Abstract
A cell’s decision to undergo meiosis is regulated by multiple signals. In budding yeast, these signals include mating type status, nutrient starvation, and respiration; the need for respiration is often manifested as a requirement for a non-fermentable carbon source. We have dissected the roles of respiration and carbon source in promoting entry into the meiotic program. This analysis revealed that respiration is needed throughout meiosis, but a non-fermentable carbon source is necessary only prior to the meiotic nuclear divisions. A non-fermentable carbon source serves several roles during the early stages of meiosis. It is required for PolII transcription, for DNA replication, and recombination. Finally, while the global down-regulation of transcription and lack of DNA replication in non-respiring cells could be due to a lack of energy, we show that the inability to induce genes initiating entry into the meiotic program is not. We propose that a separate respiration-sensing pathway governs meiotic entry.
Results
A non-fermentable carbon source is required for execution of the meiotic program
We first examined which steps of meiosis require an exogenous carbon source. Cells were grown in YPA (see Supplemental Experimental Procedures) and then transferred into standard sporulation medium containing potassium acetate (KAc), or carbon-free medium containing potassium chloride (KCl), in which cells can only oxidize internal stores of biomolecules but cannot derive energy from the medium. As expected [1], cells sporulated efficiently in KAc but not in KCl (Figure 1A). We next tested whether induction of the early meiotic genes allowed sporulation in KCl by utilizing a strain bearing an allele of the protein kinase Ime2, ime2-as, which can be reversibly inhibited by the ATP analog 1-NA-PP1 [2]. In the presence of the inhibitor, IME1, a transcription factor governing entry into meiosis [3], and other early meiotic genes are expressed, but premeiotic DNA replication and subsequent meiotic events are not initiated [2] (compare also Supplemental Figures 1A and 3A). When the inhibitor was washed out of KAc cultures, cells sporulated efficiently only when released into KAc, whereas tetranucleate formation was decreased by more than 7-fold in cells released into KCl medium (Figure 1B). These results show that a non-fermentable carbon source is needed for progression through meiosis after induction of the early meiotic genes.
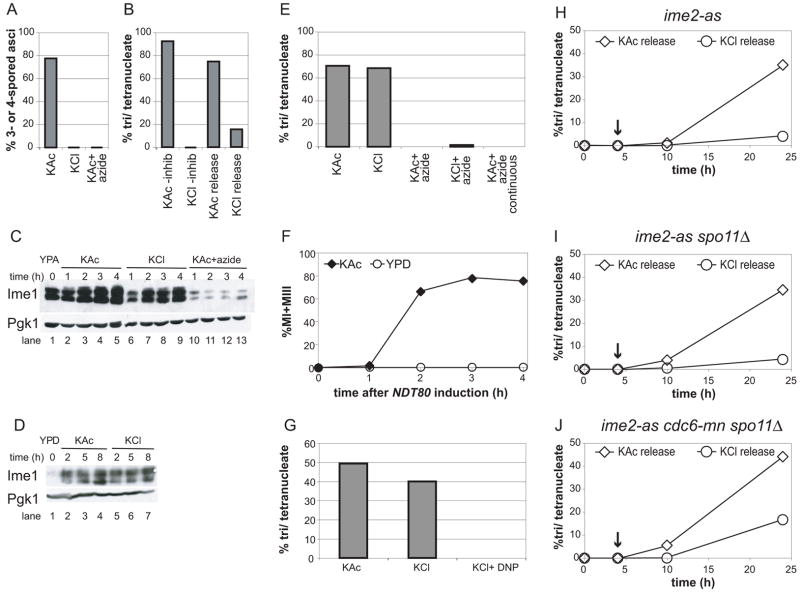
Figure 1. A carbon source is required for entry into and progression through the meiotic program but dispensable after prophase.
A) Formation of 3 and 4-spored asci of WT (A19413) cells pre-grown in YPA and then transferred to KAc, KCl, or KAc+ azide, was determined after 24 hours.
B) Nuclear divisions of the ime2-as strain (A12113) in KAc or KCl without inhibitor (-inhib), or in KAc or KCl following 4 hours’ inhibition of Ime2 in KAc (KAc release and KCl release). Nuclear divisions were analyzed after 24 hours by DAPI staining.
C) Ime1-3HA protein levels of cells shown in (A) during pre-growth in YPA (lane 1) and incubation in KAc (lanes 2-5), KCl (lanes 6–9), or KAc+azide (lanes 10–13) for the indicated times. Pgk1 is shown as a loading control.
D) Ime1-3HA protein levels of WT (A19413) cells after growth to saturation in YPD (lane 1) followed by incubation in KAc (lanes 2–4) or KCl (lanes 5–7) for the indicated times. Pgk1 is shown as a loading control.
E) Nuclear divisions were determined in GAL-NDT80 cells (A14201) incubated in KAc or KCl with or without sodium azide after 24 hours by DAPI staining. Cells were incubated continuously in 5mM azide before and after NDT80 induction (KAc+azide continuous), or incubated in KAc and then shifted to KAc or KCl with (+azide) or without azide at the time of NDT80 induction. NDT80 was induced after 6 hours’ incubation in KAc.
F) GAL-NDT80 (A14201) cells were first incubated in KAc for 6 hours. The percentage of cells having completed one or both meiotic nuclear divisions following NDT80 induction in KAc or YPD was determined by DAPI staining at the indicated times.
G) GAL-NDT80 (A14201) cells were incubated in KAc for 6 hours and then maintained in KAc or shifted to KCl with or without 10mM dinitrophenol (DNP) one hour prior to NDT80 induction as in (E). Nuclear divisions were determined after 24 hours by DAPI staining.
H–J) The percentage of tri and tetranucleate cells was determined in H) ime2-as (A12113), I) ime2-as spo11Δ or J) ime2-as cdc6-mn spo11Δ A17082) strains at the indicated times by DAPI staining. Strains were released into KAc or KCl following
Ime2 inhibition in KAc. The arrow indicates the time at which the inhibitor was washed out.
To determine whether Ime1 production was affected by the absence of a carbon source, we compared Ime1 protein levels between cells incubated in KAc and KCl. Ime1 accumulated to similar levels under both conditions (Figure 1C), indicating cells did not fail to undergo meiosis in KCl because of defects in Ime1 expression. Accordingly, premeiotic DNA replication occurred in KCl after prolonged (10 hr) incubation (Supplemental Figure 1B). Ime1 induction was also observed when cells were transferred to KAc or KCl medium after pre-growth in rich (YPD) medium, in which Ime1 expression is minimal (Figure 1D). Treatment of cells with sodium azide, an inhibitor of cytochrome C oxidase, prevented Ime1 production and sporulation (Figure 1A, C). These results indicate that respiration, but not a carbon source, is needed for Ime1 production. A non-fermentable carbon source, however, is required at the time of or following Ime2 function.
The meiotic nuclear divisions require respiration but not a carbon source
Next we determined whether a non-fermentable carbon source was needed after the completion of premeiotic DNA replication and recombination. To this end we utilized a strain bearing an estrogen-inducible allele of NDT80 [2], a meiotic transcription factor that induces the middle meiotic genes [4]. In the absence of NDT80 induction, cells arrest in prophase I prior to the first meiotic nuclear division [5, 6]. After arrest at the ndt80Δ block in KAc, cells were simultaneously transferred to carbon-free (KCl) medium and NDT80 was induced. Cells underwent both meiotic divisions (Figure 1E, KAc and KCl bars) and formed spores (data not shown) as efficiently as a control sample in which NDT80 was induced in KAc. This finding indicates that meiotic divisions can occur in the absence of an exogenous energy source. Prophase-arrested cells were not irreversibly committed to meiosis, as transfer to rich (YPD) medium at the time of NDT80 induction caused cells to resume vegetative growth and no meiotic nuclear divisions occurred (Figure 1F). This ability to “return to growth” agrees with previous findings that commitment to meiosis occurs at the time of the first nuclear division [1, 7]. Our results indicate that prophase I-arrested cells do not complete sporulation by default upon relief of arrest, and that all requirements for a non-fermentable carbon source are fulfilled by the end of prophase I.
We then monitored meiosis in the presence of sodium azide to test whether prophase-arrested cells released into KCl medium oxidized internal stores of biomolecules to fuel the two nuclear divisions. Azide blocked sporulation when added to cultures either before or after NDT80 induction (Figure 1A, E) but did not significantly affect cell viability (Supplemental Figure 2A), indicating that the failure to undergo the meiotic divisions in the presence of azide results from mitochondrial inhibition and not cell death. Furthermore, addition of dinitrophenol, an ionophore that dissipates proton gradients, to KCl cultures one hour prior to NDT80 induction mimicked the effects of azide (Figure 1G). A third inhibitor, oligomycin, which blocks mitochondrial ATP synthase, appeared only partially active. Addition of this drug at the time of (Supplemental Figure 2B) or one hour prior to (data not shown) NDT80 induction did not inhibit the meiotic nuclear divisions. Addition at the time of initial incubation in KAc (prior to NDT80 induction) drastically reduced tetranucleate formation but did not eliminate it as azide did (Figure 1E, Supplemental Figure 2B, KAc+oligomycin continuous). Tetrads produced in KAc or KCl with or without oligomycin showed greater than 90% viability (Supplemental Figure 2C), indicating that chromosome segregation occurred faithfully. These experiments reveal that a carbon source is dispensable after prophase I and even partial loss of mitochondrial activity does not interfere with the two meiotic divisions, but a minimal amount of mitochondrial function remains essential for nuclear divisions.
A non-fermentable carbon source is required for DNA replication and recombination
Having established that a carbon source is needed early in the meiotic program after Ime1 induction but before the meiotic divisions, we attempted to bypass the carbon source-dependent process following IME1 activation by eliminating DNA replication and recombination. Two alleles—cdc6-mn and spo11Δ—were introduced into the ime2-as strain (Figure 1B, Supplemental Figure 3A). The deletion of SPO11 eliminates the endonuclease which initiates meiotic recombination [8]. The cdc6-mn allele prevents premeiotic DNA replication but not subsequent meiotic events, by placing CDC6, a component of the pre-replicative complex, under a mitosis-specific promoter [9].
The ime2-as strain showed a strict dependence on a carbon source during early stages of meiosis as described above (Figure 1B). Release of ime2-as arrested cells into KAc resulted in 35% of cells forming tetranucleate cells (Figure 1H). DNA content analysis showed that relief of Ime2 inhibition was inefficient, as a large percentage of cells failed to replicate DNA (Supplemental Figure 3A). Release from the ime2-as block into KCl caused a 9-fold decrease in tetranucleate formation (4% tetranucleates), although DNA replication occurred to a similar extent as in KAc medium (Figure 1H, Supplemental Figure 3C). Nuclear divisions of the ime2-as spo11Δ strain under different media conditions were similar to that of the ime2-as strain (Figure 1I), indicating that recombination is not the sole carbon-source dependent step of meiosis.
To address the role of a carbon source in premeiotic DNA replication, we examined an ime2-as cdc6-mn strain. This strain formed tetranucleates inefficiently in KAc following relief of Ime2 inhibition (approximately 10%; Supplemental Figure 4), although this strain showed 30–40% tetranucleate formation when not pre-treated with inhibitor (data not shown). Nevertheless, it was clear that tetranucleate formation was diminished when this strain was treated with the Ime2 inhibitor and then released into KCl in the absence thereof (1% tetranucleates, Supplemental Figure 4).
The relative efficiency of nuclear divisions in the ime2-as cdc6-mn spo11Δ strain in KCl medium was different from what was observed for the ime2-as single and double mutant strains. As expected, the triple mutant never accumulated 4C DNA content (Supplemental Figure 3B, D). When this strain was released from the ime2-as block, sporulation in KCl was only 2.5-fold decreased compared to that in KAc (17% vs. 44%, respectively; Figure 1J), whereas the ime2-as mutant alone displayed a 9-fold reduction under similar conditions (4% vs. 35%; Fig. 1H). Furthermore, after 24 hours in KCl medium, 25% of cells contained meiotic spindles, suggesting that additional cells might progress to the tetranucleate stage. It was not possible to analyze cells at later time points because nuclei and spindles became difficult to visualize. In fact, we found that this strain accumulated higher levels of meiotic spindles at earlier time points in all culture conditions than could be accounted for by the number of bi- and tetra-nucleate cells at later times. This finding suggests that a larger number of cells underwent meiosis but a fraction was lost, as observed for ime2Δ cells [10]. We conclude that minimal mitochondrial function is required throughout meiosis, but a carbon source is primarily needed to execute DNA replication and recombination.
Respiration governs entry into the meiotic program independently of energy production
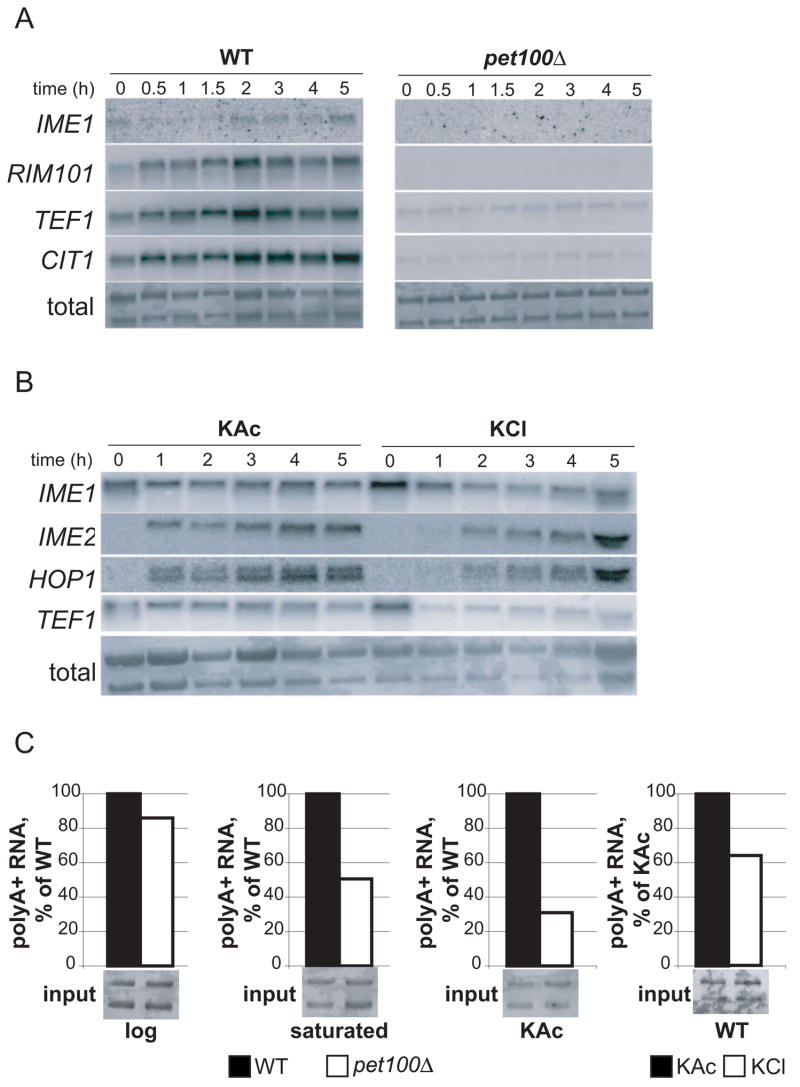
Because addition of azide nearly abolished Ime1 production (Figure 1C), we next confirmed that petite strains, which are incapable of respiration, also fail to express IME1 [11]. A pet100Δ strain, which is defective in assembly of cytochrome c oxidase [12] and therefore respiration-deficient, failed to express both tagged Ime1 and an IME1-LacZ reporter consisting of a 4.2 kb fragment including the IME1 promoter and amino acids 1–68 of the IME1 ORF [13] (Supplemental Figure 5). Northern blotting demonstrated the absence of IME1 RNA in pet100Δ cells (Figure 4A). These results confirm that flux through the mitochondrial electron transport system is necessary for IME1 expression.
Figure 4. mRNA production is decreased in the absence of respiration or carbon source.
A) RNA expression of IME1, RIM101, TEF1, or CIT1 in WT (A15869) or pet100Δ (A15871) cells incubated for the indicated times in KAc after pre-growth in YPD. Total RNA is shown as a loading control.
B) RNA expression of IME1, IME2, HOP1, or TEF1 in WT (A15869) cells incubated for the indicated times in KAc or KCl after pre-growth in YPA. Total RNA is shown as a loading control.
C) Polyadenylated RNA was isolated from WT (A15869) or pet100Δ (A15871) log phase or saturated YPD cultures, or after 2 hours’ incubation in KAc or KCl. 1/700 of the total RNA used as inputs for polyA+ RNA isolation is shown below each bar. The amount of polyA+ RNA isolated is shown normalized to WT levels in the first 3 panels or to KAc in the last panel.
It was possible that cells with reduced mitochondrial function simply lacked energy to initiate meiosis in KAc, as they cannot utilize a non-fermentable carbon source. Therefore, we tested whether pet100Δ cells could undergo meiosis in the presence of glucose upon rapamycin addition, a condition which induces sporulation in wild-type (WT) cells [14]. As expected, addition of rapamycin to WT cultures induced IME1-LacZ expression and sporulation (Figure 2A, B), with the resulting tetrads showing >95% spore viability (data not shown). In contrast, pet100Δ cells failed to express IME1-LacZ or undergo any nuclear divisions (Figure 2A, B). This inability to enter the meiotic program was not due rapamycin inhibiting glucose utilization, because cell density increased following rapamycin addition (Figure 2C) as did the amount of extracellular ethanol, a product of fermentation (Figure 2D, Supplemental Figure 6). These results reveal that the IME1 expression defects of petite cells cannot be attributed to the absence of an energy source in the medium. Rather, they indicate that a respiration-sensitive signaling pathway governs IME1 expression and entry into meiosis.
Figure 2. Respiration governs meiosis independently of energy production.
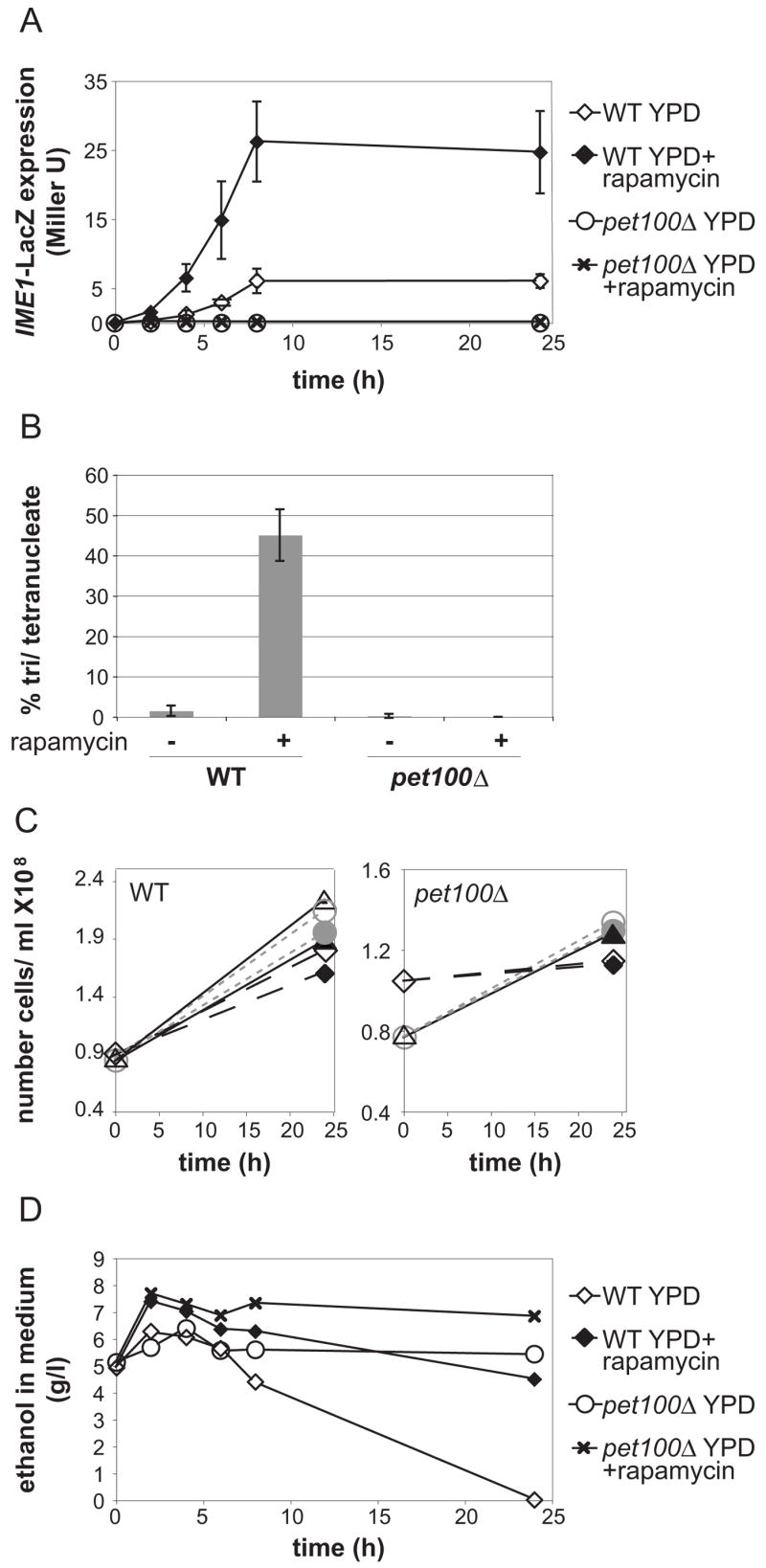
A) IME1-LacZ activity was measured in triplicate in WT (A15869) or pet100Δ (A15871) cells incubated in YPD with or without rapamycin. Cells were grown overnight to an OD600=~5.5. Rapamycin was added to half of each culture at time 0 and samples were collected for LacZ assays at the indicated times. Means and standard deviations are shown.
B) The percentage of tri and tetranucleate cells from cultures shown in (A) was determined 48 hours after rapamycin addition by DAPI staining. Means and standard deviations are shown.
C) The cell densities of WT (right) or pet100Δ (left) cultures with (filled symbols) or without (open symbols) rapamycin from (A) were measured at the time of rapamycin addition or 24 hours later. Parallel rapamycin-treated and untreated cultures are indicated with matching symbols and lines.
D) Representative trace of extracellular ethanol content in supernatants of WT (A15869) or pet100Δ (A15871) cultures from (A) treated with or without rapamycin.
Defects in RIM101 expression are not solely responsible for loss of IME1 transcription in petite cells
Because petite cells failed to transcribe IME1, we investigated whether these cells were defective in the expression or regulation of the IME1 transcriptional activators Msn2 and Rim101. Msn2 is localized to the nucleus upon glucose derepression, where it activates transcription of target genes in conjunction with Msn4 [15]. Msn2-GFP localization in pet100Δ was similar to that of WT cells (Supplemental Figure 7).
Next we analyzed expression of Rim101, a 682-residue protein, which is activated by cleavage of its carboxyl terminus [16, 17]. Rim101 has been hypothesized to govern Ime1 expression in response to respiration [18], because respiration causes alkalinization of the medium [19] and Rim101 cleavage is activated at high pH [17]. Expression and cleavage of HA-tagged Rim101 [17] were detectable in wild-type meiotic cells. In contrast, Rim101 was barely present in petΔ cells, although the small amount of detectable protein was of the faster-migrating form, indicating that cleavage was not defective (Figure 3A). Notably, Rim101 cleavage was observed prior to a significant increase in the pH of the medium in both strains (Figure 3B), confirming that respiration does not cause Rim101 cleavage via extracellular alkalinization. Furthermore, we found that while deletion of RIM101 caused significant decreases in IME1-LacZ expression and tetranucleate formation (Figures 3C, D, Supplemental Figure 8), the decreases were not as severe as in petΔ cells (Figure 3C, D). We have never observed IME1 induction or any nuclear divisions in petite strains under these conditions, whereas the rim101Δ strain consistently displayed low levels of both. Our results show that respiration does not govern meiosis solely by regulating Rim101 expression or cleavage.
Figure 3. Respiration signaling does not solely occur via Rim101 regulation.
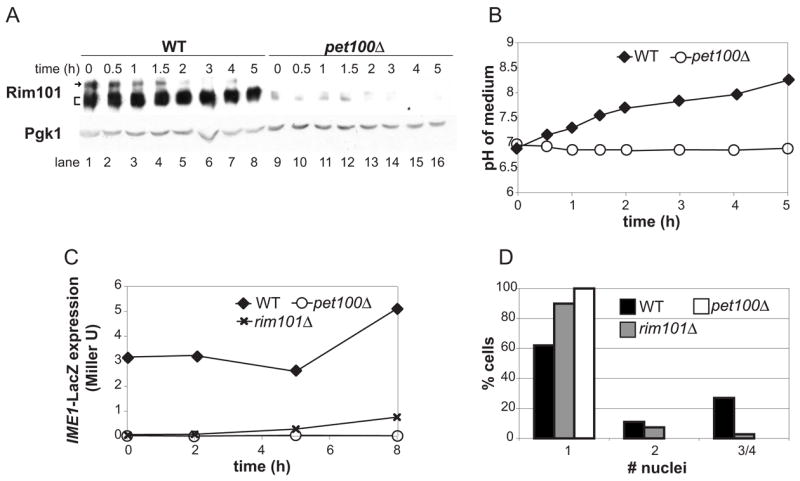
A) Rim101-3HA protein levels were determined in WT (A15869) and pet100Δ (A15871) cells incubated in KAc for the indicated times. Arrow: full-length Rim101, bracket: cleaved Rim101, which regularly appears as a doublet in KAc medium. Pgk1 is shown as a loading control.
B) Cells from (A) were pelleted and the pH of the supernatant was measured.
C) IME1-LacZ activity was measured in WT (A15869), pet100Δ (A15871), or rim101Δ (A18590) cells in saturated YPD cultures (time 0) or after incubation in KAc for the indicated times.
D) The number of nuclei per cell of cultures shown in (C) was determined by DAPI staining after 24 hours.
Global mRNA down-regulation in the absence of an utilizable carbon source
Analysis of mRNA levels in petΔ cells provided insights into the response to sporulation-inducing conditions in the absence of respiration. As expected, IME1 and RIM101 RNAs were significantly reduced in the petite cells (Figure 4A). Surprisingly, two transcripts which we intended to use as loading controls—TEF1 (whose expression is fairly constant during meiosis [4]) and CIT1 (which is induced in both wild-type and petite cells [20])—were also decreased approximately 10-fold in the petΔ strain (Figure 4A). In contrast, similar amounts of rRNA were isolated from equivalent numbers of WT and pet100Δ cells. IME1 and TEF1 levels were also decreased in WT cells incubated in KCl (Figure 4B) indicating that the decrease in these mRNAs is not specific to the PET100 deletion, but likely a consequence of decreased respiration or lack of energy. Ime1 targets IME2 and HOP1 were, however, induced in KCl, (albeit with a delay (Figure 4B)), indicating that Ime1 protein produced in KCl is functional.
To investigate whether total mRNA levels were decreased in petite cells, we isolated polyadenylated RNA (i.e. mRNA) from equal amounts of total RNA derived from WT or pet100Δ cultures. Similar amounts of mRNA were isolated from exponentially growing WT and pet100Δ cultures (Figure 4C). As cultures reached saturation in rich media, the relative amount of mRNA was decreased 2-fold in the pet100Δ strain. Northern blotting showed that levels of CIT1 and TEF1 mirrored global mRNA levels in these cultures (Supplemental Figure 9). Upon incubation of pet100Δ cells in KAc, mRNA amounts were reduced 3 to 4-fold (Figure 4C). Interestingly, the decrease in mRNA amounts seen in pet100Δ cells was not as severe as that observed by Northern blotting of specific transcripts, suggesting that a small number of genes may be strongly up-regulated (thus making a significant contribution to the total mRNA pool), or the transcripts selected for Northern blotting were disproportionately down-regulated. WT cells in KCl medium showed a 1.5-fold decrease in mRNA compared to cells in KAc, but Northern blotting suggested that the decrease might be greater at later time points (Figure 4B, 5-hour time point). These results reveal that as the energy source becomes limiting, cells globally repress mRNA transcription. While this decrease could account for the inability of petite cells to sporulate, it cannot account for the lack of IME1 expression, as wild-type cells in KCl display a decrease in mRNA levels (Figure 4B, C) yet Ime1 production is unaffected (Figure 1C, D).
Mitochondrial function is required for sporulation in S. pombe
Finally, to address whether the need for respiration to induce meiosis is conserved in other organisms, we generated S. pombe ρ0 strains, which lack mitochondrial DNA. Because this yeast does not ordinarily tolerate loss of mitochondrial DNA, the ρ0 strains were made in the ptp1 or ptp2 mutant backgrounds which permit loss of mitochondrial DNA [21]. These mutations alone do not affect sporulation when heterozygous [21]. Whereas ρ+ strains heterozygous for ptp1 or ptp2 displayed approximately 30% sporulation efficiency, no tetrads were obtained by crossing corresponding ρ0 strains to make diploids (Figure 5). Because the medium used for sporulation contains fermentable carbon sources, we conclude that respiration is also required for sporulation in S. pombe independently of its ability to produce energy.
Figure 5. Respiration is required for sporulation in S. pombe.
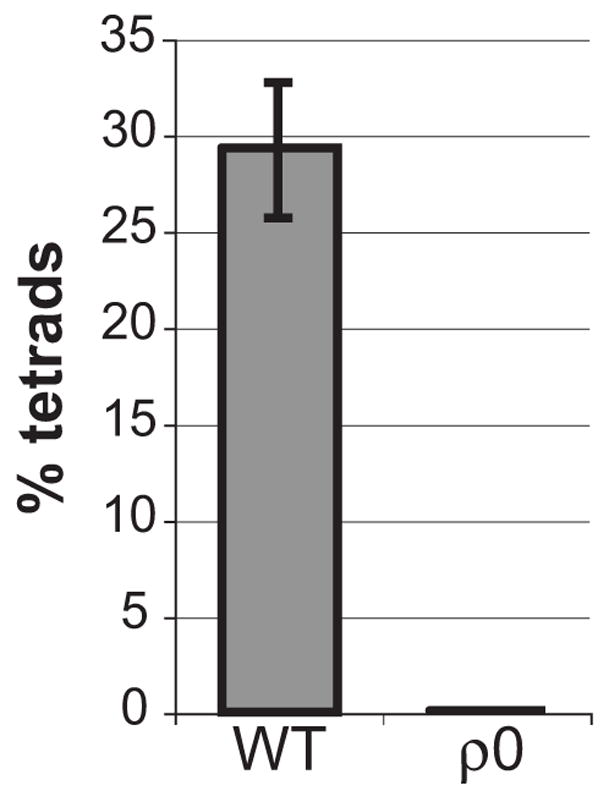
Sporulation of ρ+ or ρ0 S. pombe strains heterozygous for the ptp1 and/or ptp2 mutations was analyzed after 3 days’ incubation on malt extract plates. The average percentage of tetrads observed in 2 independent ρ0 crosses, or the average and standard deviation of 4 WT crosses, are reported.
Discussion
Our results indicate that respiration is needed to signal entry into the meiotic program and to provide energy for subsequent meiotic processes by oxidizing external and/or internal carbon sources. Surprisingly, a carbon source was not needed for Ime1 expression or function. A carbon source was needed for full DNA replication and recombination; however, it was dispensable after prophase, indicating that cells can generate energy for the two nuclear divisions by oxidizing internal stores of biomolecules only. Notably, mutants defective in autophagy fail to sporulate [22]; it is expected that autophagy would be essential for the nuclear divisions in carbon-free medium.
We have shown that respiration signals affect IME1 expression independent of energy availability, as petite cells failed to express IME1 when treated with rapamycin in rich medium. Cell size is not responsible for the meiotic entry defects of petites either, as we observed little difference between the cell sizes of WT and pet100Δ cells (Supplemental Figure 10), and sfp1Δ cells (which display reduced cell volume due to defective ribosome biogenesis [23]) are able to sporulate (A.J. unpublished observations). Global mRNA levels are reduced in petite cells under sporulation-inducing conditions; however, this decrease cannot account for the lack of Ime1, as WT cells in carbon-free medium express Ime1 despite decreased PolII transcription. It is possible, however, that the global mRNA decrease precludes induction of the full meiotic transcriptional program and hence affects steps following Ime1 expression. Interestingly, we note that this mRNA downregulation overrides mechanisms that up-regulate specific transcripts in petite cells, e.g. induction of CIT1 by the retrograde signaling (RTG) pathway [20]. Finally, we have shown that respiration signaling does not solely occur via Rim101 expression or processing, as deletion of RIM101 did not replicate the meiotic defects of petites. We were not able to test whether Rim101 overexpression promotes meiotic entry in petite cells, because overexpression of the Rim101 active cleaved segment (aa 1- 539) [17] diminished IME1 expression and sporulation in wild-type cells (data not shown). We were also unable to rescue meiosis in petites by adding bicarbonate, as reported for other strain backgrounds [19].
Identifying factors which control IME1 expression in response to respiration will reveal how cells sense and respond to their respiration status. It is likely that such respiration-sensing pathways are operative in other organisms, as respiration-deficient S. pombe strains fail to sporulate. Furthermore, infertility in humans correlates with defects in mitochondrial function [24]. Our work indicates that yeast can be used as a model system for investigating control of gametogenesis by mitochondrial activity and the basis of infertility in higher organisms.
Supplementary Material
Supplemental Expermental Procedures, 2 tables, and 10 figures are available at
Acknowledgments
We thank Tom Fox and Dan McCollum for S. pombe strains. We thank Andreas Hochwagen, Peter Chien, and members of the Amon lab for comments on the manuscript, and Amudha Panneerselvam for assistance with experiments. This work was supported by the National Institutes of Health grant GM079907 to A. J. and GM62207 to A.A. A. A is also an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Simchen G, Pinon R, Salts Y. Sporulation in Saccharomyces cerevisiae: premeiotic DNA synthesis, readiness and commitment. Exp Cell Res. 1972;75:207–218. doi: 10.1016/0014-4827(72)90538-1. [DOI] [PubMed] [Google Scholar]
- 2.Benjamin KR, Zhang C, Shokat KM, Herskowitz I. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 2003;17:1524–1539. doi: 10.1101/gad.1101503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Honigberg SM, McCarroll RM, Esposito RE. Regulatory mechanisms in meiosis. Curr Opin Cell Biol. 1993;5:219–225. doi: 10.1016/0955-0674(93)90106-z. [DOI] [PubMed] [Google Scholar]
- 4.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 5.Carlile TM, Amon A. Meiosis I is established through division-specific translational control of a cyclin. Cell. 2008;133:280–291. doi: 10.1016/j.cell.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu L, Ajimura M, Padmore R, Klein C, Kleckner N. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6572–6581. doi: 10.1128/mcb.15.12.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Honigberg SM, Esposito RE. Reversal of cell determination in yeast meiosis: postcommitment arrest allows return to mitotic growth. Proc Natl Acad Sci U S A. 1994;91:6559–6563. doi: 10.1073/pnas.91.14.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 9.Hochwagen A, Tham WH, Brar GA, Amon A. The FK506 binding protein Fpr3 counteracts protein phosphatase 1 to maintain meiotic recombination checkpoint activity. Cell. 2005;122:861–873. doi: 10.1016/j.cell.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 10.Foiani M, Nadjar-Boger E, Capone R, Sagee S, Hashimshoni T, Kassir Y. A meiosis-specific protein kinase, Ime2, is required for the correct timing of DNA replication and for spore formation in yeast meiosis. Mol Gen Genet. 1996;253:278–288. doi: 10.1007/s004380050323. [DOI] [PubMed] [Google Scholar]
- 11.Treinin M, Simchen G. Mitochondrial activity is required for the expression of IME1, a regulator of meiosis in yeast. Curr Genet. 1993;23:223–227. doi: 10.1007/BF00351500. [DOI] [PubMed] [Google Scholar]
- 12.Church C, Goehring B, Forsha D, Wazny P, Poyton RO. A role for Pet100p in the assembly of yeast cytochrome c oxidase: interaction with a subassembly that accumulates in a pet100 mutant. J Biol Chem. 2005;280:1854–1863. doi: 10.1074/jbc.M410726200. [DOI] [PubMed] [Google Scholar]
- 13.Sherman A, Shefer M, Sagee S, Kassir Y. Post-transcriptional regulation of IME1 determines initiation of meiosis in Saccharomyces cerevisiae. Mol Gen Genet. 1993;237:375–384. doi: 10.1007/BF00279441. [DOI] [PubMed] [Google Scholar]
- 14.Zheng XF, Schreiber SL. Target of rapamycin proteins and their kinase activities are required for meiosis. Proc Natl Acad Sci U S A. 1997;94:3070–3075. doi: 10.1073/pnas.94.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schuller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Futai E, Maeda T, Sorimachi H, Kitamoto K, Ishiura S, Suzuki K. The protease activity of a calpain-like cysteine protease in Saccharomyces cerevisiae is required for alkaline adaptation and sporulation. Mol Gen Genet. 1999;260:559–568. doi: 10.1007/s004380050929. [DOI] [PubMed] [Google Scholar]
- 17.Li W, Mitchell AP. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics. 1997;145:63–73. doi: 10.1093/genetics/145.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honigberg SM, Purnapatre K. Signal pathway integration in the switch from the mitotic cell cycle to meiosis in yeast. J Cell Sci. 2003;116:2137–2147. doi: 10.1242/jcs.00460. [DOI] [PubMed] [Google Scholar]
- 19.Hayashi M, Ohkuni K, Yamashita I. Control of division arrest and entry into meiosis by extracellular alkalisation in Saccharomyces cerevisiae. Yeast. 1998;14:905–913. doi: 10.1002/(SICI)1097-0061(199807)14:10<905::AID-YEA290>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 20.Liu Z, Butow RA. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol Cell Biol. 1999;19:6720–6728. doi: 10.1128/mcb.19.10.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haffter P, Fox TD. Nuclear mutations in the petite-negative yeast Schizosaccharomyces pombe allow growth of cells lacking mitochondrial DNA. Genetics. 1992;131:255–260. doi: 10.1093/genetics/131.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marston AL, Tham WH, Shah H, Amon A. A genome-wide screen identifies genes required for centromeric cohesion. Science. 2004;303:1367–1370. doi: 10.1126/science.1094220. [DOI] [PubMed] [Google Scholar]
- 23.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.May-Panloup P, Chretien MF, Jacques C, Vasseur C, Malthiery Y, Reynier P. Low oocyte mitochondrial DNA content in ovarian insufficiency. Hum Reprod. 2005;20:593–597. doi: 10.1093/humrep/deh667. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Expermental Procedures, 2 tables, and 10 figures are available at