these unlikely hypothetical cases the protein product would lack the tripartite motif, but would contain the C-terminal PHD and Bromo domains. Based on the current knowledge, it is impossible to say whether these aberrant proteins lacking the functionally important tripartite motif would posses any biological activity.
Figure 3. Development of embryos deficient in Trim33 is arrested at the early somitogenic stage.
A, PCR genotyping of embryos from the crossing between Trim33KO/WT males and females at E8.5. HE=heterozyggotes, WT=wildtype, HO=homozygotes. B, RT-PCR analysis of embryos harvested at E8.5 using primers for exons 4 and 7 demonstrates that Trim33KO/KO embryos do not display any detectable mRNA product, while wild-type and heterozygote controls show an expected 423-bp PCR product (B, upper panel). Corresponding analysis using primers for exons 1 and 6 show that the wild-type sample produces the expected 733-bp PCR product, the homozygote mutant sample produces the expected 336-bp product, while the heterozygote samples gives rise to both 733-bp and 336-bp products (second panel from the top). Corresponding analysis using primers for exons 15 and 20 shows that both the wild-type and mutant alleles produce a 684-bp amplification product (second panel from bottom). β-actin specific primers produced a 245-bp amplification product with comparable intensity from all samples (bottom panel). At E8.2–8.5, Trim33 null mutants (D, E, G) demonstrate retarded development when compared to controls (C,F) . D and E depict both lateral and frontal images of the same embryo, respectively. I, Development of the Trim33 −/− embryos is arrested at the early somitogenic stage (E9.0). J, A high power picture of a Trim33 −/− embryo shown in (I). H, Control littermate. Arrows in C, D, E, G and J point to head folds, white arrowhead in E points to the primitive streak, and black arrowhead in J points to the somites.
Table 1.
Genotype analysis of offspring from Trim33+/− intercross.
| Developmental stage | Somite pairs | Trim33−/− | Trim33+/− | Wild-type |
|---|---|---|---|---|
| Post-natal | 0 | 12 | 2 | |
| E9.5 | 1 | 8 | 0 | |
| E9.0 | 2 | 4 | 4 | |
| E8.25 – E8.5 | 5–6 | 6 | 4 | 2 |
| E8.0 | 3 | 4 | 7 | 4 |
A recent publication of Dupont and coworkers showed that in Xenopus embryos XTrim33 (Ectodermin) plays a critical role in controlling TGF-β responses during gastrulation(Dupont et al., 2005). Specifically, these authors demonstrated that XTrim33 is required for specification of the ectoderm and for restricting the mesoderm-inducing activity of TGF-βs. Our finding that Trim33 null embryos die either during somitogenesis demonstrates that Trim33 is also required for early embryogenesis in mammals. If the role of Trim33 is to negatively regulate Tgf-β signaling during embryogenesis, one would expect that Trim33 null mutants would display phenotypes that are consistent with amplified Tgf-β signaling. The observed external Trim33KO/KO phenotype is very similar to that seen in transgenic mouse embryos that demonstrate ectopic expression of the Tgf-β type II receptor (Zwijsen et al., 1999). Another study recently demonstrated that abrogation of Drap1, a negative regulator of Nodal, leads to aberrant Nodal signaling and early embryonic lethality(Iratni et al., 2002). Interestingly, similar to Trim33KOKO mutants, embryos that are heterozygous for Nodal and lack Drap1 undergo gastrulation, but die at E9.5. Subsequent studies will show whether the observed phenotype results from defects in ectodermal spefication or some other catastrophic mechanistic failure during early embryogenesis. Moreover, mice carrying the floxed Trim33 allele (Trim33FX) as described herein can be used to investigate the role of this gene later in development during organogenesis, as well as during post-natal life.
Methods
Generation of mice carrying the floxed and knockout Trim33 alleles
RP2468C16 Bac DNA was used as a template to PCR amplify both short and long arms as ClaI-XhoI and KpnI-SalI fragments, respectively. High-fidelity Supermix (Qiagen) polymerase was used for amplification, and the generated arms were sequenced to verify that no PCR-generated mutations were introduced. A single loxP site was inserted into intron 4 by replacing a small 553-bp SacI-PstI fragment with a loxP sequence flanked by SacI and PstI restriction sites. Subsequently a loxPNeoloxP cassette was inserted as a XhoI-NotI fragment into the pKODT plasmid. A long arm containing the single loxP sequence in intron 4 was inserted as a KpnI-SalI fragment into loxPNeoloxP/pKODT, and finally a short arm was added into the construct as a ClaI-XhoI fragment. The targeting vector was electroporated into TVB2 mouse ES cells, and recombinant ES cell clones were selected with G418 as described(Kaartinen et al., 2004; Kaartinen et al., 1995). Mouse chimeras were generated by injecting correctly targeted ES clones into C57BL/6J mouse blastocysts. The floxed Trim33 allele (Trim33FX) and the knockout Trim33 allele (Trim33KO) were generated by crossing female mice homozygous for the targeting vector with EIIa-Cre transgenic male mice(Xu et al., 2001), which were obtained from Jackson Laboratories (Maine).
PCR screening, clone verification and genotyping
ES cell DNAs were first screened for correct targeting by PCR using a forward primer (a′) 5′ CACGACACAAAGAACTGTAG 3′ and a reverse primer (b′) 5′ CAAGCAAAACCAAATTAAGG 3′. Subsequently, the presence of the single loxP site in intron 4 was verified by PCR using a forward primer (c) 5′ CATTGTGCTTCACCTCCTCCTCTTCG 3′ and a reverse primer (d) 5′ GGGAGGGAAAATCTGGCTGAA 3′. Trim33FX mice were genotyped using forward and reverse primers (a) 5′ GCACCTTGATGAGATCTTCCTCCTCC 3′ and (b) 5′ GACGACATACTGGACACCGTA 3′, respectively, while Trim33KO mice were genotyped using forward and reverse primers (a) 5′ GCACCTTGATGAGATCTTCCTCCTCC 3′ and (d) 5′ GGAGGGAAAATCTGGCTGAA 3′, respectively.
Timed matings and embryos analyses
Mice were mated during the dark period of the controlled light cycle. Female mice acquiring vaginal plugs were designated as day 0. At the time interval indicated in respective figures (E8 to E9), females were euthanized by CO2 and embryos were extracted in PBS (Invitrogen) followed by further analyses. All studies and procedures performed on mice were carried out at the Animal Care Facility of the Saban Research Institute, and were approved by the CHLA Animal Care and Use Committee (IACUC). The mice were maintained in mixed genetic backgrounds.
RT-PCR
Total RNA was isolated from E8 embryos using the RNeasy mini kit (Qiagen), and cDNAs were synthesized by the Omniscript reverse transcription kit (Qiagen) according to the manufacturers’ protocols. Subsequently, the cDNAs were analyzed by PCR for Trim33 expression using the following primer pairs. β-actin was used as a quality and loading control.
| Trim33-ex4-S | 5′GAGTCTGTTGGAACATCTGGTCAGCG3′ |
| Trim33-ex7-AS | 5′GGCCTGTGATATCATTCTGCTGCTGT3′ |
| Trim33-ex1-S | 5′GGTGTGTCAGCAGAGCTTGCA3′ |
| Trim33-ex6-AS | 5′GATAAGGGTGAAGATGGCCACT3′ |
| Trim33-ex15-S | 5′ACCTCATGCACAGGTCGGCAAGGAT3′ |
| Trim33-ex20-AS | 5′GCTCAAACTCTGGCAAAGGAGTGAAG3′ |
| β-actin-S | 5′GTGGGCCGGTCTAGGCACCAA3′ |
| β-actin-AS | 5′CGGTTGCCTTAGGGTTCAGG3′ |
Acknowledgments
We thank K. Chiu from the CHLA mouse genome core for technical assistance. This study was financially supported by grants from the NIH (HL074862 and DE013085 to VK).
References
- Cammas F, Mark M, Dolle P, Dierich A, Chambon P, Losson R. Mice lacking the transcriptional corepressor TIF1beta are defective in early postimplantation development. Development. 2000;127:2955–2963. doi: 10.1242/dev.127.13.2955. [DOI] [PubMed] [Google Scholar]
- Dupont S, Zacchigna L, Cordenonsi M, Soligo S, Adorno M, Rugge M, Piccolo S. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell. 2005;121:87–99. doi: 10.1016/j.cell.2005.01.033. [DOI] [PubMed] [Google Scholar]
- Fraser RA, Heard DJ, Adam S, Lavigne AC, Le DB, Tora L, Losson R, Rochette-Egly C, Chambon P. The putative cofactor TIF1alpha is a protein kinase that is hyperphosphorylated upon interaction with liganded nuclear receptors. J Biol Chem. 1998;273:16199–16204. doi: 10.1074/jbc.273.26.16199. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ., III KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massague J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- Holzenberger M, Lenzner C, Leneuve P, Zaoui R, Hamard G, Vaulont S, Bouc YL. Cre-mediated germline mosaicism: a method allowing rapid generation of several alleles of a target gene. Nucleic Acids Res. 2000;28:E92. doi: 10.1093/nar/28.21.e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iratni R, Yan YT, Chen C, Ding J, Zhang Y, Price SM, Reinberg D, Shen MM. Inhibition of excess nodal signaling during mouse gastrulation by the transcriptional corepressor DRAP1. Science. 2002;298:1996–1999. doi: 10.1126/science.1073405. [DOI] [PubMed] [Google Scholar]
- Kaartinen V, Dudas M, Nagy A, Sridurongrit S, Lu MM, Epstein JA. Cardiac outflow tract defects in mice lacking ALK2 in neural crest cells. Development. 2004;131:3481–3490. doi: 10.1242/dev.01214. [DOI] [PubMed] [Google Scholar]
- Kaartinen V, Voncken JW, Shuler C, Warburton D, Bu D, Heisterkamp N, Groffen J. Abnormal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelial-mesenchymal interaction. Nat Genet. 1995;11:415–421. doi: 10.1038/ng1295-415. [DOI] [PubMed] [Google Scholar]
- Khetchoumian K, Teletin M, Tisserand J, Mark M, Herquel B, Ignat M, Zucman-Rossi J, Cammas F, Lerouge T, Thibault C, Metzger D, Chambon P, Losson R. Loss of Trim24 (Tif1alpha) gene function confers oncogenic activity to retinoic acid receptor alpha. Nat Genet. 2007;39:1500–1506. doi: 10.1038/ng.2007.15. [DOI] [PubMed] [Google Scholar]
- Kim SS, Chen YM, O’Leary E, Witzgall R, Vidal M, Bonventre JV. A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc Natl Acad Sci USA. 1996;93:15299–15304. doi: 10.1073/pnas.93.26.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom DG, Bahary N, Niss K, Traver D, Burns C, Trede NS, Paffett-Lugassy N, Saganic WJ, Lim CA, Hersey C, Zhou Y, Barut BA, Lin S, Kingsley PD, Palis J, Orkin SH, Zon LI. The zebrafish moonshine gene encodes transcriptional intermediary factor 1gamma, an essential regulator of hematopoiesis. PLoS Biol. 2004;2:E237. doi: 10.1371/journal.pbio.0020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill C, Qutob MS, Yee SP, Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem. 2000;275:40463–40470. doi: 10.1074/jbc.M007864200. [DOI] [PubMed] [Google Scholar]
- Venturini L, You J, Stadler M, Galien R, Lallemand V, Koken MH, Mattei MG, Ganser A, Chambon P, Losson R, de TH. TIF1gamma, a novel member of the transcriptional intermediary factor 1 family. Oncogene. 1999;18:1209–1217. doi: 10.1038/sj.onc.1202655. [DOI] [PubMed] [Google Scholar]
- Xu X, Li C, Garrett-Beal L, Larson D, Wynshaw-Boris A, Deng CX. Direct removal in the mouse of a floxed neo gene from a three-loxP conditional knockout allele by two novel approaches. Genesis. 2001;30:1–6. doi: 10.1002/gene.1025. [DOI] [PubMed] [Google Scholar]
- Yan KP, Dolle P, Mark M, Lerouge T, Wendling O, Chambon P, Losson R. Molecular cloning, genomic structure, and expression analysis of the mouse transcriptional intermediary factor 1 gamma gene. Gene. 2004;334:3–13.:3–13. doi: 10.1016/j.gene.2004.02.056. [DOI] [PubMed] [Google Scholar]
- Yang LT, Kaartinen V. Tgfb1 expressed in the Tgfb3 locus partially rescues the cleft palate phenotype of Tgfb3 null mutants. Dev Biol. 2007;312:384–395. doi: 10.1016/j.ydbio.2007.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwijsen A, Goumans MJ, Lawson KA, Van Rooijen MA, Mummery CL. Ectopic expression of the transforming growth factor beta type II receptor disrupts mesoderm organisation during mouse gastrulation. Dev Dyn. 1999;214:141–151. doi: 10.1002/(SICI)1097-0177(199902)214:2<141::AID-AJA4>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]