Abstract
The transcription factor Pax-5, is vital during B lymphocyte differentiation and is known to contribute to the oncogenesis of certain cancers. The Pax-5 locus generates multiple yet structurally related mRNA transcripts through the specific activation of alternative promoter regions and/or alternative splicing events which poses challenges in the study of specific isoform function. In this study, we investigated the function of a major Pax-5 transcript, Pax-5B using an enhanced version of the Hepatitis Delta Virus ribozyme (HDV Rz) suppression system that is specifically designed to recognize and cleave the human Pax-5B mRNA. The activity of these ribozymes resulted in the specific suppression of the Pax-5B transcripts without altering the transcript levels of other closely related Pax-5 isoforms mRNAs both in vitro and in an intracellular setting. Following stable transfection of the ribozymes into a model B cell line (REH), we showed that Pax-5B suppression led to an increase of CD19 mRNA and cell surface protein expression. In response to this Pax-5B specific deregulation, a marked increase in apoptotic activity compared to control cell lines was observed. These results suggest that Pax-5B has distinct roles in physiological processes in cell fate events during lymphocyte development.
INTRODUCTION
Recent studies have demonstrated that the Pax gene family controls gene expression activity at the transcription level and regulates fundamental cellular processes such as proliferation, differentiation and cell death events (apoptosis) (1,2). Given their essential role in cell biology, deregulated Pax genes also act as oncogenes and have the potential to transform normal cells into their cancerous counterparts (2,3). A well-studied member of this family, Pax-5, is known to encode the B-cell specific activator protein (BSAP) which is expressed in the embryonic central nervous system, B lymphocytes, and adult testis (4). In the past decade, the interest in the Pax-5 transcription factor has been drawn to its pivotal role in B-cell lymphogenesis (5,6). Pax-5 requirement to B commitment is highlighted by its transcriptional control over key events that are essential for B-cell development and activation. Studies have identified a number of Pax-5 target genes several of which are known to modulate the expression of B-cell receptor (BCR) components and associated proteins (i.e. CD19, blk etc.) (7,8). Pax-5 selective pressure to B-cell commitment is also conveyed by its ability to negatively regulate non-conforming pathways of the B lineage leading to the differentiation of non-B cellular development (9,10).
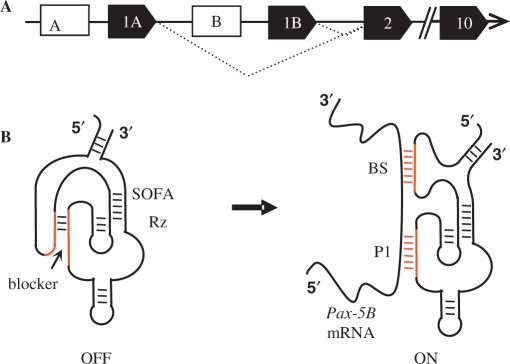
At the genetic level, human Pax-5 is mapped to chromosome 9 in region p13 where it is regulated by two distinct promoters regions: first a TATA-containing upstream promoter associated with an exon 1A, and a TATA-less downstream promoter coupled with an exon 1B. Through regulated signaling processes along with the recruitment of specific transcriptional regulators, each promoter induces the transcription of a distinct 5′UTR region and a first exon which then splices to a coding sequence (exons 2–10) common to both transcript isoforms (11,12). These two mRNAs (noted Pax-5A and Pax-5B) are discriminated only by their unique 5′UTR sequence and exon 1 of the coding region (Figure 1A).
Figure 1.
Schematic structure of the human Pax-5 gene and the SOFA-HDV Rz (A) The human Pax-5 gene located on chromosome 9 includes two alternative promoter regions (empty boxes\promoters A and B) which regulate the expression of exclusive exon regions (solid boxes) 1A and B, respectively, followed a common exons 2–10 sequence. The dotted diagonal lines indicate the spliced regions from the mature coding transcripts. (B) The structure of the SOFA-HDV Rz adopts an off (left panel) and locked configuration secured by the blocker stem in the absence of its specific mRNA target (Pax-5B). In the presence of the specific substrate (right panel) the SOFA-HDV Rz adopts a catalytically active configuration which is formed through the annealing of the ribozyme's biosensor (BS) and P1 stem (P1) to the target substrate. These structures were previously described in (18,19).
Although a great number of studies have contributed to the functional characterization of Pax-5 at the molecular and cellular level, few have discriminated between the physiological roles of Pax-5A and Pax-5B isoforms individually. Given the fact that most Pax-5 bearing cells express both Pax-5 isoforms simultaneously (almost a 1 : 1 ratio in human pre-B cells), the functional study of these alternatively spliced products has proven to be complex and very challenging. As a result, little attention has been paid to the cellular characterization of Pax-5B. Therefore, we set out to develop a Pax-5B gene suppression system using a modified ribozyme (Rz) technology to investigate the specific roles associated with Pax-5B in human B lymphocytes.
The ability of a ribozyme to specifically recognize, and subsequently catalyze the cleavage of an RNA target makes it an attractive molecular tool for the development of gene inactivation systems (13). It provides an interesting alternative to an RNA-interference (RNAi) approach in functional genomics, especially given the recent findings that RNAi specificity is limited and evidence shows that this mechanism can trigger immunological response (14,15). The ribozyme isolated from the hepatitis delta virus (HDV Rz) appears to be the most suited to develop a gene-inactivation in human cells since it has evolved within this particular cell environment (13). In support of this, results have shown this catalytic RNA to be fully active in physiological concentrations of magnesium in human cells (16). However, the target recognition mechanism of the HDV Rz is dependant on the formation of a 7 base pair (bp) between the Rz and the substrate, (namely the P1 stem/Figure 1B). It has been estimated that a minimum sequence of 15 or 16 base pairing nucleotides are required for the targeting of unique RNA species in the human transcriptome, thereby limiting non-specific cleavage (17). In order to fulfill these parameters, a new generation of HDV Rz was engineered which incorporates a Specific On/ofF Adaptor (SOFA) that switches the cleavage activity from off to on solely in the presence of the appropriate substrate (Figure 1B) (18,19). A unique feature of the SOFA-HDV Rz allows it to limit the catalytic activity via a blocking sequence that prevents the folding of the ribozyme into an active conformation. This ‘safety lock’ function prevents cleavage of a substrate that contains an appropriate P1 stem but lacks a complementary sequence to the SOFA biosensor. This new target-dependent module biosensor bearing Rz significantly reduces non-specific cleavage events.
In this study, we developed an isoform-specific SOFA-HDV Rz capable of recognizing and cleaving human Pax-5B transcripts both in vitro and in vivo. Using this gene suppression system, we report that the Pax-5B transcriptional element is associated with changes in target gene activation as well as to the balance of cell fate events in human B lymphocytes.
MATERIALS AND METHODS
Plasmids and ribozyme DNA constructs
The Pax-5A cDNA was cloned as described previously (20). The Pax-5B transcript was cloned into pBlueScript (SK) (Stratagene, Cedar Creek, Texas) through an RT-PCR amplification on RNA extracts from Ficoll-isolated PBLs (peripheral blood lymphocytes) using specific primers Pax-5B sense 5′-CCGATGGAAATACACTGTAAGC-3′ and Pax-5B antisense 5′-CTCCAAGGGTCAGTGACGG-3′. The Pax-5B insert was then subcloned into the pcDNA3.1 expression vector (Invitrogen, Burlington, Ontario, Canada) using a directional cloning technique involving the XhoI, XbaI and NotI restriction sites depending on the insert's original orientation.
Pax-5B specific SOFA-HDV Rz were constructed as described previously (18). Briefly, SOFA-HDV Rz were constructed using a PCR-based strategy that included two complementary and overlapping oligodeoxynucleotides: the antisense oligonucleotide 5′-CCAGCTAGAAAGGGTCCCTTAGCCATCCGCGAACGGATGCCC(ANNNNNN)P1ACCGCGAGGAGGTGGACCCTG(N)4-BL-3′ (where N is A, C, G or T, P1 and BL indicate the P1 strand region and the blocker sequence, respectively) and, the sense 5′-TTAATACGACTCACTATAGGGCCAGCTAGTTT(N)12-BS(N)4-BLCAGGGTCCACC-3′ (where N is A, C, G or T, BS indicates biosensor sequence and BL the blocker sequence, respectively) that permitted the incorporation of the T7 RNA promoter (underlined). It is important to note that the (N) segments vary in sequence depending on the targeted site accessible on the target RNA substrate (Table 1). The DNA coding SOFA-HDV Rz were rendered double stranded by PCR which were later purified by phenol:chloroform extraction, precipitated with ethanol and dissolved in water. The double-stranded DNA (dsDNA) fragments were either used for in vitro transcriptions (described below), or cloned without the T7 promoter region into a polymerase III-based expression vector psiRNA-hH1-GFP/Zeo (Invivogen, San Diego, CA, USA).
Table 1.
Nucleotide sequences of Pax-5B target regions
| SOFA-HDV-Rz | Targeted sequence |
|---|---|
| B75 | CCCG/ATGGAA ATACACTGTAAGCACGACCC |
| B78 | GATG/GAAATACACTGTAAGCACGACCCGTT |
| B89 | ACTG/TAAGCACGACCCGTTTGCATCCATGC |
| B93 | TAAG/CACGACCCGTTTGCATCCATGCATAG |
| B97 | CACG/ACCCGT TTGCATCCATGCATAGACAT |
(/) indicates the cleavage site of ribozyme.
The complementary sequence to the P1 binding site of the SOFA-HDV-Rz is indicated in bold.
The complementary sequence to the biosensor of the SOFA-HDV-Rz is underlined
RNA synthesis
Both SOFA-HDV Rz and substrates were synthesized by run-off transcription from PCR products and HindIII linearized pcDNA3.1-Pax-5B templates, respectively, as described previously (18). Briefly, transcriptions were performed in the presence of purified T7 RNA polymerase (10 µg), RNA Guard (24 U, Amersham Biosciences), pyrophosphatase (0.01 U, Roche Diagnostics, Laval, QC, Canada) and either linearized plasmid DNA (5 µg) or PCR product (2–5 µM) in a buffer containing 80 mM HEPES-KOH pH 7.5, 24 mM MgCl2, 2 mM spermidine, 40 mM DTT, 5 mM of each NTP in a final volume of 100 µl at 37°C for 2–4 h. Upon completion, the reaction mixtures were treated with DNase RQ1 (Amersham Biosciences, Piscataway, NJ) at 37°C for 20 min, the RNA purified by phenol:chloroform extraction and then precipitated with ethanol. The RNA products were fractionated by denaturing (6 or 10%) polyacrylamide gel electrophoresis (PAGE; 19 : 1 ratio of acrylamide to bisacrylamide) in buffer containing 45 mM Tris-borate pH 7.5, 7 M urea and 1 mM EDTA. The reaction products were visualized either by UV shadowing or autoradiography. The bands corresponding to the correct sizes for both the SOFA-HDV Rz and substrate RNA molecules were cut out and the transcripts eluted overnight at room temperature. The transcripts were desalted on Sephadex G-25 (Amersham Biosciences) spun-columns, and were then ethanol precipitated and quantified by absorbance at 260 nm.
Ribozyme cleavage assays
The conditions for the SOFA-HDV Rz cleavage assays have been described previously (18). Briefly, in all reactions the ribozymes and substrate were preincubated together at 37°C for 2–5 min; thereafter, the magnesium was added to initiate the cleavage reaction. Reactions were performed using 1 µM HDV Rz and trace amounts of 5′-terminal [32P]-labeled substrate at 37°C in a final volume of 10 μl containing 50 mM Tris–HCl pH 7.5 and 5 mM MgCl2. Experiments were incubated at 37°C for 2.5 h, stopped by the addition of ice-cold formamide dye buffer (5 µl of 97% formamide, 10 mM EDTA pH 8.0, 0.025% xylene cyanol and 0.025% bromophenol blue), electrophoresed on an 8% PAGE gel and exposed onto a PhosphorImager screen (Molecular Dynamics, Sunnyvale, CA, USA).
Cells lines and transfections
HEK293 and 293T cells (human embryonic kidney) were obtained from the American Type Culture Collection (Rockville, MD, USA) and the REH (acute lymphocytic leukemia cells) cell line was kindly provided by Dr Edward A. Clark, University of Washington Medical Center, Seattle. We also made of HEK293 stably transfected with the full length human Pax-5B coding region, the HEK 293-B2 cell line, which was created in our laboratory. Cell lines were maintained in complete culture medium made of DMEM (HEK) or RPMI-1640 (REH) supplemented with 10% fetal bovine serum (FBS; Hyclone Laboratories, Logan, UT, USA), l-glutamine (2 mM), penicillin G (100 U/ml), and streptomycin (100 µg/ml) (Invitrogen). Cells were either left untreated or treated (as described in figure legends) with 2 µM of Actimomycin D (Sigma, St Louis, Missouri).
Transient transfections of the HEK cell lines and stably transfected REH cells were conducted using the Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Briefly, cells were seeded into 6-well plates (2 × 105/well) where they received a total of 2 μg (or indicated otherwise) of plasmid DNA that was complexed with the provided transfection reagent.
Northern Blot hybridizations
Northern blot analyses were performed as described previously (18) using 10 µg of RNA isolated by TrizolTM (Invitrogen). The Pax-5B probe was obtained by a T3 RNA polymerase-based transcription in the presence of [α-32P]UTP on a pBlueScript Pax-5B construct. The β-actin RNA probe used was prepared using the Strip-EZTM RNA T7/T3 kit (Ambion, Austin, TX, USA). All hybridizations were carried out for 16–18 h at 65°C. The membranes were then exposed to PhosphorImager screens for 2–24 h and thereafter analyzed on a radioanalytic scanner (Storm, Amersham Biosciences).
Preparation of nuclear extracts and electrophoretic mobility shift assays (EMSA)
Nuclear extracts were prepared according to the previously described microscale preparation protocol and analyzed by gel shift assays previously described (20). Briefly, cells (5 × 106) were washed with ice-cold PBS and resuspended in 400 μl of cold buffer A [10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES; pH 7.9), 10 mM KCl, 0.1 mM EGTA, 0.1 mM EDTA, 1 mM DTT, and 0.5 mM phenylmethylsulfonyl fluoride (PMSF)]. The cells were allowed to swell on ice for 15 min, after which 25 μl of a 10% solution of Nonidet P-40 was added and the tube was vigorously vortexed for 30 s. The homogenate was centrifuged for 10 s at 12 000g. The supernatant fraction was discarded and the pellet was resuspended in 50 μl of cold buffer C [20 mM HEPES-KOH (pH 7.9), 0.4 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT and 1 mM PMSF] and incubated at 4°C for 15 min on a shaking platform. Cellular debris were removed by centrifugation at 12 000g for 20 min at 4°C and the supernatant fractions were stored at −270°C until used. Ten micrograms of nuclear extracts were used to perform EMSA as determined by the bicinchoninic acid assay (BCA) with a commercial protein assay reagent (Pierce, Rockford, IL, USA). Nuclear extracts were incubated for 20 min at room temperature in 20 μl of the binding buffer (100 mM HEPES (pH 7.9), 40% glycerol, 10% Ficoll, 250 mM KCl, 10 mM dithiothreitol, 5 mM EDTA, 250 mM NaCl, 2 μg poly (dI-dC) and 10 μg nuclease-free bovine serum albumin fraction V) containing 1 ng of 32P-5′-end-labeled double-stranded (dsDNA) oligonucleotide. Double-stranded DNA (100 ng) was labeled with [γ-32P]ATP and T4 polynucleotide kinase in a kinase buffer (New England Biolabs, Beverly, MA, USA). This mixture was incubated for 30 min at 37°C and the reactions were stopped with 5 μl of 0.2 M EDTA. The labeled oligonucleotides were extracted with phenol/chloroform and passed through a G-50 spin column. The dsDNA oligonucleotides, which were used as probes, contained the consensus NF-κB and the BSAP sites corresponding to the sequences 5′-ATGTGAGGGGACTTTCCCAGGC-3′ and 5′-GAATGGGGCACTGAGGCGTG-3′, respectively. The DNA-protein complexes were migrated on a 4% (w/v) polyacrylamide gel containing Tris-Borate-EDTA. The gels were then subsequently dried and autoradiographed. Cold-competition assays were carried out by adding a 100-fold molar excess of unlabeled dsDNA oligonucleotide simultaneously with the labeled probe.
Quantitative RT–PCR and mRNA half-life analysis
Levels of gene expression were verified by quantiative PCR. Reaction mixtures were composed of 20 nM each sense and antisense primers and 0.1 µg of human total cDNA, diluted in water to a total of 12.5 µl and mixed with 12.5 µl of 2X iQ SYBR Green Supermix (BioRad, Mississauga, Ontario, Canada) where they were run in a Smart Cycler II real-time PCR apparatus (Cepheid, Sunnyvale, CA, USA) with the following parameters: 2 min initial denaturation, 35 cycles of 1 min at 94°C, 1 min at 55°C (optics on), 1 min at 72°C, followed by melt curve analysis from 65°C to 95°C. Quantitative PCR applications were conducted on Pax-5B sense 5′-TTGGCACGAGGTAGACAC-3′ and antisense 5′-AGCCCTCCAACACTGAGA-3′; SOFA-HDV Rz sense 5′-ATCCATCGGGTACCGGGCCAGCTAGTTT-3′ and antisense 5′-CCAGCTAGAAAGGGTCCCTTAGCCATCCGCG-3′; and finally CD19 sense 5′-CCACCTGGAGATCACTG-3′ and antisense 5′-ATAAGCCAAAGTCACAGC-3′. Comparative expression levels were calculated using the ▵▵Ct method of Livak and Schmittgen (21), with the endogenously expressed gene hypoxanthine ribosyltransferase (HPRT) using sense 5′-TGACACTGGCAAAACAATGCA-3′ and antisense 5′-GGTCCTTTTCACCAGCAAGCT-3′ primers as the internal control. Amplification efficiencies were also considered and calculated for all real-time amplification products (data not shown). Where applicable, transcript decay rates were performed through the use of quantitative RT–PCR on RNA extracts from REH cells treated with actimomycin-D (2 μM) as previously described (22).
Flow cytometry analysis
Cells (106) were incubated for 30 min on ice with saturating concentrations (1 μg/106 cells) of PE-conjugated monoclonal antibodies directed against CD19 or HLA-DR (BD Bioscience, Mississauga, Ontario, Canada). Finally, cells were resuspended in 500 μl of phosphate-buffered saline (PBS) before flow cytometry analysis (FACS Calibur, BD Biosciences, Miami, FL, USA). Controls consisted of commercial isotype-matched irrelevant murine monoclonal antibodies (Sigma, Oakville, Ontario, Canada) (10 000 events/sample).
Cell proliferation and apoptosis assays
Cells (107) were seeded in 96 well plates and analyzed in time for cellular viability and apoptotic events using a multiplexed assay of CellTiter Blue (Promega, Madison, WI, USA) and Apo-One (Promega) assay kits, respectively, according to the manufacturer's instructions. Briefly, 20 μl of the CellTiter Blue substrate was added to 100 μl of cell suspension and incubated for 1 h at room temperature on a plate shaker. Thereafter, the plate was subjected to a colorimetric analysis and read on a fluorescence microplate reader (560Ex/590Em). Apoptotic events were then measured on the same microplate by the addition of 120 µl of Apo-ONE, incubated for another hour and analyzed by fluorescence (485Ex/527Em).
RESULTS
In order to investigate the functional role of the human Pax-5B transcription factor, we have developed a RNA-based gene suppression system which has the ability to specifically recognize, bind and cleave an RNA substrate in a sequence-specific manner. The specific SOFA-HDV Rz were designed to target sequences within the exon 1B that includes non-translated and coding sequence of the human Pax-5 gene (Figure 2). Target sequences were identified considering the substrate preference of the SOFA-HDV Rz (i.e. a guanosine residue in position +1 of the cleavage site, no guanosine in position –1, and no two consecutive pyrimidines in positions –1 and –2 (23). Moreover, in order to ensure the substrate specificity of the SOFA-HDV Rz, the sequences targeted were analyzed using RiboSubstrates, an integrated bioinformatics software that enables a search of a cDNA database for all potential substrates for a given ribozyme (24). This includes the mRNAs that perfectly match the specific requirements of a given SOFA-HDV Rz, as well those including wobble and mismatches base pairings. These analyses revealed that the designed SOFA-HDV Rz should solely cleave the Pax-5B mRNAs from the human transcriptome (data not shown).
Figure 2.
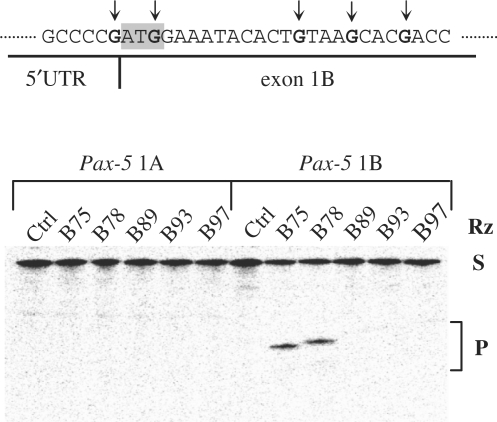
Autoradiogram of in vitro cleavage assays showing the cleavage specificity of the SOFA-HDV Rz directed against the human Pax-5B transcript. SOFA-HDV Rz were tailored against the human Pax-5B mRNA (above sequence) to target nucleotide positions 75, 78, 89, 93 and 97 of the exon 1 region (black arrows). In vitro cleavage assays were performed using 5′ terminal [32P]-labeled RNA regions of the human Pax-5A and Pax-5B transcript incubated with Pax-5B-specific SOFA-HDV Rz-B75, -B78, -B89, -B93 and -B97 and then migrated on to a 5% polyacrylamide gel and autoradiographed. On the right of the panel, ‘S’ indicates the substrates (∼600 bases), ‘Rz’ indicates the SOFA-HDV Rz and ‘P’ indicates the 5′ terminal cleaved products (∼75 and 78 bases).
SOFA-HDV Rz specifically cleave their appropriate human Pax-5 RNA substrate in vitro
In vitro cleavage assays were first conducted to evaluate the catalytic potential of each SOFA-HDV Rz directed against a partial region of the human Pax-5B transcript (Figure 2). SOFA-HDV Rz directed against the sequence position 75, 78, 89, 93 and 97 of the human Pax-5B mRNA sequence (labeled SOFA-HDV Rz-B75, -B78, -B89, -B93 and -B97, respectively) were incubated with a specific (Pax-5B) as well as with a non-specific (Pax-5A) RNA substrate. Following an incubation period, SOFA-HDV Rz-B75 and B78, efficiently recognized and cleaved their appropriate Pax-5B substrate (i.e. with 25.1 and 25.4% cleavage, respectively) while no observable cleaved products appeared in the reactions containing the Pax-5A RNA fragments. In the three other tests, SOFA-HDV Rz exhibited only minimal cleavage activity (∼1–2%) while no detectable cleavage of the Pax-5A mRNA was observed with Pax-5B specific ribozymes. The absence of cleavage activity of the Pax-5A-derived transcripts is an illustration that the SOFA-HDV Rz do not elaborate off-target cleavage activity in the presence of non-specific substrates. Importantly, the in vitro experiments led to the identification of two ribozymes possessing the greatest catalytic activity (SOFA-HDV Rz-B75 and HDV Rz-B78).
SOFA-HDV Rz suppressed Pax-5B intracellular levels in a dose-dependent manner
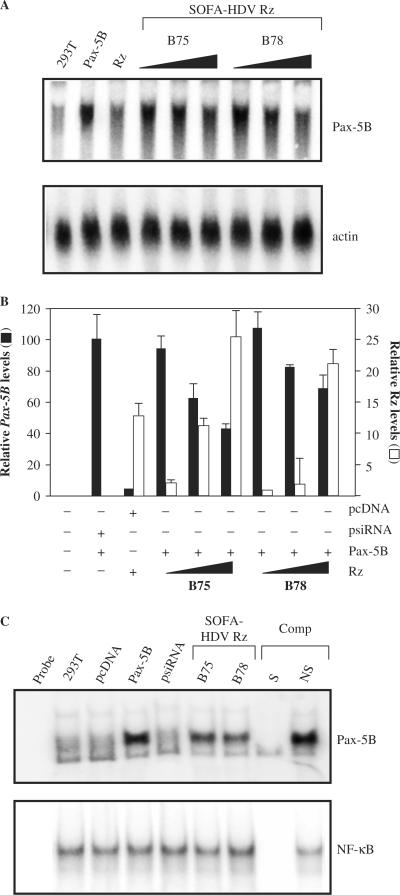
In order to determine the intracellular efficiency and catalytic potential of the two selected SOFA-HDV Rz to target Pax-5B, Northern blot hybridization on Pax-5 negative 293T cells co-transfected with a recombinant Pax-5B construct and increasing amounts of SOFA-HDV Rz DNA (10, 100 and 1000 ng) were performed (Figure 3A). The insert encoding the ribozymes were cloned in a Pol III-driven vector (psiRNA hH1). An increasing reduction of the intracellular levels of human Pax-5B transcripts was observed concomitant to increasing amounts of transfected specific ribozymes in all cases (Figure 3A and B). The highest concentration of SOFA-HDV Rz yielded a decrease of the Pax-5B substrate over 50% (Figure 3B). Control samples including untreated 293T cells, empty expression vectors, and individual Pax-5B and SOFA-HDV Rz transfectants were also included to evaluate ribozyme catalytic activity (Figure 3A). Thereafter, the same blots were subsequently hybridized with a human actin probe (Figure 3A, lower panel) to demonstrate that the decreasing levels observed in Pax-5B transcript expression were in fact due to specific ribozyme cleavage and not a general outcome of messenger RNA degradation following treatment. Evidence shows that SOFA-HDV Rz are capable of cleaving their specific substrate in a dose-dependant manner in an intracellular environment. Finally, Northern blot hybridizations were also performed using a probe for the specific detection of the Pax-5A isoform and no variations of the mRNA level was detected in the presence of Pax-5B specific ribozymes (data not shown). The latter results also supported the notion that SOFA-HDV Rz do not elicit off-target cleavage.
Figure 3.
In vivo evaluation of genetically engineered SOFA-HDV Rz to suppress the expression levels of the human Pax-5B gene products. (A) Northern analysis were conducted on RNA extracts from 293T cells transiently cotransfected with Pax-5B (500 ng); increasing amounts of SOFA-HDV Rz (1, 100 and 1000 ng) and finally filler DNA to totalize 1.5 μg. Individual controls are indicated as the untransfected cells (293T); cells cotransfected with Pax-5B along with the SOFA-HDV Rz empty vector (identified Pax-5B); and cells cotransfected with SOFA-HDV Rz along with the Pax-5B empty vector (identified Rz). Radiolabeled RNA probes were then hybridized onto the membrane to reveal transcript expression levels from intracellular Pax-5B (top panel) and actin (bottom panel) as an internal reference. (B) The radioactivity from the hybridized Pax-5B probes was then calculated and normalized on actin transcripts densities using a radioanalytic scanner and plotted with error bars in relation to expression ratios (in black). SOFA-HDV Rz RNA levels are also plotted accordingly (right of the panel, in white). (C) Electrophorectic Mobility Shift Assays (EMSA) were conducted on nuclear extracts from 293T cells cotransfected with Pax-5B and SOFA-HDV Rz. Nuclear extracts were incubated with either with Pax-5 (top panel) or NF-κB-specific (bottom panel) dsDNA oligonucleotides labeled at the 5′ end with [γ-32P]. DNA–protein complexes were resolved and analyzed on a 4% polyacrylamide gel. Specific (S) and non-specific (NS) cold competition (Comp) assays were carried out by adding a 100-fold molar excess of unlabeled dsDNA oligonucleotides corresponding to the Pax-5 or the NF-κB consensus-binding motif. Control lanes were loaded with free probe (Probe) and extracts from untouched 293T cells (293T). Other controls include transfections of the Pax-5B empty vector (identified pcDNA), the Pax-5B expression vector (identified Pax-5B) and finally the SOFA-HDV Rz empty vector (identified psiRNA).
In order to confirm the previous data, we then investigated the effect of mRNA reduction by SOFA-HDV Rz cleavage on protein expression levels. Since, there are no known anti-Pax-5B antibodies available, we analyzed Pax-5B protein levels via its DNA-binding activity by electrophoretic mobility shifts assays (EMSA). Using 293T cells co-transfected with recombinant Pax-5B and SOFA-HDV Rz plasmids, the levels of Pax-5B DNA-binding activity was measured using radiolabeled dsDNA consensus-binding sites for the Pax-5B transcription factor. As shown in figure 3C, a noticeable shifted complex in 293T expressing recombinant Pax-5B was observed while no protein-DNA complexes were detected in control samples (lanes Probe, 293T, pcDNA and psiRNA-hH1). More importantly, nuclear extracts from cells co-expressing Pax-5B with either SOFA-HDV Rz-B75 or SOFA-HDV Rz-B78, revealed a significant reduction in shifted complex intensity. Control EMSA experiments were also performed with the same nuclear extracts incubated with a radiolabeled NF-κB (Figure 3C, lower panel) DNA consensus-binding site. NF-κB binding efficiencies were not altered in any of the samples tested. Moreover, the shifted complexes in both assays were subjected to both specific and non-specific 100-fold excess cold competitions, thereby demonstrating the identity of the protein/DNA complexes.
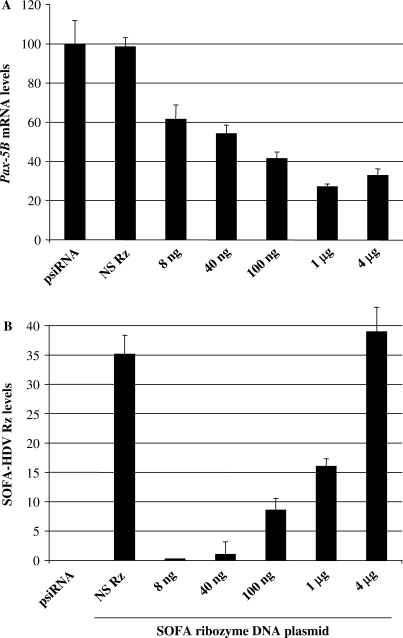
Next, we wanted to assess SOFA-HDV Rz activity in cells expressing physiological levels of intracellular Pax-5B substrates. To accomplish this, a 293-B2 cell clone which stably expresses the full coding region of human Pax-5B was generated. Subsequently, these cells were transiently transfected with increasing amounts of the most effective ribozyme, SOFA-HDV Rz-B75, (8 ng to 4 μg) and the level of Pax-5B mRNA was monitored by quantitative RT-PCR using primers amplifying a region spanning the 5′UTR and the first exon of the target substrate (Figure 4A). We observed a decrease of 76% in Pax-5B levels as intracellular levels of SOFA-HDV Rz-B75 increased following transfection treatments (Figure 4B). Conversely, no suppression was observed in control extract of 293-B2 cells transfected with 4 µg of either the expression vector alone (psiRNA-hH1) or with a non-specific functional SOFA-HDV Rz (NS Rz) containing a mismatched recognition motif unable to recognize the Pax-5B substrate. As expected, no ribozymes were detected in cells transfected with the empty vector alone, whereas increasing amounts of ribozyme expression was observed in other samples, which corroborated with the dose-response transfections of SOFA-HDV Rz in the 293-B2 cell line. The levels of Pax-5B were normalized in relation to the HPRT reference gene.
Figure 4.
Pax-5B tailored SOFA- HDV Rz cleave their intracellular target substrate in a dose-dependent manner. Quantitative RT–PCR was performed on the 293-B2 cell line (293 HEK cells stably expressing Pax-5B) transiently transfected with increasing amounts of SOFA-HDV Rz-B75 DNA to determine (A) Pax-5B transcripts and (B) SOFA-HDV Rz expression levels. Pax-5B and SOFA-HDV Rz levels were normalized using HPRT mRNA and plotted in relation to the relative percentage of the control 293-B2 cell line. Control samples are also included being the 293-B2 cell line transfected with the SOFA-HDV Rz empty vector (identified psiRNA) and a non-specific SOFA-HDV Rz (identified NS Rz) were also tested. Error bars are indicative of two independent sets of experiments.
Using nuclear protein extracts from the cell described above, we again analyzed Pax-5B DNA-binding suppression through EMSA analyses. We observed a strong Pax-5B shifted complex from 293-B2 nuclear extracts in the absence of SOFA-HDV Rz, while its presence led to a significant reduction of the amount of shifted Pax-5B complex (data not shown). Moreover, the shift was shown to be specific to Pax-5B protein since NF-κB DNA complexes did not vary in intensity following SOFA-HDV Rz transfections. Together, these data demonstrate that the intracellular suppression of Pax-5B expression is mediated through specific ribozyme activity which can be observed at both the transcriptional and translational levels.
Ribozyme-mediated suppression decreases target Pax-5B mRNA stability in B lymphocytes
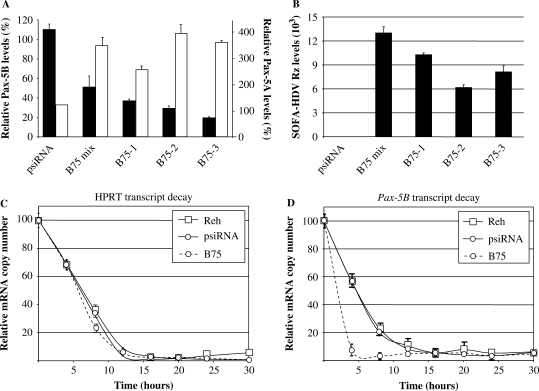
In order to investigate the gene suppression activities of the ribozyme on endogenously expressed Pax-5B, again the SOFA-HDV Rz-B75 construct was stably transfected in the REH B-cell line. Zeocin-resistant clones as well as a mixed population of transfectants were then analyzed by quantitative RT–PCR to assess SOFA-HDV Rz-mediated Pax-5B suppression. All amplified elements were normalized to HPRT expression. Stably transfected cell clones demonstrated a marked suppression up to 79% (clone B75-3) of Pax-5B mRNA levels in comparison with the parental REH cell line (Figure 5A). These REH B75 cell clones were also compared with a REH cell population stably transfected with the empty vector alone (psiRNA-hH1), which did not demonstrate any Pax-5B suppression activity when compared with the control parental REH cell line. The fact that Rz-mediated suppression of Pax-5B was observed in all screened REH B75 cell clones and in the mixed population of transfected cells demonstrates the consistent activity of the ribozyme in a B lymphocyte intracellular environment and rules out a clonal effect. Stable REH B75 clones were also analyzed for Pax-5A expression by real-time PCR. As predicted, we observed no cross-reactive suppression from our Pax-5B-specific SOFA-HDV Rz on Pax-5A mRNAs (Figure 5A). Interestingly, the SOFA-HDV Rz-mediated knockdown of Pax-5B resulted in an induction of Pax-5A mRNA expression. These results suggest that Pax-5B plays a negative role in the trans-activity of other Pax-5 isoforms (i.e. Pax-5A).
Figure 5.
SOFA-HDV Rz decrease Pax-5B intracellular levels by increasing Pax-5B mRNA decay rates in B lymphocytes. The REH B-cell line was stably transfected with either the empty vector (identified psiRNA) or Pax-5B specific SOFA-HDV Rz-B75 and antibiotic resistant clones were selected (clones 1, 2, 3 and mix populations). Quantitative RT–PCR was then performed on (A) Pax-5A (in white) and Pax-5B (in black) as well as (B) SOFA-HDV Rz expression levels in each cell population. Pax-5B and Rz transcript levels were normalized using the HPRT reference mRNA and plotted in relation to the percentage of their respective transcript levels (except for the Rz) in the non-transfected REH parental cell line. mRNA decay rates for (C) HPRT and (D) Pax-5B were also analyzed by quantitative RT-PCR and compared in time (hours) between REH (square symbols/solid line) cell clones stably transfected with psiRNA (circle symbols/solid line) and the SOFA-HDV Rz-B75 (circle symbols/dotted line) following actinomycin D (2 μM) treatment. The transcript half-life was then calculated in percentage ratio to respective mRNA levels in untreated cell clones and plotted error bars are indicative of two independent experiments.
Clones of REH cells stably transfected with SOFA-HDV Rz-B75 were then subjected to quantitative RT–PCR analysis to examine ribozyme expression in every REH clone in comparison to the REH parental cell line. Relatively high expression levels of SOFA-HDV Rz were observed in every cell population except for REH cells stably transfected with the empty vector (psiRNA-hH1) alone (Figure 5B). Since the REH B75-3 cell clone demonstrated the highest attenuation in Pax-5B levels, this latter cell clone was used in subsequent cellular and molecular analyses for the functional investigation of the Pax-5B gene product.
Initially, the specific mRNA decay rates of Pax-5B targeted by SOFA-HDV Rz in the REH cell line were examined. More specifically, Pax-5B mRNA stability in SOFA-HDV Rz-B75 bearing REH cells was monitored and compared with control cell populations (REH and REH/psiRNA-hH1) in time following a treatment of 2 μM actinomycin-D to inhibit mRNA synthesis. This was done to verify whether or not there were any differences in global transcript decay rates between the REH parental cell line and REH transfectants (psiRNA-hH1 or the SOFA-HDV Rz-B75). For this experiment, relative amounts of the HPRT mRNA from the different cell line clones were analyzed at various time points by quantitative RT–PCR analysis (Figure 5C). Following actinomycin-D treatment, a steady decline of HPRT transcripts was registered up to 12 h and these decay rates were identical in all cell lines tested. The half-lives of HPRT mRNA were calculated according to the slope which represented 2.9, 2.2 and 2.6 h for the REH, REH/psiRNA-hH1 and REH B75 cell lines, respectively. However, in the case where Pax-5B degradation rates were plotted in time following actinomycin-D treatment, a significant decrease in Pax-5B stability was observed in B cells containing the SOFA-HDV Rz-B75 construct in comparison to the control cell lines (Figure 5D). Interestingly, Pax-5B mRNA levels in the REH B75 clones were nearly undetectable after only 4 h of treatment, whereas these transcripts could still be observed up to 14 h in the other control cell lines (REH and REH/psiRNA-hH1. The Pax-5B mRNA half-life in REH B cells was calculated at 3.2, 3.3 and 0.7 h for the REH, REH/psiRNA-hH1 and REH B75 cell lines, respectively. These results strongly suggest that this suppression occurs through specific SOFA-HDV Rz activity which accelerates 4-fold the intracellular Pax-5B transcript degradation, which, in turn, leads to a reduction in Pax-5B protein activity.
The human Pax-5B transcriptional element negatively regulates CD19 expression
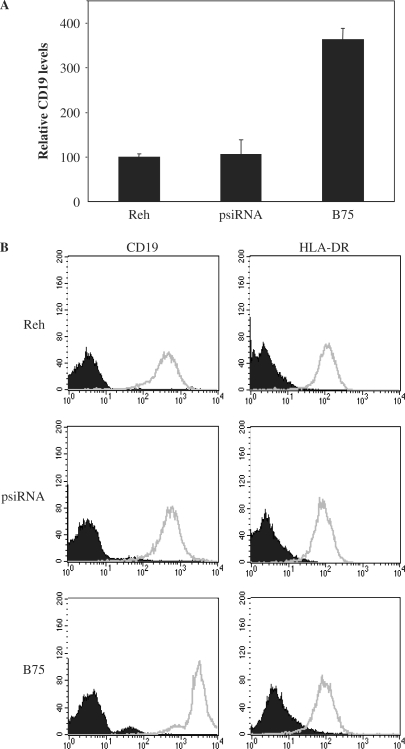
A different variant, Pax-5A (BSAP), has been consistently associated as a transcriptional activator of the CD19 surface antigen and therefore we set out to evaluate the transactivation potential of Pax-5B on CD19 expression. Using REH cells stably expressing SOFA-HDV Rz-B75, the influence of Pax-5B on CD19 mRNA expression were assessed through quantitative RT–PCR (Figure 6A). Unexpectedly, we observed a 3.6-fold induction of CD19 mRNA expression in the Pax-5B suppressed REH cell line when compared to both control cell populations (REH parental and the psiRNA-hH1 transfected cells). To confirm this finding, we measured the cell surface expression of the CD19 protein in the same REH cell systems (Figure 6B). Using flow cytometry, we were able to detect expression of the CD19 antigen in all B cells tested. Concomitant to mRNA levels, the REH B75 cells expressed higher levels of cell surface CD19 up to 6-fold over the REH parental cell line. In order to properly demonstrate that the induction of CD19 surface expression was not a result of a generalized increase in total cell surface receptors, the expression of a cell surface major histocompatibility complex class II element (HLA-DR) was examined on all three cellular settings. We found no variation in the expression of cell surface HLA-DR in all cell populations tested (Figure 6B). As a further confirmation, flow cytometry for CD19 expression was also performed on other REH B75-1 and -2 clones and again, an increase in CD19 expression was observed following Pax-5B suppression (data not shown). These results strongly suggest that the human Pax-5B protein negatively regulates the expression of CD19 in B lymphocytes.
Figure 6.
Pax-5B is a negative transcriptional regulator of the CD19 cell surface antigen. (A) Quantitative RT–PCR was conducted on extracts from the REH, the psiRNA-hH1 and the B75 cell clones to reveal CD19 intracellular transcript levels. CD19 mRNA values were normalized using HPRT levels and plotted in relation to the percentage of CD19 expression levels of the parental untreated REH cell line. Error bars are indicative of two independent experiments. (B) The cells were evaluated for cell surface protein expression of CD19 (left panels) and HLA-DR (right panels) using cell-flow cytometry. The plotted fluorescence from the direct labeling of the specific antibodies (empty graphs) is overlaid on the non-specific signal from the control isotype matched antibodies (solid graphs). The presented data represents one of two independent experiments.
Pax-5B suppression increases B-cell susceptibility to apoptotic events
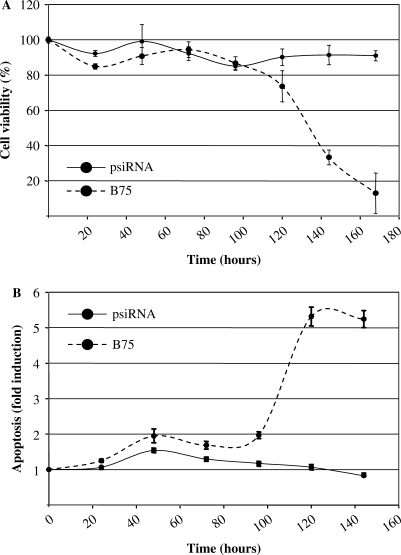
Previous studies have shown that both Pax-5 and CD19 are important regulatory components in B-cell proliferation and differentiation (25,26), therefore we sought to determine if the suppression of Pax-5B expression would impact cell growth and apoptosis in B lymphocytes. Cellular metabolic activities were thus examined in time for REH cells stably expressing the SOFA-HDV Rz-B75 in comparison with the REH parental and the vector-transfected control. Initially, the various cell lines proliferated at similar rates (Figure 7A). However, after 72 h, the Pax-5B attenuated REH cells ceased to proliferate. To assess the cause of this growth arrest, we determined cell death events in these cell lines using a microscale procedure to measure the activity of apoptosis effectors, CASPASES 3 and 7 (Figure 7B). We detected no significant apoptosis activity in the control REH cells (REH and vector-transfected) up to 168 h (7 days) of growth in appropriate conditions. Conversely, Pax-5B-suppressed cells initiated apoptotic signaling cascades after 72 h. The onset of apoptotic events observed in the REH B75 cell line correlated with the proliferation arrest observed after 72 h. To our knowledge, this is the first indication that the human Pax-5B transcriptional element is involved in the cell fate outcomes of B lymphocytes.
Figure 7.
Pax-5B promotes REH cell survival by suppressing apoptotic events. (A) Cellular viability was evaluated in time (hours) in the B75 (dotted line) and the control psiRNA-hH1 cell (solid line) clones and plotted in relation to the percentage of cell growth events from the REH parental cell line using a colorimetric test (Cell titer Blue/Promega). (B) In the same microplates, cells were then submitted to the analysis of cellular apoptotic events through the evaluation of CASPASE 3/7 activity (Apo-ONE/Promega). Cellular apoptosis for each cell clone was then plotted in fold induction over the events measured in the REH parental cell line. Error bars are indicative of three independent experiments.
DISCUSSION
Many recent studies have contributed to the better understanding of general Pax-5 function in various biological processes (5,6); however, few have attempted to discriminate between the physiological roles of the individual Pax-5A and Pax-5B variants. The inherent difficulty is largely due to the fact that both genes and proteins are nearly identical differing only by their first exon. Most studies that examined the role of Pax-5 have relied mainly on antibodies (27,28), nucleic probes (29,30), RNAi (31), antisense oligonucleotides (32) and/or site-directed genetic alteration (33) that target common regions present in both Pax-5A and Pax-5B. It is, therefore, not possible to draw conclusions on the specific roles of the individual genes and/or proteins from these previous works. Recently, the development of gene inactivation tools has focused on the attenuation or silencing of target mRNA for the study of gene function. Reliable and selective gene suppression systems are a valuable tool to create loss and gain of function models which provide new insights for intracellular signaling discovery and pathway analysis. In this context, the findings of this study are significant. Here, we validate a novel gene-suppression system based on a genetically modified viral ribozyme for the functional characterization of highly similar gene products transcribed from the Pax-5 gene locus. This new gene-silencing approach has allowed us to reveal the important contribution of the Pax-5B gene during apoptotic events in human B lymphocytes.
We have recently modified the native HDV Rz by the addition of a biosensor for the recognition of its specific substrate (18,19). The SOFA-HDV Rz is the only RNA-based gene suppression system of its kind to function as a riboswitch and therefore cleave only when the specific target mRNA is present. It has been shown previously that most Pax-5 expressing cells simultaneously produce multiple Pax-5 isoforms (11,20,34). Therefore, to study the unique contributions of the different gene products, we developed several SOFA-HDV Rz designed to target unique regions found in the human Pax-5B exon 1 mRNA sequence. When tested in vitro and in vivo, these Pax-5B specific SOFA-HDV Rz displayed a high level of target specificity. In all assays, we observed specific recognition and cleavage of the Pax-5B substrate with no suppressive off-target effects on the levels of the other major isoform Pax-5A. More interestingly, in B lymphocytes co-expressing Pax-5 isoforms, Pax-5A mRNA expression levels were up-regulated following Pax-5B down-regulation. These results suggest that Pax-5B plays a negative role in the transactivity of other Pax-5 isoforms. These observations are not surprising given the fact that autoregulation and negative transregulation activities between Pax-5 variants have been previously reported (35,36). As a control, we also designed a functional yet non-specific SOFA-HDV Rz and when tested in similar conditions, our results show that this ribozyme does not affect levels of Pax-5B RNA. Furthermore, the catalytic activity of the SOFA-HDV Rz was not cell-type dependent since Pax-5B transcript cleavage was observed in both HEK 293 and REH cells lines.
It is important to note that substrate cleavage occurs via direct ribozyme activity and is not mediated by RNA interference since the longest complementary region between a SOFA-HDV Rz and its Pax-5B substrate is 12 bp. We further ruled out the involvment of RNAi pathway components by gene expression analysis of mRNA from SOFA-HDV Rz transfected cells which revealed no activity from any RNAi elements following SOFA-HDV Rz mediated cleavage of a specific substrate (unpublished data; F. Brière, M. Laflamme, J.P. Perreault and R. Ouellette). The dose-dependent catalytic activity of the SOFA-HDV Rz observed in our transient co-transfections was not as pronounced when studying transfected B lymphocytes. In certain cases using B lymphocytes, the highest level of gene expression knock-down was observed in transfected clones with lower cellular concentration of ribozyme expression. These observations may be due to substrate accessibility and competition between promoter elements in our co-transfection settings (23,37).
The reduction of Pax-5B transcript levels had a subsequent effect on Pax-5B protein levels which was most likely due to decreases in synthesis levels. Our findings using EMSA assays revealed that ribozyme-mediated gene silencing led to a concomitant attenuation of the target protein expression as observed by a decrease in Pax-5B DNA-binding activity. This reduced binding was not associated with a general decrease in DNA-binding activity as revealed by NF-κB control experiments. Our findings suggest that high levels of Pax-5B suppression are incompatible with survival of B cells (REH). Following selection of stably-transfected REH clones, the maximum level ribozyme-mediated suppression of Pax-5B was 85% of control cells. (Figure 5A). Despite numerous attempts, we were unable to identify any stably-transfected SOFA-HDV Rz clones with a greater reduction of Pax-5B expression. These findings are consistent with our results of Pax-5B knock-down experiments using RNAi (unpublished data; Robichaud, G.A. and R. Ouellette). In these experiments, we found that with Pax-5B-directed RNAi sequences we were able to completely suppress Pax-5B expression. However, total suppression of Pax-5B led to complete cell death of REH cells compared to control REH cells that received a scrambled RNAi sequence. Taken together, these results strongly suggest that a minimal requirement of Pax-5B expression is required for cell survival. It also reveals the usefulness of the SOFA-HDV Rz system as a tool to selectively suppress and study vital cellular elements.
We then used our Pax-5B-attenuated B-cell system to study the effects on a known downstream target of Pax-5A, in this case, the CD19 gene. In previous studies, the DNA-binding motif of the Pax-5A protein or BSAP has been shown to bind the CD19 promoter region via a cluster of proximal Pax-5 recognition sites (7). Subsequently, this protein–DNA interaction induces CD19 gene expression which acts as a developmental checkpoint for early B-lineage commitment (38). Surprisingly, we observed a marked increase in CD19 expression both at the mRNA and protein levels in our Pax-5B knockdown B cells which, taken together, imply that Pax-5B is a negative regulator of CD19. This finding is contrary to the positive regulatory effect of Pax-5A on CD19 observed in previous studies (37). Interestingly, CD19 is a key molecule in the B lymphocyte signal transduction complex, where it associates with other proteins including the B-cell antigen complex (BCR) which has been shown to regulate B-cell fate and function (25,26). The CD19 surface protein modulates basal signaling thresholds and accelerates BCR signals thus serving as a general ‘rheostat’ that defines downstream signaling critical for B-cell responses, growth and autoimmunity (26,39). Furthermore, in certain instances, CD19 has also been shown to attenuate B-cell activation and proliferation events (40,41). It is precisely these CD19 events that could explain the lower proliferation rates observed in Pax-5B attenuated B lymphocytes.
Previous studies have also demonstrated that a decrease in the expression of the human Pax-5 gene (with no discrimination to Pax-5A or Pax-5B) is associated with decreased B-cell growth rates (28,32). In addition to the lower proliferation rates seen in Pax-5B knockdown cells, we observed that these B lymphocytes undergo apoptotic events that are mediated through the activity of CASPASES 3\7. Apoptosis is a key cellular pathway that mitigates the production of defective cells that occur during aberrant cellular processes such as hypersensitivity, autoimmunity, infection, mutation, anergy and oncogenesis (42). Apoptosis is particularly important during the development of lymphocytes when it removes both ineffective and potentially harmful cells via the withdrawal of survival signals. Interestingly, Pax-5 expression has been primarily associated with the development and maintenance of B-cell progenitors, then subsequently decreases and ceases in terminally differentiated plasma cells (4,43). Our findings suggest that Pax-5B may be closely involved in the differentiation checkpoints that ensure the precise regulation of the development and proliferation programs. This is consistent with data from Rahman et al. (28), which show that the suppression of Pax-5 results in the growth arrest of pre-B cells—results that were not observed in either pro-B or mature B-cell lines.
Apoptosis also plays a role in the prevention of oncogenic transformation associated with the development of cancer. Numerous studies have shown that aberrant expression of Pax-5 has been associated with the onset of different types of cancers (3,44), whereas others have also shown that Pax-5 is overexpressed in many types of B cell lymphoma and lymphocytic leukemia (20,30). Therefore, it appears that Pax-5 gene products are responsible on the one hand for the promotion B-cell commitment, and on the other, for the homeostasis of cell fate events. As a key switch in the balance between proliferation and apoptosis, Pax-5B could represent a potential target for therapeutic intervention. Further analysis of other downstream cell-fate regulators and known Pax-5 pathway components [i.e. p53 (45), Ets-1 (46), PD-1 (38) etc.] will help elucidate the complex networks and cell-fate events involved in B-cell development and oncogenesis. We are presently addressing some of these issues.
More recently, we and others have newly identified and characterized Pax-5 isoforms which result from the alternative splicing of the Pax-5 coding regions (20,34). On a functional basis, these protein variants possess different transactivation potential and have the ability to interact with different protein partners, and in certain cases, specific Pax-5 isoforms have been correlated with different types of cancers (20,47). These findings begin to address the complexity of the Pax-5 transcript and protein isoform function in B lymphocytes. Through the use of isoform-specific gene-silencing tools such as SOFA-HDV Rz, we are now able to better interrogate these regulators involved in B-cell development, cell-fate events and oncogenesis.
ACKNOWLEDGEMENTS
The work in the laboratory of JPP was supported by a grant from the Canadian Institutes of Health Research (CIHR; EOP-39322). R.J.O. was supported by a grant from the Canadian Institutes of Health Research (CIHR; 142650). The RNA Group is supported by grants from both the CIHR (PRG-80169) and the l’Université de Sherbrooke. G.A.R. held a post-doctoral fellowship from the National Cancer Institute of Canada\Terry Fox Foundation. J.P.P. holds the Canada Research Chair in Genomics and Catalytic RNA. Funding to pay the Open Access publication charges for this article was provided by CIHR (EOP-39322).
Conflict of interest statement. None declared.
REFERENCES
- 1.Chi N, Epstein JA. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 2002;18:41–47. doi: 10.1016/s0168-9525(01)02594-x. [DOI] [PubMed] [Google Scholar]
- 2.Robson EJ, He SJ, Eccles MR. A PANorama of PAX genes in cancer and development. Nat. Rev. Cancer. 2006;6:52–62. doi: 10.1038/nrc1778. [DOI] [PubMed] [Google Scholar]
- 3.Maulbecker CC, Gruss P. The oncogenic potential of Pax genes. EMBO J. 1993;12:2361–2367. doi: 10.1002/j.1460-2075.1993.tb05890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams B, Dorfler P, Aguzzi A, Kozmik Z, Urbanek P, Maurer-Fogy I, Busslinger M. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Dev. 1992;6:1589–1607. doi: 10.1101/gad.6.9.1589. [DOI] [PubMed] [Google Scholar]
- 5.Hagman J, Wheat W, Fitzsimmons D, Hodsdon W, Negri J, Dizon F. Pax-5/BSAP: regulator of specific gene expression and differentiation in B lymphocytes. Curr. Top. Microbiol. Immunol. 2000;245:169–194. doi: 10.1007/978-3-642-57066-7_5. [DOI] [PubMed] [Google Scholar]
- 6.Rolink AG, Schaniel C, Busslinger M, Nutt SL, Melchers F. Fidelity and infidelity in commitment to B-lymphocyte lineage development. Immunol. Rev. 2000;175:104–111. [PubMed] [Google Scholar]
- 7.Kozmik Z, Wang S, Dorfler P, Adams B, Busslinger M. The promoter of the CD19 gene is a target for the B-cell-specific transcription factor BSAP. Mol. Cell Biol. 1992;12:2662–2672. doi: 10.1128/mcb.12.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zwollo P, Desiderio S. Specific recognition of the blk promoter by the B-lymphoid transcription factor B-cell-specific activator protein. J. Biol. Chem. 1994;269:15310–15317. [PubMed] [Google Scholar]
- 9.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 10.Wallin JJ, Rinkenberger JL, Rao S, Gackstetter ER, Koshland ME, Zwollo P. B cell-specific activator protein prevents two activator factors from binding to the immunoglobulin J chain promoter until the antigen-driven stages of B cell development. J. Biol. Chem. 1999;274:15959–15965. doi: 10.1074/jbc.274.22.15959. [DOI] [PubMed] [Google Scholar]
- 11.Busslinger M, Klix N, Pfeffer P, Graninger PG, Kozmik Z. Deregulation of PAX-5 by translocation of the Emu enhancer of the IgH locus adjacent to two alternative PAX-5 promoters in a diffuse large-cell lymphoma. Proc. Natl Acad. Sci. USA. 1996;93:6129–6134. doi: 10.1073/pnas.93.12.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu ML, Rahman M, Hirabayashi Y, Sasaki T. Sequence analysis of 5'-flanking region of human pax-5 gene exon 1B. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2002;10:100–103. [PubMed] [Google Scholar]
- 13.Asif-Ullah M, Levesque M, Robichaud G, Perreault JP. Development of ribozyme-based gene-inactivations; the example of the hepatitis delta virus ribozyme. Curr. Gene Ther. 2007;7:205–216. doi: 10.2174/156652307780859008. [DOI] [PubMed] [Google Scholar]
- 14.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 15.Svoboda P. Off-targeting and other non-specific effects of RNAi experiments in mammalian cells. Curr. Opin. Mol. Ther. 2007;9:248–257. [PubMed] [Google Scholar]
- 16.Wu HN, Lin YJ, Lin FP, Makino S, Chang MF, Lai MM. Human hepatitis delta virus RNA subfragments contain an autocleavage activity. Proc. Natl Acad. Sci. USA. 1989;86:1831–1835. doi: 10.1073/pnas.86.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peracchi A. Prospects for antiviral ribozymes and deoxyribozymes. Rev. Med. Virol. 2004;14:47–64. doi: 10.1002/rmv.415. [DOI] [PubMed] [Google Scholar]
- 18.Bergeron LJ, Perreault JP. Target-dependent on/off switch increases ribozyme fidelity. Nucleic Acids Res. 2005;33:1240–1248. doi: 10.1093/nar/gki278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergeron LJ, Reymond C, Perreault JP. Functional characterization of the SOFA delta ribozyme. RNA. 2005;11:1858–1868. doi: 10.1261/rna.2112705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robichaud GA, Nardini M, Laflamme M, Cuperlovic-Culf M, Ouellette RJ. Human Pax-5 C-terminal isoforms possess distinct transactivation properties and are differentially modulated in normal and malignant B cells. J. Biol. Chem. 2004;279:49956–49963. doi: 10.1074/jbc.M407171200. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Leclerc GJ, Leclerc GM, Barredo JC. Real-time RT-PCR analysis of mRNA decay: half-life of Beta-actin mRNA in human leukemia CCRF-CEM and Nalm-6 cell lines. Cancer Cell Int. 2002;2:1. doi: 10.1186/1475-2867-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergeron LJ, Ouellet J, Perreault JP. Ribozyme-based gene-inactivation systems require a fine comprehension of their substrate specificities; the case of delta ribozyme. Curr. Med. Chem. 2003;10:2589–2597. doi: 10.2174/0929867033456486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lucier JF, Bergeron LJ, Briere FP, Ouellette R, Elela SA, Perreault JP. RiboSubstrates: a web application addressing the cleavage specificities of ribozymes in designated genomes. BMC Bioinformatics. 2006;7:480. doi: 10.1186/1471-2105-7-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ulivieri C, Baldari CT. The BCR signalosome: where cell fate is decided. J. Biol. Regul. Homeost. Agents. 2005;19:1–16. [PubMed] [Google Scholar]
- 26.Tedder TF, Poe JC, Fujimoto M, Haas KM, Sato S. The CD19-CD21 signal transduction complex of B lymphocytes regulates the balance between health and autoimmune disease: systemic sclerosis as a model system. Curr. Dir. Autoimmun. 2005;8:55–90. doi: 10.1159/000082087. [DOI] [PubMed] [Google Scholar]
- 27.Hirokawa S, Sato H, Kato I, Kudo A. EBF-regulating Pax5 transcription is enhanced by STAT5 in the early stage of B cells. Eur. J. Immunol. 2003;33:1824–1829. doi: 10.1002/eji.200323974. [DOI] [PubMed] [Google Scholar]
- 28.Rahman M, Hirabayashi Y, Ishii T, Watanabe M, Maolin L, Sasaki T. Prednisolone sodium succinate down-regulates BSAP/Pax5 and causes a growth arrest in the Nalm6 pre-B cell line. Tohoku J. Exp. Med. 2001;193:237–244. doi: 10.1620/tjem.193.237. [DOI] [PubMed] [Google Scholar]
- 29.Qiu G, Stavnezer J. Overexpression of BSAP/Pax-5 inhibits switching to IgA and enhances switching to IgE in the I.29 mu B cell line. J. Immunol. 1998;161:2906–2918. [PubMed] [Google Scholar]
- 30.Hamada T, Yonetani N, Ueda C, Maesako Y, Akasaka H, Akasaka T, Ohno H, Kawakami K, Amakawa R, Okuma M. Expression of the PAX5/BSAP transcription factor in haematological tumour cells and further molecular characterization of the t(9;14)(p13;q32) translocation in B-cell non-Hodgkin's lymphoma. Br. J. Haematol. 1998;102:691–700. doi: 10.1046/j.1365-2141.1998.00842.x. [DOI] [PubMed] [Google Scholar]
- 31.Baumann Kubetzko FB, Di Paolo C, Maag C, Meier R, Schafer BW, Betts DR, Stahel RA, Himmelmann A. The PAX5 oncogene is expressed in N-type neuroblastoma cells and increases tumorigenicity of a S-type cell line. Carcinogenesis. 2004;25:1839–1846. doi: 10.1093/carcin/bgh190. [DOI] [PubMed] [Google Scholar]
- 32.Max EE, Wakatsuki Y, Neurath MF, Strober W. The role of BSAP in immunoglobulin isotype switching and B-cell proliferation. Curr. Top. Microbiol. Immunol. 1995;194:449–458. doi: 10.1007/978-3-642-79275-5_52. [DOI] [PubMed] [Google Scholar]
- 33.Kovac CR, Emelyanov A, Singh M, Ashouian N, Birshtein BK. BSAP (Pax5)-importin alpha 1 (Rch1) interaction identifies a nuclear localization sequence. J. Biol. Chem. 2000;275:16752–16757. doi: 10.1074/jbc.M001551200. [DOI] [PubMed] [Google Scholar]
- 34.Zwollo P, Arrieta H, Ede K, Molinder K, Desiderio S, Pollock R. The Pax-5 gene is alternatively spliced during B-cell development. J. Biol. Chem. 1997;272:10160–10168. doi: 10.1074/jbc.272.15.10160. [DOI] [PubMed] [Google Scholar]
- 35.Lowen M, Scott G, Zwollo P. Functional analyses of two alternative isoforms of the transcription factor Pax-5. J. Biol. Chem. 2001;276:42565–42574. doi: 10.1074/jbc.M106536200. [DOI] [PubMed] [Google Scholar]
- 36.Pridans C, Holmes ML, Polli M, Wettenhall JM, Dakic A, Corcoran LM, Smyth GK, Nutt SL. Identification of Pax5 target genes in early B cell differentiation. J. Immunol. 2008;180:1719–1728. doi: 10.4049/jimmunol.180.3.1719. [DOI] [PubMed] [Google Scholar]
- 37.Akashi H, Matsumoto S, Taira K. Gene discovery by ribozyme and siRNA libraries. Nat. Rev. Mol. Cell Biol. 2005;6:413–422. doi: 10.1038/nrm1646. [DOI] [PubMed] [Google Scholar]
- 38.Nutt SL, Morrison AM, Dorfler P, Rolink A, Busslinger M. Identification of BSAP (Pax-5) target genes in early B-cell development by loss- and gain-of-function experiments. EMBO J. 1998;17:2319–2333. doi: 10.1093/emboj/17.8.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujimoto M, Fujimoto Y, Poe JC, Jansen PJ, Lowell CA, DeFranco AL, Tedder TF. CD19 regulates Src family protein tyrosine kinase activation in B lymphocytes through processive amplification. Immunity. 2000;13:47–57. doi: 10.1016/s1074-7613(00)00007-8. [DOI] [PubMed] [Google Scholar]
- 40.Fearon DT, Carroll MC. Regulation of B lymphocyte responses to foreign and self-antigens by the CD19/CD21 complex. Annu. Rev. Immunol. 2000;18:393–422. doi: 10.1146/annurev.immunol.18.1.393. [DOI] [PubMed] [Google Scholar]
- 41.Mahmoud MS, Fujii R, Ishikawa H, Kawano MM. Enforced CD19 expression leads to growth inhibition and reduced tumorigenicity. Blood. 1999;94:3551–3558. [PubMed] [Google Scholar]
- 42.Callard R, Hodgkin P. Modeling T- and B-cell growth and differentiation. Immunol. Rev. 2007;216:119–129. doi: 10.1111/j.1600-065X.2006.00498.x. [DOI] [PubMed] [Google Scholar]
- 43.Barberis A, Widenhorn K, Vitelli L, Busslinger M. A novel B-cell lineage-specific transcription factor present at early but not late stages of differentiation. Genes Dev. 1990;4:849–859. doi: 10.1101/gad.4.5.849. [DOI] [PubMed] [Google Scholar]
- 44.Kozmik Z, Sure U, Ruedi D, Busslinger M, Aguzzi A. Deregulated expression of PAX5 in medulloblastoma. Proc. Natl Acad. Sci. USA. 1995;92:5709–5713. doi: 10.1073/pnas.92.12.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stuart ET, Haffner R, Oren M, Gruss P. Loss of p53 function through PAX-mediated transcriptional repression. EMBO J. 1995;14:5638–5645. doi: 10.1002/j.1460-2075.1995.tb00251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fitzsimmons D, Hodsdon W, Wheat W, Maira SM, Wasylyk B, Hagman J. Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes Dev. 1996;10:2198–2211. doi: 10.1101/gad.10.17.2198. [DOI] [PubMed] [Google Scholar]
- 47.Oppezzo P, Dumas G, Lalanne AI, Payelle-Brogard B, Magnac C, Pritsch O, Dighiero G, Vuillier F. Different isoforms of BSAP regulate expression of AID in normal and chronic lymphocytic leukemia B cells. Blood. 2005;105:2495–2503. doi: 10.1182/blood-2004-09-3644. [DOI] [PubMed] [Google Scholar]