Abstract
The intracellular part of the Rel signal transduction pathway in Drosophila is encoded by Toll, tube, pelle, dorsal, and cactus, and it functions to form the dorsal–ventral axis in the Drosophila embryo. Upon activation of the transmembrane receptor Toll, Dorsal dissociates from its cytoplasmic inhibitor Cactus and enters the nucleus. Tube and Pelle are required to relay the signal from Toll to the Dorsal–Cactus complex. In a yeast two-hybrid assay, we found that both Tube and Pelle interact with Dorsal. We confirmed these interactions in an in vitro binding assay. Tube interacts with Dorsal via its C-terminal domain, whereas full-length Pelle is required for Dorsal binding. Tube and Pelle bind Dorsal in the N-terminal domain 1 of the Dorsal Rel homology region rather than at the Cactus binding site. Domain 1 has been found to be necessary for Dorsal nuclear targeting. Genetic experiments indicate that Tube–Dorsal interaction is necessary for normal signal transduction. We propose a model in which Tube, Pelle, Cactus, and Dorsal form a multimeric complex that represents an essential aspect of signal transduction.
The Rel pathway is one of the best understood signal transduction pathways and is evolutionarily conserved in invertebrates and vertebrates. In mammals, it controls the activity of immune and inflammatory response genes as well as viral genes (1). In Drosophila, the Rel pathway functions in the immune response and in the establishment of dorsal–ventral polarity in the early embryo. Five genes, Toll, tube, pelle, cactus, and dorsal, form the intracellular part of the Drosophila Rel pathway that is remarkably homologous to the mammalian interleukin-1 receptor–NF-κB pathway (2) .
The Drosophila Toll gene encodes a transmembrane receptor for the ventrally localized extracellular signal (3, 4). The intracellular domain of the receptor shows sequence similarity to the cytoplasmic domain of the interleukin-1 receptor (5). A human homologue of the Drosophila Toll, hToll, has been identified and shown to function in the activation of adaptive immunity (6). Although the activation of Toll triggers the intracellular signaling cascade and culminates in the selective nuclear translocation of Dorsal, the exact mechanism by which the Toll receptor transmits its signal is not well understood.
Dorsal belongs to the Rel family of transcription factors (7). All members of this family share a conserved 300-amino acid region, the Rel homology region (RHR) (1). Structure and function analyses have revealed two distinct subdomains of the RHR. The N-terminal domain 1 contains the motif RXXRXRXXC that is important for DNA binding, and the C-terminal domain 2 contains the dimerization and Cactus binding sequences (8–11). Like other Rel proteins, Dorsal is regulated at the level of nuclear translocation (12–15). Dorsal is initially retained in the cytoplasm by its inhibitor Cactus, a member of the IκB family (12, 16). At the syncytial blastoderm stage, the Dorsal–Cactus complex is disrupted in response to the activated spatial signal transmitted through Toll, resulting in the graded nuclear import of Dorsal. In the nucleus, Dorsal regulates the asymmetric expression of a set of zygotic genes (17, 18). Cactus and other IκB family members associate with Rel proteins via the ankyrin repeats (12, 16, 19, 20). Immunoprecipitation and cross-linking experiments revealed that two molecules of Dorsal are complexed with one molecule of Cactus (21, 22).
The transduction of the signal from the Toll receptor to the Dorsal–Cactus complex requires at least two other cytoplasmic components, Pelle and Tube (23). Tube is a novel intracellular protein (24). Analysis of Tube defined two distinct domains. The N-terminal domain is the activation domain that is sufficient for partial function in transient rescue experiments. The C-terminal half contains five highly conserved 8-amino acid repeats (25). pelle encodes a cytoplasmic serine/threonine kinase homologous to the mammalian IRAK kinase, a component of the interleukin 1 receptor–NF-κB pathway (26, 27). tube and pelle have been shown to interact genetically, and epistasis experiments using an activated form of pelle and tube have suggested that pelle functions downstream from tube. Direct interaction between the two proteins has been demonstrated in a yeast two-hybrid assay and in an in vitro co-immunoprecipitation experiment. Both proteins interact in the N-terminal half, which contains a “death domain” in both proteins (28–30).
Although Tube and Pelle are essential for nuclear translocation of Dorsal, the mechanism of signal transduction from the Toll receptor through the Tube–Pelle complex to the Dorsal–Cactus complex remains unclear. When four N-terminal serines in Cactus are mutated to alanines, Cactus degradation is blocked and nuclear targeting of Dorsal is presumably inhibited (31, 32). Dorsal is a phosphoprotein. The phosphorylation of Dorsal correlates with its dissociation from Cactus (33, 34). However, there is no evidence indicating that Pelle directly phosphorylates either Cactus or Dorsal (29).
The intracellular events of the Rel pathway have been the subject of numerous studies. Further proteins that either modify Dorsal or function in concert with Dorsal remain to be identified. To this end, and to test for interactions between the known components of the pathway, we have taken advantage of the yeast two-hybrid system. In this paper, we establish that Tube and Pelle specifically interact with Dorsal in yeast and in vitro. We mapped the Dorsal binding region to the C terminus of Tube. This domain of Tube is essential for full function. We also find that in contrast to Cactus, Tube and Pelle interact with Dorsal in domain 1 of the RHR. This domain has been shown to be essential for Dorsal nuclear targeting. We propose that Tube, Pelle, Cactus, and Dorsal form a complex, the formation of which is essential for signal transduction.
MATERIALS AND METHODS
Plasmid Constructions.
All LexA fusion plasmids were constructed by inserting coding sequences in frame into the pEG202 vector (35). LexA-DL-RHR contains the MunI restriction fragment from the Dorsal cDNA (amino acids 35–356). The LexA-DL222–342 fusion was tagged with the hemagglutinin (HA) epitope at its C terminus because the untagged protein was unstable in yeast. LexA-DL35–246 was made by inserting a PCR-produced MunI-SalI fragment into pEG202 at the EcoRI/XhoI site. LexA-cactus and the LexA fusion for the intracellular domain of the transforming growth factor-β receptor (LexA-TGFB-R) are gifts from P. H. Lin and X. Ting, respectively. All constructs were checked for expression in yeast by Western blot analysis. The activation fusion proteins were expressed using pJG4-5 (36).
The constructs expressing the full-length Tube, Tube-(296–462), and Tube-(1–198) were made by cloning the corresponding PCR products into pJG4-5 as EcoRI/XhoI fragments. The pJGTube1–257 was generated by cloning an EcoRI/SalI fragment excised form pJGTube full-length. Pelle full-length, Pelle-(1–209), and Pelle-(210–501) were constructed by cloning corresponding PCR fragments into the pJG vector.
The proteins used in the gel-shift assays were expressed using pET28a, pET21a, or pGEX plasmids. The construct expressing the cactus protein contains an ApoI cDNA fragment, deleting the N-terminal 64 amino acids. Tube and Pelle proteins were expressed using pET28a recombinant constructs.
The Yeast Two-Hybrid Screen.
The yeast two-hybrid screen was performed according to the procedures described (36). The strain containing the bait was transformed with a Drosophila cDNA ovarian library, which was constructed in the pJG4-5 plasmid and was a gift from J. Grosshans (Max Plank Institute, Tübingen, Germany). Approximately 2 × 107 transformed cells were screened, and a total of 72 colonies were collected as positives on days 3 and 4. These positives were further tested for galactose dependence and specificity using the interaction mating assay (35). Of 72 positives, 56 showed galactose-dependent expression and specific interactions with LexA–DL-RHR fusion. The library plasmids were rescued from these yeast cells and sequenced.
The individual pair-wise interactions were tested using the LacZ reporter plasmid. The β-galactosidase assays were performed following standard procedures (37). For each pair of interactions, three or more individual colonies were assayed. The results are normalized to the basal level obtained with vector only, which is routinely 1–2 Miller units.
Preparation of Dorsal, Cactus, Tube, and Pelle Proteins from Escherichia coli.
Proteins were expressed in E. coli as described by Hoey and Levine (38). The proteins were purified from [Ni2+] resin according to the manufacturer (Novagen). The elutes were collected and renatured by stepwise dialysis, containing a decreasing amount of guanidine⋅HCl. As a negative control, the protein encoded by the D-Lis gene was expressed as a glutathione S-transferase fusion and purified. The DL47–244 protein was expressed as a glutathione S-transferase fusion protein from a pGEX construct (a gift from Michael Levine) (11). The protein was prepared exactly as described (39).
Gel-Shift Assay.
The double-stranded zenB κB site TAACTGGGAGAAACCCAATCA was synthesized and end labeled with 32P, and the binding assay was performed as described (12, 16, 39).
Transgenic Flies.
The flies containing wild-type p-tub462 and p-tubΔC205 transgenes were generous gifts from Steven Wasserman (25). The transgenes were crossed into the null allele tubR5.6 background. Cuticle preparations were made as described by Wieschaus et al. (40).
RESULTS
Tube Interacts with Dorsal Specifically.
To identify new proteins that function as cofactors or modifers of Dorsal in either the cytoplasm or the nucleus, we screened 20 million clones from a Drosophila ovarian cDNA library using a lexA–DL-RHR fusion protein as bait. We recovered 56 positives representing 20 different cDNA clones. Among them, 10 clones contained cactus cDNA, and five contained Tube cDNAs. The five Tube clones encoded three different N-terminally deleted Tube proteins: three isolates were identical and encoded Tube-(34–462), one isolate encoded Tube-(184–462), and one encoded Tube-(245–462). Most of the other positive clones coded for novel proteins and will be described later.
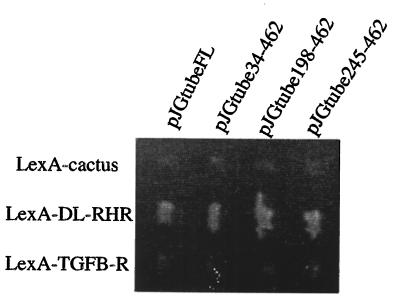
To test if Tube binds specifically to Dorsal, we performed an interaction mating assay (35). Our results show that full-length Tube-(1–462) and all three different tube clones recovered from the screen interact with the lexA–DL-RHR fusion, but not with other proteins, including Cactus, and the transforming growth factor-β receptor cytoplasmic domain, indicating that the interaction between Tube and Dorsal is specific (Fig. 1).
Figure 1.
Tube interacts with Dorsal specifically. The yeast-mating interaction assay shows specific interaction between DL-RHR and full-length and three N-terminally deleted Tube proteins. Cells expressing Tube and DL-RHR grow only on Leu−/galactose plates (shown here) but not on Leu−/glucose plates.
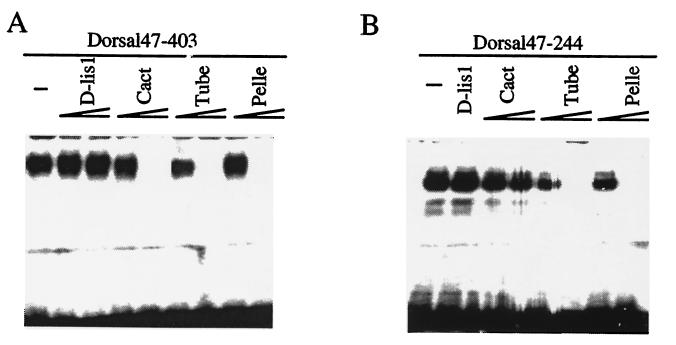
To confirm the Dorsal–Tube interaction, we performed in vitro binding assays. It had been shown previously that Dorsal binding to the consensus κB site in the zerknüllt (zen) promoter can be inhibited by Cactus (12, 16). Using the same assay, we tested if Tube had a similar effect on Dorsal DNA binding. Fig. 2A (lane 1) shows the shift observed when no Tube protein is added (lane 1). However, bacterially expressed Tube can inhibit Dorsal binding, and increasing the amount of Tube protein enhances the inhibition (lanes 6 and 7). Cactus was used as a positive control (lanes 3 and 4). As a negative control, a protein encoded by the D-Lis1 gene, thought to function in nuclear migration (Z. Liu and R.S., unpublished observations), was used, and it did not affect the Dorsal-DNA shift (lanes 2 and 3), indicating the effect on DNA binding of Dorsal is specific to Tube.
Figure 2.
In vitro association between the Dorsal RHR and Dorsal RHR domain 1 with Tube and Pelle. (A) Dorsal RHR binding to DNA is inhibited by Tube and Pelle. Gel-shift assays were performed with 0.2 μg of partially purified Dorsal RHR (Dorsal47–403) and a labeled zenB oligonucleotide containing a κB site (lane 1). Each protein was purified and assayed at about 0.1 μg (lanes 2, 4, 6, 8) and 0.5 μg (lanes 3, 5, 7, 9). (B) Dorsal RHR domain 1 binding to DNA is also inhibited by Tube and Pelle but not by Cactus. The amount of all proteins used was the same as in A, except that only one dose of 0.5 μg of D-Lis1 protein was assayed.
Tube Interacts with Dorsal via Its C-Terminal Repeats.
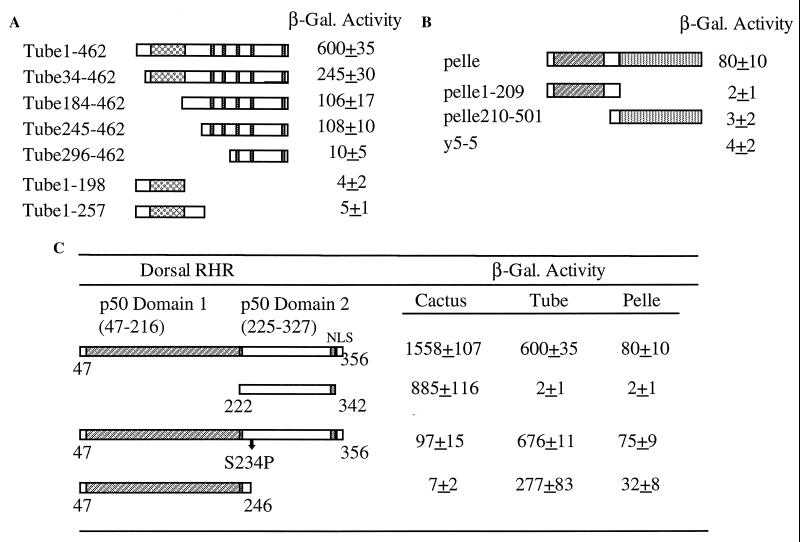
The Tube protein has no known homologs and contains two domains. The N-terminal half has been shown to contain a protein–protein interaction “death domain” and is essential for its interaction with Pelle, whereas the C-terminal half contains five 8-amino acid repeats that are evolutionarily conserved (25). Two tube isolates from our screen, Tube-(184–462) and Tube-(245–462), contain only the C-terminal half of the protein, showing that the C terminus of Tube is sufficient to bind to Dorsal. To map the interaction region more precisely, we made a series of deletion constructs of Tube and tested their interaction with Dorsal by using lacZ as a reporter gene. Three N-terminally deleted proteins, Tube-(34–462), Tube-(184–462), and Tube-(245–462), retain substantial if reduced Dorsal binding activity as reflected by β-galactosidase activity (Fig. 3A). Two C-terminally truncated constructs that still contain the “death domain” and interact with Pelle, Tube-(1–198) and Tube-(1–257), failed to interact with Dorsal. In addition, Tube 296–462, lacking the first two repeats, reduced the Dorsal binding activity significantly (Fig. 3A). These results indicate that Tube depends on its C-terminal domain, or on at least the first two repeats, for its interaction with Dorsal.
Figure 3.
Mapping the Dorsal interaction regions in Tube and Pelle. (A) Tube interacts with Dorsal via its C-terminal repeats. The pJG plasmids expressing full length Tube (Tube1–462) and different Tube deletions were transformed into EGY48 yeast cells containing the LexA-DL-RHR and LacZ reporter plasmid, and β-galactosidase activity was determined. The N-terminal “death domain” and C-terminal five repeats are indicated by boxes. (B) Both the N-terminal “death domain” and C-terminal kinase domain of Pelle are required for interaction with Dorsal. pJG plasmids expressing full length Pelle, Pelle1–209 and Pelle210–501, were transformed into yeast containing lexA-DL-RHR and the LacZ reporter plasmid. A novel protein y5-5 was used as negative control. (C) The Dorsal RHR domain 1 is required for Tube and Pelle binding. The positions of RHR domain 1 (amino acids 47–216) and domain 2 (amino acids 225–327) are indicated. The LexA fusion of Dorsal RHR, two of its derivatives containing only one domain, and a mutant construct DS234P were tested for their interactions with Cactus, Tube, and Pelle. The arrow indicates the position of serine-234.
The C-Terminal Repeats Are Required for Wild-Type Activity of Tube.
Our finding that the C-terminal repeats of Tube are required for the interaction with Dorsal is unexpected because it had been shown that the function of Tube mainly depends on its N-terminal domain (25). To understand the possible role of the interaction of Tube with Dorsal, we compared the Tube function in two different transgenic lines. One line contained a transgene expressing full length Tube under the control of the endogenous tube promoter. The second line contained the same minigene encoding a C-terminally deleted Tube protein (both gifts from S. Wasserman). We find that although the wild-type tube transgene rescued the tube null phenotype completely, the truncated transgene tubΔC205 only rescued part of the function even though both proteins appear to be expressed at equal levels (results not shown). The cuticular phenotype of embryos from tubΔC205 females shows a hypomorphic D2 phenotype as compared with the D0 phenotype seen in tube null embryos. Less than 1% and 1.6% of the embryos hatched at 25 and 18°C (Table 1). These results agree with those obtained in mRNA injection rescue experiments and with a previous less detailed study of the transgene function (25). Together, these results show that the C terminus is essential for the full activity of Tube.
Table 1.
The C-terminal half of Tube is essential
| Transgene* | n | % hatch | Phenotype |
|---|---|---|---|
| p-tub462 | 520 | 100 | WT |
| p-tubΔC205 (25°C) | 182 | 0.6 | D2 |
| p-tubΔC205 (18°C) | 920 | 1.6 | D2 |
Genetic background: tubR5.6 homozygous.
Pelle Also Directly Interacts with Dorsal but Not with Cactus.
According to epistatic analyses, pelle acts downstream from tube and functions upstream from cactus and dorsal (29). Tube–Pelle interaction has been demonstrated in both yeast two-hybrid assay and in vitro immunoprecipitation (29, 30). Although we did not identify a Pelle cDNA in our two-hybrid screen, because our results indicate that Dorsal interacts with Tube, we tested whether there is also a direct interaction between Pelle and Dorsal.
Using LacZ as a reporter, we demonstrate that Pelle interacts with Dorsal in the pair-wise yeast interaction assay, although this interaction is considerably weaker than that of Dorsal with Cactus and that of Tube with Dorsal. Transformation of lexA-DL-RHR and activator-tagged Pelle together results in an 80-fold increase of the β-galactosidase activity over the basal level, as compared with a 600-fold increase for Tube and Dorsal and a 1550-fold increase for Cactus and Dorsal (Table 2). The activation of the reporter gene was not observed when the activator-tagged Pelle was transformed with other LexA fusion proteins, including Cactus, and two novel gene products, y5-5 and y3-9, both isolated as strong Cactus interactors in a two-hybrid screen (P.-H. Lin and R.S., unpublished results), suggesting that the interaction between Dorsal and Pelle is specific.
Table 2.
Pelle interacts with Dorsal specifically
| LexA fusion | β-gal activity/pJGpelle |
|---|---|
| LexA-DL-RHR | 80 ± 10 |
| LexA-Cactus | 3 ± 2 |
| LexA-y3-9 | 2 ± 1 |
| LexA-y5-5 | 1 ± 1 |
pJGpelle was cotransformed into yeast expressing LexA fusions and a lacZ reporter gene. For each interaction, β-galactosidase (β-gal) activity was determined for at least three independent clones. Values were normalized to the basal level seen with LexA fusions and the pJG4-5 vector.
The Dorsal-Pelle interaction was also confirmed in vitro by gel-shift assay. Fig. 2A (lanes 8 and 9) shows that Pelle competes with Dorsal binding to the zenB κB site in a dose-dependent manner. The gel-shift assay does not show a significant difference between Pelle and Dorsal binding, Tube and Dorsal binding, and Cactus and Dorsal binding. In that assay, similar amounts of each protein were used for the experiments. The discrepancy between the weak interaction observed in yeast and the strong one in vitro may be due to the Pelle protein being less highly expressed or less stable than Tube in yeast.
The Pelle protein consists of two domains, the N-terminal “death domain” and the C-terminal kinase domain. To map the domain of Pelle required for the interaction with Dorsal, we generated activator-tagged N-terminal Pelle-(1–209), which has been shown to interact with Tube, and Pelle-(209–501), which corresponds to the C-terminal domain. Our results show that neither the N-terminal nor the C-terminal domain alone could activate the lacZ reporter gene in the presence of lexA-DL-RHR (Fig. 3B), suggesting that either the interaction domain maps to the middle of the protein or the whole Pelle protein is necessary for Dorsal binding.
Dorsal Binds to Tube and Pelle in Domain 1 of the RHR.
The structural analysis of the p50 RHR bound to DNA defines two domains (9, 10). These domains can also be subdivided functionally. In Dorsal, as in all Rel proteins, the sequences required for DNA binding are located in domain 1 (11). Domain 1 was also found to contain sequences important for the nuclear import of Dorsal (8). The Cactus binding site is located in domain 2. One region of six amino acids (amino acids 230–236) is essential for Cactus binding (39), and the mutation of a serine residue at position 234 to proline strongly reduces the interaction between Dorsal and Cactus (41).
To determine which domain of the Dorsal RHR mediates Tube and Pelle binding, we assayed the two RHR domains for their interaction with Tube and Pelle in the yeast interaction assay. Fig. 3C shows that domain 2 of the DL-RHR from amino acids 222–342 interacts with Cactus strongly, whereas it fails to bind to either Tube or Pelle. Furthermore, we find that the Cactus binding site is not involved in the binding of Tube and Pelle. The known Cactus-binding defective mutant DLS234P does not affect the interaction of the Dl-RHR with either Tube or Pelle, whereas its interaction with Cactus is strongly reduced. These results indicate that the interaction domain of the DL-RHR with Tube and Pelle maps to domain 1. To test if domain 1 is sufficient to mediate Tube and Pelle binding, we generated a construct deleting domain 2. This construct, DL35–246, fails to interact with Cactus; however, it retains substantial binding activity with Tube and Pelle (Fig. 3C). Thus, the region essential for Tube and Pelle binding is located in the N-terminal domain of DL-RHR.
These findings above were further confirmed by gel-shift assays. The DL47–244 protein contains the DNA binding domain and was shown previously to be capable of binding to the zenB κB site (11). Our results show that both bacterially expressed Tube and Pelle can inhibit DNA binding of domain 1 (Fig. 2B, lanes 5–8). In comparison, the negative control, D-Lis1 protein, did not show any inhibition of Dorsal binding (lane 2). When Cactus is used in this same assay, there is at most a weak inhibition (lane 3) that does not change significantly with increasing amounts of Cactus (lane 4).
DISCUSSION
In our two-hybrid screen for proteins interacting with the Dorsal RHR, we identified Tube as a strong Dorsal interactor. This result was unexpected because in injection rescue experiments, mRNA encoding an activated form of Pelle rescued the tube phenotype, suggesting that the signal in the intracellular part of the pathway is transmitted from Toll via Tube to Pelle and then to the Dorsal–Cactus complex (29, 30). Our results do not contradict this sequence of events but suggest that the components of the pathway form a complex that is essential for the transmission of the signal. Consistent with this idea, we found that Pelle also interacts with Dorsal and hence is part of the complex.
In the absence of Tube or Pelle, Dorsal does not dissociate from Cactus and remains cytoplasmic (13, 15). Injection rescue experiments have shown that when an mRNA encoding only the N-terminal domain of Tube is injected into embryos from tube homozygous mutant females, the encoded protein can partially rescue the mutant phenotype. However, even at very high doses, at which wild-type RNA rescues 50% of embryos to hatching, this truncated mRNA could not fully rescue embryos (25). To confirm this observation, we retested the function of a minigene encoding only the N-terminal half of tube (25). The minigene has also only partial function. The injection rescue and the results from the transgenic experiments together strongly indicate that the C-terminal half is essential for full function of Tube. We have mapped the Dorsal interaction in Tube to the C-terminal half of the protein. Together, these results suggest that the signal transduction is compromised when Tube does not directly interact with Dorsal.
It has been shown in Drosophila tissue culture cells that Tube targets to the nucleus in concert with Dorsal (42), leading to the conjecture that Tube may function as a chaperone of Dorsal and possibly function with Dorsal in the nucleus. Our results do not support any function of Tube with Dorsal in the nucleus. The presence of Tube inhibits Dorsal DNA binding in our in vitro binding experiment.
The formation of the Dorsal nuclear gradient is the result of two distinct events. The first event is the signal-dependent Cactus dissociation from Dorsal, coupled with the degradation of Cactus (31, 32, 43). In a complete loss of Cactus function background, Dorsal targets to nuclei all along the dorsal–ventral axis and still forms a gradient, albeit a shallow one (32). A second event controls the formation of this weak gradient. It is also dependent on the ventral signal transmitted via Toll, because in the absence of Cactus and the ventral signal, the distribution of Dorsal is uniform all along the axis (44).
Our finding of Tube and Pelle interacting with Dorsal shows that there is a direct connection between Dorsal and the ventral signal. It could be that Dorsal functions as an adaptor that recruits Tube and Pelle so they can ultimately modify Cactus. But in addition to this function on Cactus, Pelle and Tube also control the high level of Dorsal import seen on the ventral side of the embryos in the absence of Cactus. This could be achieved through specific phosphorylation of “free” Dorsal.
Consistent with the observation that Tube and Pelle control nuclear targeting of Dorsal, we find that both Tube and Pelle interact with Dorsal in domain 1 of the RHR. This domain has been shown to be essential for nuclear targeting. A Dorsal protein consisting only of domain 2 of the RHR is structurally sound enough to form dimers and to interact with Cactus normally, but it fails to translocate to the nucleus either in the presence or absence of Cactus (8). The failure of Tube–Dorsal interaction results in a partial loss of function, indicating that at least some Dorsal protein targets to the nucleus. In the absence of domain 1 of the Dorsal RHR, the protein does not target to the nucleus at all. This phenotype might possibly be explained by the failure of both Tube and Pelle complexing with Dorsal.
The establishment of the dorsal–ventral axis in the early embryo is thoroughly dependent on the nuclear concentration of the Dorsal protein. This concentration seems to be indirectly dependent on the graded activation, by the Spätzle ligand, of the Toll transmembrane receptor (45). A close connection has been suggested between the signal transmitted via Toll and the amount of the Dorsal–Cactus complex (46). Reduction of the amount of the complex to one-half in females, by elimination of one copy of the dorsal gene, often results in a weak dorsalized embryonic phenotype. On the other hand, this same weak dorsalized phenotype is also found when the signal is reduced. This is the case in transheterozygote combinations of certain Toll and Spaetzle alleles. However, if in this reduced signaling situation the amount of the Dorsal–Cactus complex is increased, the dorsalized phenotype is rescued. Our results suggest that the tight regulation of the transmission of the signal may be mediated by the formation of a multimeric complex upon activation of the Toll receptor. This complex is expected to be present only transiently because it disintegrates when Dorsal dissociates from Cactus.
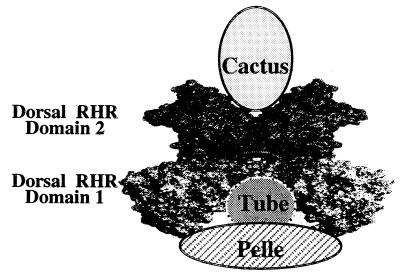
Our experiments show that the four proteins interact with one another through distinct sequences and that the proteins do not compete for binding sites. This should allow the formation of a complex. A model of how this complex may look is shown in Fig. 4. Based on the structure of the p50 homodimer bound to DNA, it has been proposed that Cactus binds between the two domain 2s of the RHR and that this binding would result in dislocating Dorsal from DNA (41). Similarly, it may be possible that either one or two molecules of the Tube–Pelle complex bind between the two domain 1s of the Dorsal dimer, also effecting the dissociation of Dorsal from DNA.
Figure 4.
A model of the proposed Dorsal-Cactus–Tube-Pelle complex. The Dorsal RHR is shown as a dimer. Domain 1 is shaded in light gray, and domain 2 is darker.
Our results show that protein–protein interaction is an essential aspect of signal transduction and suggest that a signal is most efficiently transduced through the formation of a multimeric complex. In addition to the four proteins, Cactus, Dorsal, Tube, and Pelle, the complex may well contain other kinases and other unidentified proteins.
Acknowledgments
We thank H. S. Huang for assistance in this project and Le Nguyen for fly food. We are especially grateful to Jörg Grosshans for the ovarian cDNA library, and we thank Steven Wasserman for the ptub462 and ptubC205 transgenic lines and Michael Levine for the pGEXdl244 plasmid. We thank P. H. Lin for help and Eric Drier, Kim McKim, David Norris, and Drew Vershon for suggestions on the manuscript. This work was supported by the Horace W. Goldsmith Foundation, a Waksman Institute fellowship (J.Y.), and a grant from the National Institutes of Health.
ABBREVIATION
- RHR
Rel homology region
References
- 1.Verma I M, Stevenson J K, Schwarz E M, Antwerp D V, Miyamoto S. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 2.Drier E, Steward R. Semin Cancer Biol. 1997;8:83–92. doi: 10.1006/scbi.1997.0059. [DOI] [PubMed] [Google Scholar]
- 3.Hashimoto C, Hudson K L, Anderson K V. Cell. 1988;52:269–279. doi: 10.1016/0092-8674(88)90516-8. [DOI] [PubMed] [Google Scholar]
- 4.Hashimoto C, Gerttula S, Anderson K V. Development. 1991;111:1021–1028. doi: 10.1242/dev.111.4.1021. [DOI] [PubMed] [Google Scholar]
- 5.Schneider D S, Hudson K L, Lin T Y, Anderson K V. Genes Dev. 1991;5:797–807. doi: 10.1101/gad.5.5.797. [DOI] [PubMed] [Google Scholar]
- 6.Medzhitov R, Preston-Hurlburt P, Janeway C A. Cell. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 7.Steward R. Science. 1987;238:692–694. doi: 10.1126/science.3118464. [DOI] [PubMed] [Google Scholar]
- 8.Govind S, Drier E, Huang L H, Steward R. Mol Cell Biol. 1996;16:1103–1114. doi: 10.1128/mcb.16.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh G, Duyne G V, Ghosh S, Sigler P B. Nature (London) 1995;373:303–310. doi: 10.1038/373303a0. [DOI] [PubMed] [Google Scholar]
- 10.Muller C W, Rey F A, Sodeoda M, Verdine G L, Harrison S C. Nature (London) 1995;373:311–317. doi: 10.1038/373311a0. [DOI] [PubMed] [Google Scholar]
- 11.Tatei K, Levine M. Mol Cell Biol. 1995;15:3627–3634. doi: 10.1128/mcb.15.7.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kidd S. Cell. 1992;71:623–635. doi: 10.1016/0092-8674(92)90596-5. [DOI] [PubMed] [Google Scholar]
- 13.Roth S, Stein D, Nusslein-Volhard C. Cell. 1989;59:1189–1202. doi: 10.1016/0092-8674(89)90774-5. [DOI] [PubMed] [Google Scholar]
- 14.Rushlow C A, Han K, Manley J L, Levine M. Cell. 1989;59:1165–1177. doi: 10.1016/0092-8674(89)90772-1. [DOI] [PubMed] [Google Scholar]
- 15.Steward R. Cell. 1989;59:1179–1188. doi: 10.1016/0092-8674(89)90773-3. [DOI] [PubMed] [Google Scholar]
- 16.Geisler R, Bergmann A, Hiromi Y, Nusslein-Volhard C. Cell. 1992;71:613–621. doi: 10.1016/0092-8674(92)90595-4. [DOI] [PubMed] [Google Scholar]
- 17.Jiang J, Levine M. Cell. 1993;72:741–752. doi: 10.1016/0092-8674(93)90402-c. [DOI] [PubMed] [Google Scholar]
- 18.Ferguson E L, Anderson K V. Curr Top Dev Biol. 1991;25:17–43. doi: 10.1016/s0070-2153(08)60410-x. [DOI] [PubMed] [Google Scholar]
- 19.Hatada E N, Nieters A, Wulczyn F G, Naumann M, Meyer R, Nucifora G, McKeithan T W, Scheidereit C. Proc Natl Acad Sci USA. 1992;89:2489–2493. doi: 10.1073/pnas.89.6.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaffray E, Wood K M, Hay R T. Mol Cell Biol. 1995;15:2166–2172. doi: 10.1128/mcb.15.4.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Govind S, Whalen A M, Steward R. Proc Natl Acad Sci USA. 1992;89:7861–7865. doi: 10.1073/pnas.89.17.7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isoda K, Nüsslein-Volhard C. Proc Natl Acad Sci USA. 1994;91:5350–5354. doi: 10.1073/pnas.91.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hecht P M, Anderson K V. Genetics. 1993;135:405–417. doi: 10.1093/genetics/135.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Letsou A, Alexander S, Orth K, Wasserman S A. Proc Natl Acad Sci USA. 1991;88:810–814. doi: 10.1073/pnas.88.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letsou A, Alexander S, Wasserman S. EMBO J. 1993;12:3449–3458. doi: 10.1002/j.1460-2075.1993.tb06019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shelton C A, Wasserman S A. Cell. 1993;72:515–525. doi: 10.1016/0092-8674(93)90071-w. [DOI] [PubMed] [Google Scholar]
- 27.Cao Z, Henzel W J, Gao X. Science. 1996;271:1128–1131. doi: 10.1126/science.271.5252.1128. [DOI] [PubMed] [Google Scholar]
- 28.Feinstein E, Kimchi A, Wallach D, Boldin M, Varfolomeev E. Trends Biochem Sci. 1995;20:342–344. doi: 10.1016/s0968-0004(00)89070-2. [DOI] [PubMed] [Google Scholar]
- 29.Grosshans J, Bergmann A, Hafftner P, Nusslein-Volhard C. Nature (London) 1994;372:563–566. doi: 10.1038/372563a0. [DOI] [PubMed] [Google Scholar]
- 30.Galindo R L, Edwards D N, Gillespie S K H, Wasserman S A. Development. 1995;121:2209–2218. doi: 10.1242/dev.121.7.2209. [DOI] [PubMed] [Google Scholar]
- 31.Reach M, Galindo R L, Towb P, Allen J L, Karin M, Wasserman S. Dev Biol. 1996;180:353–364. doi: 10.1006/dbio.1996.0308. [DOI] [PubMed] [Google Scholar]
- 32.Bergmann A, Stein D, Geisler R, Hagenmaier S, Schmid B, Fernandez N, Schnell B, Nusslein-Volhard C. Mech Dev. 1996;60:109–123. doi: 10.1016/s0925-4773(96)00607-7. [DOI] [PubMed] [Google Scholar]
- 33.Whalen A M, Steward R. J Cell Biol. 1993;123:523–534. doi: 10.1083/jcb.123.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gillespie S K H, Wasserman S. Mol Cell Biol. 1994;14:3559–3568. doi: 10.1128/mcb.14.6.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finley R L, Brent R. Proc Natl Acad Sci USA. 1994;91:12980–12984. doi: 10.1073/pnas.91.26.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gyuris J, Golemis E A, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 37.Ausubel F M, Brent R, Kingston R E, Moore D D. Current Protocols in Molecular Biology. New York: Wiley; 1996. [Google Scholar]
- 38.Hoey T, Levine M. Nature (London) 1988;332:858–861. doi: 10.1038/332858a0. [DOI] [PubMed] [Google Scholar]
- 39.Ip Y T, Park R E, Koosman D, Bier E, Levine M. Genes Dev. 1992;6:1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- 40.Wieschaus E, Nüsslein-Volhard C, Jurgens G. Roux’s Arch Dev Biol. 1984;193:296–307. doi: 10.1007/BF00848158. [DOI] [PubMed] [Google Scholar]
- 41.Lehming N, McGuire S, Brinkman J M, Ptashne M. Proc Natl Acad Sci USA. 1995;92:10242–10246. doi: 10.1073/pnas.92.22.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Norris J L, Manley J L. Genes Dev. 1995;9:358–369. doi: 10.1101/gad.9.3.358. [DOI] [PubMed] [Google Scholar]
- 43.Belvin M P, Jin Y, Anderson K V. Genes Dev. 1995;9:783–793. doi: 10.1101/gad.9.7.783. [DOI] [PubMed] [Google Scholar]
- 44.Isoda K, Roth S, Nüsslein-Volhard C. Genes Dev. 1992;6:619–630. doi: 10.1101/gad.6.4.619. [DOI] [PubMed] [Google Scholar]
- 45.Morisato D, Anderson K. Cell. 1994;76:677–688. doi: 10.1016/0092-8674(94)90507-x. [DOI] [PubMed] [Google Scholar]
- 46.Govind S, Brennan L, Steward R. Development. 1993;117:135–148. doi: 10.1242/dev.117.1.135. [DOI] [PubMed] [Google Scholar]