Abstract
The spatial configuration of the chicken α-globin gene domain in erythroid and lymphoid cells was studied by using the Chromosome Conformation Capture (3C) approach. Real-time PCR with TaqMan probes was employed to estimate the frequencies of cross-linking of different restriction fragments within the domain. In differentiated cultured erythroblasts and in 10-day chick embryo erythrocytes expressing ‘adult’ αA and αD globin genes the following elements of the domain were found to form an ‘active’ chromatin hub: upstream Major Regulatory Element (MRE), −9 kb upstream DNase I hypersensitive site (DHS), −4 kb upstream CpG island, αD gene promoter and the downstream enhancer. The αA gene promoter was not present in the ‘active’ chromatin hub although the level of αA gene transcription exceeded that of the αD gene. Formation of the ‘active’ chromatin hub was preceded by the assembly of multiple incomplete hubs containing MRE in combination with either −9 kb DHS or other regulatory elements of the domain. These incomplete chromatin hubs were present in proliferating cultured erythroblasts which did not express globin genes. In lymphoid cells only the interaction between the αD promoter and the CpG island was detected.
INTRODUCTION
The α-globin gene domain in chicken is one of the popular models that has been used over years to study the mechanisms controlling transcription in higher eukaryotes (1–3). Besides three α-type globin genes (π, αD and αA) and their promoters the domain contains several regulatory elements located both upstream (4–7) and downstream (8) of the gene cluster. The Locus Control Region (LCR)-like Major Regulatory Element (MRE) of the chicken α-globin gene domain is located in the intron of a housekeeping gene ∼20 kb upstream of π, the first of the α-globin genes (4,7). It is not clear how this regulatory element works. From studies of other vertebrate globin gene domains (9,10) one may infer that it interacts directly with the promoters of the α-globin genes. Naturally, it cannot interact simultaneously with two or more promoters. Thus, the interaction should be short-term and alternating as postulated by the so-called ‘flip-flop’ model (11,12). Another possibility is that a complex chromatin hub (13–15) is formed including several regulatory elements, promoters of all active globin genes and promoters of neighboring housekeeping genes. Complex interactions of such kind were described in the mouse α-globin gene domain (16). In chicken, the promoter of a housekeeping gene is located within the CpG island just 4.1 kb upstream of the embryonic α-type globin gene π (4,17). Further upstream several erythroid-specific DNase I hypersensitive sites (DHS) were mapped at distances of 9.1, 12.8, 14.9 and 20.5 kb upstream of the π gene (4,18,19). Some of these DHS colocalized roughly with previously characterized regulatory elements such as MRE (4) and enhancer-blockers (20). The significance of the others is presently unknown. In the downstream area of the chicken α-globin gene domain there is an erythroid-specific enhancer located at a distance of 0.9 kb after the αA gene (8).
In order to understand how the expression of chicken α-globin genes is regulated it is important to know if and how different regulatory elements interact with each other and with the promoters of the globin genes. Furthermore, assuming that such interactions exist it is important to know if they are modified with the beginning of globin gene expression. To clarify the above questions, we have studied the spatial organization of the chicken α-globin gene domain in 10-day chick embryo red blood cells (10-day RBC) and in cultured AEV-transformed pre-erythroblasts before and after induction of differentiation of these cells resulting in the beginning of productive expression of globin genes. The study was carried out using the Chromosome Conformation Capture (3C) approach (9,21,22), and real-time PCR with TaqMan probes was employed to estimate the frequencies of cross-linking between different restriction fragments of the domain. The results obtained have demonstrated that in 10-day RBC and in differentiated pre-erythroblasts MRE, −9 kb DHS, CpG island, αD gene promoter and the downstream enhancer interact with each other.
MATERIALS AND METHODS
Cell culture
The avian erythroblastosis virus-transformed chicken erythroblast cell line HD3 [clone A6 of line LSCC (23,24)] and the chicken lymphoid cell line DT40 (CRL-2111, ATCC, London, UK) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 2% chicken serum and 8% fetal bovine serum at 37°C in 5% CO2 atmosphere. In the case of DT40 cells the medium additionally contained 50 μM β-mercaptoethanol.
To induce erythroid differentiation, HD3 cells at a density of 8 × 105 cells/ml were incubated in the above medium additionally containing 10 mM HEPES (pH 8.0) and 20 μM iso-H-7 (1-(5-Isoquinolinylsulfonyl)-3-methylpiperazine dihydrochloride, Fluka, Seelze, Germany) at 42°C in 100% air atmosphere (25).
To estimate the percentage of dead cells, seven parts of the cell suspension in growth medium were mixed with one part of 0.4% trypan-blue solution (Gibco, Carlsbad, CA, USA) and the percentage of stained blue cells was determined under a light microscope.
To estimate the percentage of cells containing hemoglobin, 25 μl of 0.4% (w/v) solution of benzidine (Sigma, St. Louis, MO, USA) in 4% (v/v) acetic acid was mixed with 1 μl of 30% H2O2 and then 25 μl of the cell suspension in growth medium was added. After 10 min staining, the percentage of stained dark blue cells was determined under a light microscope (26).
Chicken red blood cells were collected from 10-day old embryos.
Northern blot analysis
Total RNA was isolated by using the RNAgents Total RNA Isolation System (Promega, Madison, WI, USA). After electrophoretic separation on denaturing MOPS-formaldehyde agarose gel, RNA was transferred to a Hybond N + nylon membrane (Amersham, Piscataway, NJ, USA) and hybridized with P32-labeled DNA probes specific to π, αD or αA globin genes. The probes were prepared by Random Prime labeling of PCR-amplified fragments of chicken genomic DNA.
3C analysis
3C analysis was performed as described (27) with minor modifications. Briefly, 2 × 107 cells were fixed in DMEM/serum with 2% formaldehyde for 10 min at room temperature. The reaction was stopped by adding glycine to a concentration of 0.125 M and placing the samples on ice. Cross-linked cells were washed with cold PBS and lysed by incubation for 10 min in ice-cold lysis buffer [10 mM Tris (pH 8.0), 10 mM NaCl, 0.2% NP-40, a protease inhibitors cocktail tablet (Complete Mini, Roche, Basel, Switzerland)]. The nuclei were harvested, and aliquots containing 107 nuclei were frozen in liquid nitrogen and stored at −70°C.
About 107 nuclei were suspended in 0.25 ml of 1.2× restriction buffer (NEB buffer 3), SDS was added to a final concentration of 0.3% and the solution was incubated with shaking at 37°C for 1 h. The solution was then diluted 2-fold by the same restriction buffer, and triton X-100 was added to a final concentration of 1.8% (to sequester the SDS) and incubation in the same conditions for another hour followed. After incubation, 1000 U of BglII and 1500 U of BamHI or 1000 U of MboI (highly concentrated, NEB, Hitchin, UK) were added and digestion was carried out for 16 h at 37°C with continuous shaking. The reaction was stopped by addition of SDS to a final concentration of 1.3% and incubation at 65°C for 20 min. The solution was diluted by adding 7 ml of 1 × ligation buffer [50 mM Tris (pH 7.5), 10 mM MgCl2, 10 mM DTT, 1 mM ATP]. Triton X-100 was added to a final concentration of 1% and the solution was incubated at 37°C for 1 h while shaking. Quantity of 100 U of T4 DNA Ligase (Fermentas, Vilnius, Lithuania) was added, and DNA was ligated at 16°C for 4.5 h and then at room temperature for 30 min with slow agitation. Cross-links were reversed by incubation at 65°C for 16 h in the presence of Proteinase K (40 μg/ml). After cross-link reversion, RNase A was added to a final concentration of 40 μg/ml, and RNA was digested at 37°C for 45 min. DNA was purified by extraction with phenol, phenol–chloroform and chloroform followed by precipitation with ethanol. To generate a random-ligation control, 5 μg of a bacterial artificial chromosome containing the chicken α-globin gene domain along with the flanking areas (clone CH261-75C12, CHORI BACPAC Resources Center) was digested with BamHI and BglII or with Sau3A and then ligated at a high DNA concentration.
The real-time PCR technology was employed to analyze the ligation products. In each PCR set one primer from a pair (anchor primer) and a TaqMan probe annealed closer to the end of the corresponding restriction fragment (anchor fragment) were always the same, whereas other primers were complementary to successive restriction fragments walking 5′ or 3′ or both directions in the locus. For each primer pair six successive 5-fold dilutions of the random ligation standard were used to make a calibration curve for determining a relative quantity of the corresponding ligation product in a 3C template. To be sure that the calibration curves are reliable for determining the DNA quantity, the 3C templates were always tested in 2-fold dilutions to verify that the values obtained indeed differed 2-fold. For more accurate calculations, each PCR reaction was carried out in quadruple repetition and the corresponding results were averaged. Once a resulting 3C curve representing the spectrum of interactions between an anchor fragment and other fragments throughout the domain was obtained, experiments starting with living cells were repeated twice more in order to check the reproducibility of the results. Primers and TaqMan probes for PCR analysis were designed using the DNA sequence AY016020 (GeneBank) (4) and Primer Premier 5 computer software (PRIMER Biosoft International, Palo Alto, CA, USA). The sequences of the primers and TaqMan probes are present in Tables 1 and 2, respectively. The size of target PCR products that can be obtained using as a template a mixture of randomly ligated BamHI/BglII fragments or MboI fragments of the area under study was within the range of 120–230 bp. Annealing temperature for the primers was 55–58°C. Annealing temperature for the TaqMan probes was 67–68°C. A real-time PCR in a volume of 20 μl included 30 or 60 ng of a 3C DNA template or 30 ng of digested and religated chicken genomic DNA along with 0.4, 2, 10, 50, 250 or 1250 pg of a control BAC template, 1 × PCR buffer [50 mM Tris (pH 8.6), 50 mM KCl, 1.5 mM MgCl2, 0.1% Tween 20], 0.5 μM of each primer, 0.25 μM of a TaqMan probe (5′-FAM dye, inside BHQ-1 quencher), 0.2 mM of each dNTP, 0.75 U of Hot start Taq DNA pol (Sibenzyme, Novosibirsk, Russia), and was performed as follows: initial denaturation for 5 min at 94°C; 58 cycles of 15 s at 94°C, 60 s at 60°C, plate read. All real-time PCR components reaction were aliquoted and stored at −20°C till required for PCR with each new primer pair.
Table 1.
Primers used for 3C and PCR-stop analysis
| 2977 R | GTCCTTCCAGAATCGACTTGA |
| 4356 R | GCAAACTCATAGAGCAGAAAACA |
| 5153 R | AAGCGATCCTCAATGTCCC |
| 9818 F* | CAGGAAAGGGAAGAGAGAACAG |
| 9938 R (anchor)* | CTTGTGTCCTTCAGTAGGCAGA |
| 23578 F | GGATAACCACCTAAGGAACAG |
| 25621 F* | GCTGCCTCATGTTTGTTAAGATA |
| 25741 R* | GTGACTCAGCAAGAACAGCAGA |
| 26233 F (anchor)* | CTTTTGAAGCCAATGTCTCTC |
| 26397 R* | AACAGTTATCCCAGCAATCAG |
| 26757 F | AAATTAGCCGAGTCAGGATCT |
| 27922 F | TGTTAGAAAAATCACAGCAGG |
| 30783 F | ACAGCAATCCGATCCTCTAA |
| 31453 F | GTGAAGAATCAGAAATCAGGAA |
| 34941 F | TTTGGTCATTGCTTGTCAGA |
| 36533 F | TTTTCTGAGCACCTCTCTGTT |
| 36940 F (anchor)* | GCGATATTGAATGTTCTCTAGG |
| 37110 R* | TGGCAAGTTTGATAATGTTCTC |
| 39275 F | AAGAACCAGCTTACATTTTGC |
| 39471 R | ACGGACATGGACCCTCTAACT |
| 42419 F | CACTCAACACTCTTCAGCCAA |
| 45195 F (anchor)* | TTCACAGCACAAGGGATAACT |
| 45356 R (anchor)* | TGCTTCCTGAGATGGTAACA |
| 46806 F | CTCACAGCAGTTTGAAGACCT |
| 49492 F | ACAGGACAGTGACTGCCAAC |
| 49602 R | CTCACAGGCAGCTCCACACT |
| 50108 F* | TGTTCACCACCTATCCCCA |
| 50268 R* | GTTGCTCAGCTCAGCCATG |
| 50273 F (anchor)* | ATGCCTACAACCTGCGTG |
| 50481 R* | TGGCCTCTGGCTCCTGAT |
| 51282 F | GTGGAAATGTCACAGGCAG |
| 51891 F | CTCACCTTCCTCATCACCTT |
| 52112 R (anchor) | TCCCTCTTTGCCAGCCA |
| 52915 F (anchor) | AGGGCATCTTCACCAAAATC |
| 53618 F (anchor)* | AGCCAAATGAGATGAAATAAAA |
| 53786 R* | CCACCTGAGCCCATATCCT |
| 53935 F | CAGCAGAGCCACATACTCATAC |
| 54448 F | AGGCTCTCCTCCAGCTCAC |
| 54946 F | TGCCAAGCACTGGTAAGAG |
| 55977 F | GAGTTTAGCAAGAATTTCCCAT |
| 58058 F | ATACACTACAATGGGAAGCCT |
| ERCC3 2449 R | CTGCTGGCTCATGTTAGGTTT |
| ERCC3 4695 R (anchor) | TCTAAGCGGCTGTCAGATTTG |
5′–3′ sequences are presented. The figures in primer names correspond to the position of primer 5′-ends on the AY016020 sequence (GeneBank) (4) or, for ERCC3 primers, on the NC_006094 sequence (GeneBank); F or R designate forward or reverse primers (as compared to the direction of globin gene or ERCC3 gene transcription). *Denotes the marked primers used for PCR-stop analysis.
Table 2.
TaqMan probes used for 3C and PCR-stop analysis
| 9889 R* | (FAM)TCTTCAT(BHQ-1)GCAGAGAGAAAC ACCAGGC |
| 25713 R* | (FAM)AAGTGTTGACT(BHQ-1)CATGGTTTG CTAGTTTGC |
| 26297 F* | (FAM)CCTCCTAACCT(BHQ-1)AACAATAAC CCACAGCAC |
| 36990 F* | (FAM)CTGAAGGCAGT(BTQ-1)CATCCAGTA CAAAGCA |
| 45235 F* | (FAM)CCACAAAT(BHQ-1)CAAAGCGATGC GGTAT |
| 45289 F | (FAM)ATCAGGGAGAAT(BHQ-1)TGCAGAT TAAGGAAACA |
| 50215 F* | (FAM)AACGCCGT(BHQ-1)GAAGAACGTG GACAAC |
| 50309 F* | (FAM)AGGCAAGCAAAGGCT(BHQ-1)GGG GTCT |
| 52045 R | (FAM) CAGCAGAGCCACGGGGT(BHQ-1) CAGT |
| 52974 F | (FAM)AGGTAGGTGT(BHQ-1)CCTTCTCTG TCCTCCG |
| 53663 F* | (FAM)CTGGTGTCCT(BHQ-1)GCTCTGGTT TCTGC |
| ERCC3 4573 F* | (FAM)TGTCTTGCCATGT(BHQ-1)GGTAAA TTACTGCG |
5′–3′ sequences are presented. FAM, a fluorescent dye at the 5′-ends of the probes; BHQ-1, a quencher dye inside the probes at T. Probe names are assigned as in Table 1. *Denotes the marked probes used for PCR-stop analysis.
RESULTS
Cellular model for comparative 3C analysis
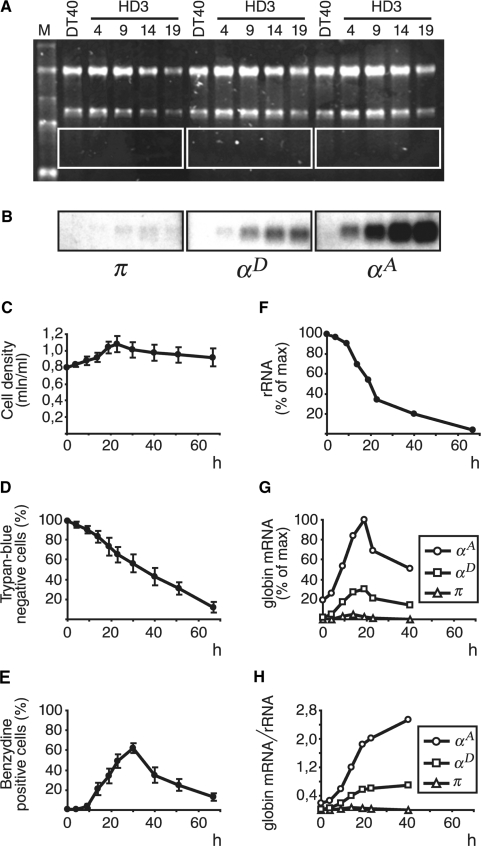
AEV-transformed chicken proerythroblasts (line HD3, clone A6 of line LSCC) correspond to chicken hemopoietic cells of the red lineage arrested at early stages of differentiation (23,24). They do not express globin genes, although the α-globin gene domain resides in an active configuration supported by low-level transcription of the whole domain (28). After induction of terminal erythroid differentiation, HD3 cells stop proliferation and start production of globins. To induce differentiation, proliferating HD3 cells were incubated with the iso-H-7 inducer at 42°C and pH 8.0 (see Materials and Methods section for details). The percentage of hemoglobin-producing cells was calculated using benzidine staining. Hemoglobin-containing (benzydine-positive) cells were present in small proportion (≤1%) already in non-induced culture. A slight increase in the percentage of benzydine-positive cells was detected already between 4 and 9 h after induction; at 9 h benzydine-positive cells comprised 4%, followed by 21% at 14 h and 62% at 30 h (Figure 1E). The percentage of living cells was calculated using trypan-blue staining and was found to be 90, 65, 43 and 12%, respectively, at 10, 23, 40 and 67 h after induction (Figure 1D). Total cellular RNA isolated at different time intervals after the beginning of induction was analyzed by Northern hybridization with probes recognizing mRNAs encoded by π, αD and αA genes. The results of the hybridizations clearly show that the levels of mRNA encoded by adult αD and αA genes start to increase already during the first 4 h after induction, reach their maximum at 19 h and then gradually decrease thus vividly demonstrating how the wave of RNA synthesis precedes the wave of protein synthesis (Figure 1B and G). The kinetics of αA and αD mRNA accumulation was especially evident when the results were normalized to the amount of rRNA in the samples (Figure 1H) which decreased with the time after induction (Figure 1A and F) reflecting the decrease in the percentage of living cells. The embryonic α-type gene π was practically not expressed at any time after induction (Figure 1B and G). No expression of the α-globin genes was detected in lymphoid DT40 cells (Figure 1B).
Figure 1.
Induction of erythroid differentiation of HD3 cells. (A) and (B) Northern-blot analysis of RNA from DT40 and HD3 cells at different time intervals after the beginning of induction. (A) Ethidium bromide staining of RNA after gel electrophoresis. M: RNA marker (0.24, 1.35, 2.37, 4.40, 7.46 kb); DT40: RNA from DT40 cells; HD3: RNA from HD3 cells; the figures indicate the time after the beginning of induction. (B) Autoradiographs showing the results of hybridization of probes recognizing π, αD and αA mRNAs with blots representing the framed areas in section A. (C–E) Kinetics of changes in cell density (C), percentage of living cells (D) and percentage of hemoglobin-containing cells (E) with time after induction. The data shown represent the average of the results of three independent experiments; error bars represent SEM. (F) Relative amount of rRNA (18S + 28S) in the slots. (G) Relative amount of globin mRNAs in the slots. (H) Relative amount of globin mRNAs normalized to the relative amount of rRNA. The density of bands was quantified using TotalLab 2 computer software (Nonlinear Dynamics, Newcastle, UK).
Whatever regulatory events are required for activation of αD and αA genes in induced HD3 cells, they must occur before the start of transcription. Once assembled, the activator complexes are likely to remain in this form as long as the transcription continues. Taking into account these considerations, we decided to perform a comparative 3C analysis of the α-globin gene domain in non-induced HD3 cells and in induced HD3 cells at 12 h after induction. At this moment the total level of α-globin gene transcription in culture (judging from the rate of mRNA accumulation) is close to its maximum but still continues to grow. The increase in the transcription level most likely reflects the fact that there are cells in the population which are just starting transcription of the globin genes at this time because induction of differentiation does not occur absolutely synchronously. Correspondingly, at 12 h after induction the configuration of the α-globin gene domain is likely to be typical for cells which actively transcribe the globin genes or are starting to. Considering the fact that transformed cells may possess some special features, the spatial configuration of the α-globin gene domain in 10-day chick embryo normal erythrocytes was studied by the 3C analysis. It was shown previously that 10-day chicken RBC express adult α-globin genes (αD and αA) but do not express the embryonic α-type globin gene π (29). To understand better the significance of the results obtained with erythroid cells, the 3C analysis was also applied to lymphoid DT40 cells where the α-globin gene domain is not potentiated for transcription.
Quantitative 3C analysis of the chicken α-globin gene locus: an overall experimental approach and control experiments
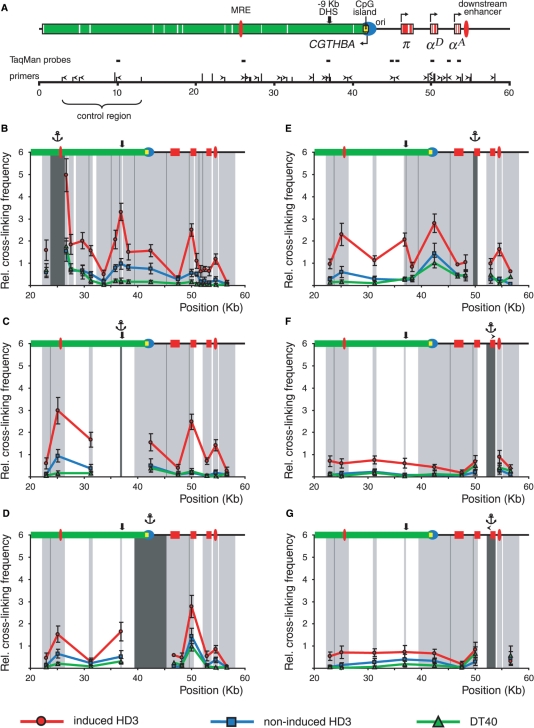
The Chromosome Conformation Capture (3C) technique was described previously (9,21,22). The combination of BglII and BamHI restriction enzymes was used for 3C analysis in the first set of experiments. These enzymes recognize different consensuses (AGATCT and GGATCC) but produce compatible DNA ends which can be cross-ligated. The BamHI/BglII restriction map of the chicken α-globin gene domain is shown in Figure 2A along with the scheme demonstrating positions of the α-globin genes and important regulatory elements of the domain. It is evident that important functional elements of the domain, such as MRE, replication origin, promoter of the housekeeping gene CGTHBA, promoters of the α-globin genes and the erythroid-specific downstream enhancer, are located within different restriction fragments. In the second set of experiments MboI restriction enzyme was used. This enzyme cuts DNA at the GATC site and thus recognizes both BamHI and BglII restriction sites as well as 14 other hexanucleotides. The MboI restriction map of the domain is shown in Figure 3A. Utilization of MboI permits certain regulatory elements to be localized more precisely within smaller restriction fragments. The positions of the primers and TaqMan probes used for real-time PCR analysis of the ligation products are shown above the restriction maps. They were designed in such a way as to compose two unidirectional arrays for analysis of the α-globin gene locus and a control locus, respectively (see below).
Figure 2.
BamHI/BglII 3C analysis of the chicken α-globin gene domain in non-erythroid and transformed erythroid cells. (A) A BamHI/BglII restriction map of the chicken α-globin gene domain and a scheme showing positions of important functional elements. Red boxes: α-globin genes, green box: CGTHBA gene (the white lines represent exons, the arrows show the directions of transcription); filled red ovals: MRE and the downstream enhancer; black bold arrow: −9 kb DHS; filled yellow rectangle: CpG island; blue circle: origin of replication. The long and short vertical lines above the scale show positions of BamHI and BglII restriction sites, respectively. The ‘0’ point in the map corresponds to the start of the AY016020 sequence (GeneBank) (4). Primers and TaqMan probes used for the 3C analysis are shown respectively by black horizontal tailless arrows and rectangles. (B–G) Relative frequencies of cross-linking between the anchor fragments bearing (B): MRE, (C): −9 kb DHS, (D): CpG island, (E): αD gene promoter, (F) and (G): αA gene and other fragments of the locus. The ‘x’ axis shows fragment positions according to the restriction map scale. On the top of each graph a scheme of the domain with the same symbols as in (A) is shown. The results of 3C analysis for induced HD3, cycling HD3 and DT40 cells are shown by the red, blue and green lines, respectively. The dark gray rectangle in the background of each graph with the anchor drawn above indicates a fixed (anchor) DNA fragment, and the light gray rectangles—test-fragments. The borders between the neighboring fragments are indicated by dark gray lines. The relative cross-linking frequencies (‘y’ axis) were calculated as described in the second paragraph of the ‘Results’ section. Error bars represent SEM for three independent experiments.
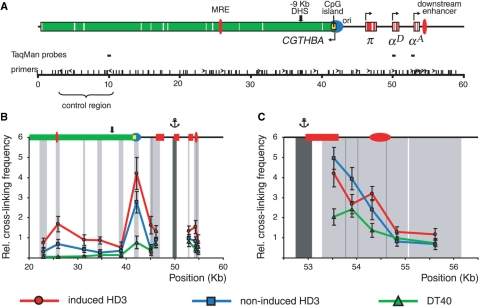
Figure 3.
MboI 3C analysis of the chicken α-globin gene domain in non-erythroid and transformed erythroid cells. (A) A MboI restriction map of the chicken α-globin gene domain and a scheme showing positions of functional elements of the domain. The short vertical lines above the scale show positions of MboI restriction sites. The other designations are as in Figure 2. (B and C) Relative frequencies of cross-linking between the anchor fragments bearing (B): αD gene promoter, (C): αA gene promoter and other fragments of the locus. The results of 3C analysis for induced HD3, cycling HD3 and DT40 cells are shown by the red, blue and green lines, respectively. The other notations are as in Figure 2.
A number of control experiments were done to rule out potential artifacts and ensure stringency of the technique. First, the efficiency of digestion with the restriction enzymes of cross-linked chromatin was checked by the PCR-stop analysis. It was found to be about the same (∼80% of complete digestion) for different sites throughout the locus and for different types of cells used (the results are shown in Supplementary Figure S1). Second, a standard for correction of PCR amplification efficiency for each primer set was generated. A bacterial artificial chromosome harboring the whole chicken α-globin gene domain with the bordering areas was digested with BamHI and BglII or with Sau3A (isoschisomer of MboI insensitive to dam methylation) and ligated at a high DNA concentration allowing intermolecular ligation. The resulting DNA was used for PCR assay as a template in which all possible ligation products are present in equimolar amounts. Third, a standard taking into account variations in the efficiency of cross-linking/restriction/ligation taken together and in the quantity of DNA in a 3C template was used for normalization of the data because cross-linking frequencies were planned to be measured in different types of cells. Usually, a locus that does not contain tissue-specific genes and is located far from the area under study is selected as such a standard as it can reasonably be assumed to adopt a similar spatial organization in different cell types (27). We used as such a standard a region of the chicken genome situated sufficiently close to the α-globin gene domain (13–23 kb upstream of MRE) but within the body of the housekeeping gene (4) (indicated as ‘control region’ in Figures 2A and 3A). For more accurate normalization in each cell type used we measured the cross-linking frequency for three pairs of control restriction fragments (one anchor fragment and three test fragments) in the case of BamHI/BglII restriction and for two pairs of control restriction fragments in the case of MboI restriction (the results are shown in Supplementary Figure S2A). The data for each restriction were then averaged and the result was arbitrarily considered as the cross-linking frequency value of 1. Although there are no reasons to suggest that the body of a house-keeping gene may have different spatial organizations in lymphoid and erythroid (either cycling or differentiated) cells, such a possibility can not be excluded because of the neighborhood of a tissue-specific gene domain (27). Thus we carried out another control 3C experiment on a house-keeping gene situated on another chromosome in house-keeping gene surroundings (the ERCC3 gene). The results obtained were very similar to those for the control region within the CGTHBA gene (supplementary Figure S2B). It is essential to note that the normalization of the data did not influence the profile of the 3C curves although slightly changed the absolute values (the original data are shown in supplementary Figures S3, S4 and S5). Fourth, we verified that all primer pairs worked equally well on both BAC random ligation and 3C DNA matrixes resulting in appearing of specific PCR products which were detected by agarose gel electrophoresis (typical results are present in Supplementary Figure S6). Finally, it was checked that a positive 3C PCR signal was totally dependent both on formaldehyde cross-linking and ligation (Supplementary Table S1).
Comparison of the spatial organization of the chicken α-globin gene domain in non-erythroid cells and transformed erythroid cells before and after the beginning of globin gene transcription
We first investigated the interaction of the LCR-like MRE with each of the downstream restriction fragments (with the exception of very short fragments). In lymphoid DT40 cells the frequency of cross-linking of the anchor fragment with each of the downstream fragments decreased sharply with an increase in the distance between the test-fragment and the anchor fragment and then remained very low (Figure 2B, green curve). This is typical for chromatin regions that are not organized in loops (9). In contrast, already in non-differentiated HD3 cells a clear increase was observed in the frequency of cross-linking of the anchor fragment with most of the downstream fragments with apparent peaks in the region containing the αD gene promoter and a broad region containing the CpG island and −9 kb (9 kb upstream of the π gene) DNase I hypersensitive site (Figure 2B, blue curve). The frequency of interaction of MRE with −9 kb DHS, CpG island and the promoter region of the αD gene (estimated on the basis of the cross-linking frequency for the corresponding restriction fragments) increased upon induction of terminal erythroid differentiation of HD3 cells (about 3.5, 2 and 4.5 times for each of the three above-mentioned elements, respectively) (Figure 2B, red curve). In addition, a clear interaction between the MRE element and the downstream enhancer was detected. Interestingly, no interaction of MRE with π and αA genes was observed either in non-induced or in induced HD3 cells. In the case of the αA gene it was an unexpected result, as this gene is actively transcribed in HD3 cells (Figure 1B, G and H). To verify this important observation we analyzed the ligation frequency for the opposite end of the restriction fragment containing the αA gene. With this aim the ‘opposite direction’ PCR primer located at the beginning of the restriction fragment under study was designed (Figure 2A). The frequencies of ligation of MRE to both ends of the restriction fragment containing the αA gene were found to be practically the same.
In the next series of experiments, the anchor primer was fixed sequentially on restriction fragments containing −9 kb DHS, CpG island, the promoter region of the αD gene and the αA gene. The results of these experiments shown in Figure 2C–G basically confirmed the conclusions made in experiments with the anchor fixed on MRE. In addition, it became clear that in cycling HD3 cells −9 kb DHS occasionally formed a complex with MRE but did not interact with either the αD gene promoter or the downstream enhancer (Figure 2C and E). In differentiated HD3 cells the frequency of association of −9 kb DHS with MRE increased about 3 times. At the same time, in these cells the interactions of −9 kb DHS with the αD gene promoter and the downstream enhancer were established (compare the red and blue curves in Figure 2C).
The 3C analysis with the anchor fixed either at the CpG island or at the promoter region of the αD gene demonstrated that they interacted in all three cellular models. The frequency of interaction of the above two elements increased in the following order: DT40 < cycling HD3 < differentiated HD3 (Figure 2D and E). However, a supposition that such an interaction exists (at least in DT40 cells) depends on the data showing sufficiently low frequencies of interaction of the CpG island and αD gene promoter with the π gene. In order to verify that the π gene indeed associates weakly with these two elements, we designed three oppositely directed primers at the beginnings of restriction fragments containing the above-mentioned elements and performed 3C analysis with an anchor fixed at the π gene. The results of this analysis confirmed the low degree of association of the π gene with both the CpG island and αD gene promoter (Figure 2D and E, points without error bars).
In agreement with the results obtained with the anchor fixed at MRE (Figure 2B), the results of the experiments with the anchor fixed at the CpG island demonstrated that the frequency of association of MRE with the CpG island was about 2 times higher in differentiated than in proliferating HD3 cells (Figure 2D, blue and red curves).
Experiments with the anchor fixed either on the right or the left end of the restriction fragment containing the αA gene (Figure 2F and G, respectively) demonstrated that this gene was not located in close proximity to any of the upstream regulatory elements.
In order to increase the resolution of the 3C analysis, experiments with the anchor fixed at the αD gene promoter were repeated after digestion of fixed chromatin with MboI. Utilization of this restriction enzyme permitted us to separate the 6 kb BglII-BglII fragment containing the CpG island into several smaller fragments. As a result, it became possible to demonstrate that the αD gene promoter interacted preferentially with the CpG island per se (more precisely, with the about 1 kb DNA fragment containing the downstream part of the CpG island with the promoter of the housekeeping gene CGTHBA) (Figure 3B). The frequency of association of the ∼0.8 kb DNA fragment containing the αD gene promoter and the 1 kb DNA fragment containing the downstream part of the CpG island was even more pronounced than that observed when larger fragments obtained by BamHI/Bgl II digestion were used for the 3C analysis (Figure 2D and E). Possibly, in this particular case the positions of the shorter restriction fragments within the DNA–protein complex are more favorable for cross-ligation. In a similar way, by using MboI we divided 1 kb BamHI-BglII fragment containing the downstream enhancer into two subfragments one of which carried the essential part of the enhancer (8). The latter subfragment demonstrated a high frequency of association with the αD gene promoter while the other, ‘empty’ one, did not. Most important, the differences between DT40, proliferating and differentiated HD3 cells observed in the experiment with BamHI/Bgl II restriction were well reproduced in the experiment with MboI restriction. Unfortunately, MboI cuts the region containing −9 kb DHS into a number of very small pieces. Thus this region was not analyzed. On the other hand, MboI digestion permitted the αA gene promoter to be localized within a much shorter ∼0.35 kb DNA fragment as compared to the 1.7 kb fragment in the case of BamHI/BglII restriction. The ligation frequencies for short fragments containing the αD and αA gene promoters were again relatively small (considering a close proximity of the corresponding fragments, which, by itself, may provide an elevated level of cross-ligation). Furthermore, no significant differences in the frequencies of association of the αD and αA gene promoters were observed when different cellular models (including lymphoid DT40 cells) were compared (Figure 3B).
Taking into consideration the apparent absence of interactions of the αA gene promoter with all tested upstream regulatory elements, it was important to check whether this promoter interacted with the closely located downstream enhancer (see the map in Figure 3C). Thus we have carried out a detailed 3C analysis of the ∼3 kb region including the αA gene promoter and the downstream enhancer. MboI cuts this area into a number of fragments five of which are of sufficient sizes to place primers for the 3C analysis. The results of the analysis of interaction between the anchor fragment containing the αA gene promoter and each of the five downstream fragments tested are shown in Figure 3C. A slight increase in the frequency of interaction between the fragment containing the αA gene promoter and the fragment containing the downstream enhancer was observed only in differentiated HD3 cells.
Analysis of the spatial organization of the chicken α-globin gene domain in 10-day chick embryo erythrocytes
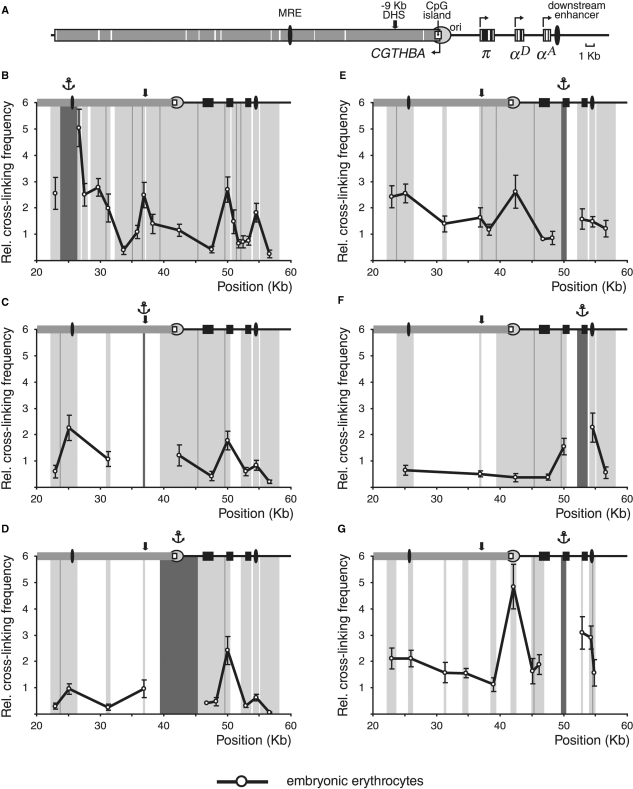
In order to find out if the configuration of the α-globin gene domain in differentiated HD3 cells resembles that in normal embryonic erythrocytes, 3C experiments were carried out with 10-day chick embryo normal erythrocytes (10-day red blood cells, RBC). In these cells the π gene is not active while both αA and αD genes are intensively transcribed (29–31). The results of the experiments with the use of Bam HI/Bgl II restriction are represented in Figure 4B–F. One can see that the relative frequencies of association between MRE and the downstream regulatory elements are very similar in differentiated HD3 cells (Figure 2B, red curve) and in 10-day RBC (Figure 4B). Of particular importance are low frequencies of association of MRE with the promoters of the π and αA genes as compared to the frequencies of association of MRE with −9 kb DHS, αD gene promoter or the downstream enhancer. The 3C analysis with the anchor fixed at the −9 kb DHS (Figure 4C) and at the CpG island (Figure 4D) also gave the results very similar to those obtained in experiments with differentiated HD3 cells. With the anchor fixed at the promoter region of the αD gene we observed a relatively high frequency of association with the αA gene. This result was confirmed in the experiments with the anchor fixed at the αA gene (compare Figure 4F and the red curve in Figure 2F). Furthermore, the results presented in Figure 4F also suggest a certain degree of association between the αA gene and the downstream enhancer. To verify these observations, we carried out the 3C analysis using digestion with MboI restriction enzyme and the anchor fixed at the αD gene promoter. The results obtained (Figure 4G) show that in 10-day RBC the αD gene promoter may interact both with the αA gene promoter and with the downstream enhancer.
Figure 4.
3C analysis of the chicken α-globin gene domain in 10-day chick embryo erythrocytes. (A) A scheme showing positions of functional elements of the domain. Black boxes: α-globin genes; grey box: CGTHBA gene; black ovals: MRE and the downstream enhancer; open rectangle: CpG island; circle: origin of replication. The other designations are as in Figure 2. (B–G) Relative frequencies of cross-linking between the anchor fragments bearing (B): MRE, (C): −9 kb DHS, (D): CpG island, (E) and (G): αD gene promoter, (F): αA gene and other fragments of the locus [3C was performed with (B–F): BamHI and BglII restriction enzymes, (G): MboI restriction enzyme]. The other notations are as in Figure 2.
DISCUSSION
To interpret adequately the results of the present study it is necessary to keep in mind that the 3C analysis makes it possible to estimate only the relative probabilities of interaction of different restriction fragments within the area under study. The possibility of existence of an alternating spatial configuration of the α-globin gene locus in the same cells should be taken into account. In addition, the spatial organization of this locus can differ in two copies of homologous chromosomes in the same cells and also in different cells present in the population. To facilitate the discussion we shall consider only the most interesting spatial configurations typical for each of the cell types studied. Yet, we realize that in all cell types the most complex spatial configurations discussed below may coexist with others including even the simplest linear configuration. That is why it is essential to quantify all interactions rather than to draw conclusions from the presence or absence of PCR products. On the other hand, it is equally necessary to control the specificity of PCR products, which is only possible when real-time PCR with TaqMan probes is employed to estimate the ligation frequencies for different restriction fragments (32–35).
The most obvious conclusion from our studies is that the spatial organization of the chicken α-globin gene domain is drastically different in erythroid versus non-erythroid cells. Even in proliferating HD3 cells the frequencies of association of the MRE element with most of the downstream fragments of the α-globin gene domain are noticeably elevated as compared to those in non-erythroid cells. The three exceptions are the region containing CTCF-dependent enhancer-blocking elements (20) which is located 11–16 kb upstream of the π gene, the promoter of the π gene and the promoter of the αA gene. In differentiated HD3 cells and in 10-day RBC the frequencies of association of MRE with all the downstream restriction fragments except the one containing the π gene promoter increase as compared to proliferating HD3 cells.
Careful analysis of the ligation frequencies for different fragments of the chicken α-globin gene locus in erythroid and non-erythroid cells permitted us to distinguish several characteristic spatial configurations of this gene locus. The first one (Figure 5A) is typical for non-erythroid DT40 cells. In these cells the promoter area of the αD gene interacts with the upstream CpG island of the α-globin gene domain which contains a replication origin (36,37) and the promoter of the house-keeping gene (4,17). This association is not likely to be stable yet as the frequency of association between the αD promoter and the CpG island increases in erythroid and especially in globin-producing cells. Thus in non-erythroid cells the liner configuration of the α-globin gene domain exists in a certain equilibrium with a loop formed due to the association of the αD promoter with the CpG island. This result is in contrast with observations for the mouse α-globin gene domain where promoters of globin genes interacted with the CpG islands surrounding the domain only in cells transcribing globin genes (16). Apparently the spatial organization of the α-globin gene domain in cells not-expressing globins differs in mouse and chicken cells. These differences may reflect different long-range organizations of the domains. Indeed, the conserved synteny between chicken and mouse α-globin gene domains extends over a relatively short genomic region in contrast to the long regions of conserved synteny between α-globin gene domains in different mammals (38).
Figure 5.
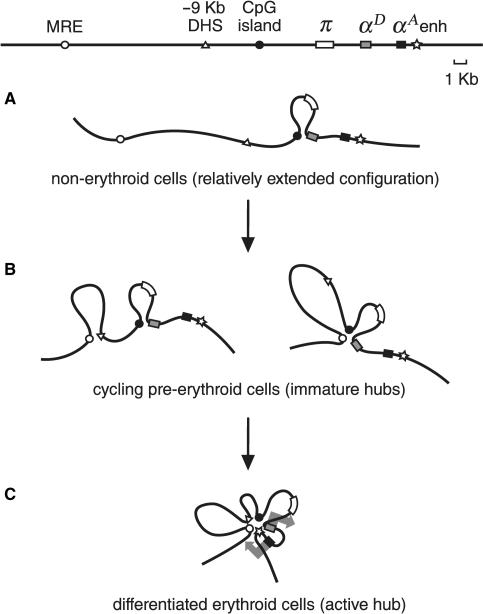
A scheme representing spatial configurations of the chicken α-globin gene domain in different cell types. (A) DT40 cells. (B) Cycling HD3 cells. (C) Induced to terminal erythroid differentiation HD3 cells and 10-day RBC. Different domain elements are shown by the following symbols: MRE: open circle, −9 kb DHS: triangle, CpG island: black circle, π gene: white rectangle, αD gene: gray rectangle, αA gene: black rectangle, downstream enhancer: asterisk. Large gray arrows in (C) symbolize active transcription statuses of αD and αA genes.
In proerythroblasts (proliferating HD3 cells) the interactions between the αD gene promoter and the CpG island become more frequent or occur in a larger portion of cells. In addition, various ‘immature’ chromatin hubs started to appear in these cells due to the associations of different regulatory elements. Analysis of the frequencies of interaction of different restriction fragments located downstream of MRE demonstrates that the most complex ‘immature’ chromatin hub includes MRE, CpG island and the αD gene promoter. It is important to underline that experiments with the anchor fixed at different restriction fragments have clearly demonstrated that all the above-mentioned elements interact with each other. Thus, they really form a chromatin hub. However, this does not exclude the possibility that this chromatin hub exists in a dynamic equilibrium with different less complex associates of the above-mentioned regulatory elements. Interestingly, in cycling HD3 cells the −9 kb DHS does not participate in the formation of the chromatin hub with the αD gene promoter and the CpG island, although it may form an independent complex with MRE (Figure 2C). Thus in cycling HD3 cells the α-globin gene domain may adopt two mutually exclusive configurations due to the association of MRE with either −9 kb DHS or with the CpG island and the αD gene promoter (Figure 5B).
It is interesting that in mouse proerythroblasts activator complexes are assembled on the upstream regulatory elements but the promoters of adult α-globin genes do not interact with these regulatory elements before induction of differentiation (32). In contrast, in chicken proerythroblasts (cycling HD3 cells) associations of MRE, CpG island and the αD gene promoter are clearly detectable.
After induction of terminal erythroid differentiation of HD3 cells the frequency of interaction between MRE, CpG island and the αD gene promoter increases. Furthermore, the downstream enhancer and −9 kb erythroid-specific DHS are also recruited to the same chromatin hub. A very similar configuration of the α-globin gene domain is typical also for chicken embryonic red blood cells (10-day RBC) producing αA and αD globin chains. Thus the involvement of the downstream enhancer and −9 kb DHS is a distinctive feature of the chicken α-globin gene domain ‘active’ chromatin hub. Unfortunately, nothing particularly interesting is known about −9 kb DHS. It was mapped long ago and found to roughly colocalize with a MAR element (18). Although the general organization of α-globin gene domains in different vertebrates is conserved in evolution (38,39), this particular DHS is not located in ‘multispecies conserved sequences’ and there is no obvious analogs of this DHS in human and mouse genomes (39). Apparently, the fragment harbors binding sites for some erythroid-specific transcriptional factors participating in the assembly of an ‘active’ chromatin hub typical for mature erythroblasts expressing globins (Figure 5C). Computer analysis of a DNA fragment spanning the DHS revealed four potential binding sites for the erythroid-specific transcription factors GATA 1 and 2.
In all cellular models studied we failed to detect associations of the embryonic α-type gene π with any of either upstream or downstream regulatory elements. It seems logical as the π gene is not expressed in differentiated HD3 cells and in 10-day RBC [Figure 1B, G and H and (29)]. Much more unexpected was the failure of detecting the interaction of the αA gene promoter with any of the upstream regulatory elements, and, first of all, with MRE (Figures 2B, F, G, 4B, F and G). In this context, it is important to remind that in differentiated HD3 cells the level of αA gene expression exceeds the expression of the αD gene (Figure 1B, G and H). The same is true for 10-day RBC (40). Although the frequency of association of MRE with the αA gene promoter was not exactly at the zero level in differentiated HD3 cells and in 10-day RBC, it was much less prominent than the frequency of association of MRE with the αD gene promoter (Figures 2B and 4B). The same was observed for −9 kb DHS (Figures 2C and 4C) and for the CpG island (Figures 2D and 4D). Taking into account the fact that the αA gene promoter is located ∼3 kb downstream of the αD gene promoter and ∼1.5 kb upstream of the enhancer which both strongly interact with MRE and the other above-mentioned upstream regulatory elements, one can hardly expect the apparent association frequency of the αA gene promoter to attain the zero level even if this promoter does not interact by itself with MRE and other upstream regulatory elements.
Of course, our analysis of cross-linking frequencies for different restriction fragments is not sensitive enough to exclude the possibility of infrequent short-term interactions of the αA gene promoter with any of the above-mentioned elements. Nevertheless is seems clear that the activities of the αA and αD gene promoters are regulated by different mechanisms. The situation is obviously not similar to the one observed in the human β-globin gene domain where the LCR interacts alternatively with all active promoters (9,11,41). One could propose that the chicken αD gene promoter directs the synthesis of a bi-cistronic pre-mRNA which is then processed to give rise to individual αD and αA mRNAs. However, the pattern of α-globin gene domain transcription was extensively studied in the past and no signs of such bi-cistronic pre-mRNA were detected (2). Furthermore, the promoter of the αA gene is well characterized and there are no reasons to suppose that this promoter is not active in living cells (8,42,43). A more attractive model is that the αA gene promoter is activated by the enhancer located <1 kb downstream of the end of the αA gene (8). There is no similar enhancer in human and mouse α-globin gene domains. The association, if any, of the downstream enhancer with the αA gene promoter is difficult to demonstrate by the 3C analysis since these elements are located too close to each other. Nevertheless, some of our results suggest that this association indeed exists in 10-day RBC (Figure 4F) and possibly in differentiated HD3 cells (Figure 3C). The possibility should also be considered that an enhancer located so close to the gene promoter may activate this promoter via a distinct mechanism that does not involve any looping of a short intermediate DNA fragment. What we know for sure it is that the downstream enhancer interacts with the αD gene promoter participating in the formation of an ‘active’ chromatin hub (Figure 5C). The possibility may thus be suggested that besides activation of the αD gene promoter the formation of an active chromatin hub in cells transcribing αA and αD genes is necessary for activation of the ‘adult’ sub-domain of the α-globin gene cluster. Among other molecular events necessary for this activation one may consider inactivation of the silencer element located upstream of the αD gene and active in respect of both αD and αA gene promoters (43).
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by the Presidium of the Russian Academy of Sciences [a grant from the Program on Molecular and Cellular Biology (MCB)], by RFBR (08-04-91970 and 08-04-00048) and by the FEBS Collaborative Experimental Scholarship for Central and Eastern Europe grant. We are grateful to Dr de Laat and Dr Splinter for helpful advices concerning utilization of the 3C procedure. Funding to pay the Open Access publication charges for this article was provided by the Russian Academy of Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1.Higgs DR, Vernimmen D, De Gobbi M, Anguita E, Hughes J, Buckle V, Iborra F, Garrick D, Wood WG. How transcriptional and epigenetic programmes are played out on an individual mammalian gene cluster during lineage commitment and differentiation. Biochem. Soc. Symp. 2006;73:11–22. doi: 10.1042/bss0730011. [DOI] [PubMed] [Google Scholar]
- 2.Recillas-Targa F, Razin SV. Chromatin domains and regulation of gene expression: familiar and enigmatic clusters of chicken globin genes. Crit. Rev. Eukaryot. Gene Expr. 2001;11:227–242. [PubMed] [Google Scholar]
- 3.Razin SV, Ioudinkova ES. Mechanisms controlling activation of the alpha-globin gene domain in chicken erythroid cells. Biochemistry (Mosc) 2007;72:467–470. doi: 10.1134/s000629790705001x. [DOI] [PubMed] [Google Scholar]
- 4.Flint J, Tufarelli C, Peden J, Clark K, Daniels RJ, Hardison R, Miller W, Philipsen S, Tan-Un KC, McMorrow T, et al. Comparative genome analysis delimits a chromosomal domain and identifies key regulatory elements in the alpha globin cluster. Hum. Mol. Genet. 2001;10:371–382. doi: 10.1093/hmg/10.4.371. [DOI] [PubMed] [Google Scholar]
- 5.Razin SV, Ioudinkova ES, Scherrer K. Extensive methylation of a part of the CpG island located 3.0-4.5 Kbp upstream to the chicken alpha-globin gene cluster may contribute to silencing the globin genes in non-erythroid cells. J. Mol. Biol. 2000;209:845–852. doi: 10.1006/jmbi.2000.3775. [DOI] [PubMed] [Google Scholar]
- 6.Razin SV, Shen K, Ioudinkova E, Scherrer K. Functional analysis of DNA sequences located within a cluster of DNase I hypersensitive sites colocalising with MAR element at the upstream border of the chicken a-globin gene domain. J. Cell Biochem. 1999;74:38–49. [PubMed] [Google Scholar]
- 7.Vyas P, Vickers MA, Picketts DJ, Higgs DR. Conservation of position and sequence of a novel, widely expressed gene containing the major human alpha-globin regulatory element. Genomics. 1995;29:679–689. doi: 10.1006/geno.1995.9951. [DOI] [PubMed] [Google Scholar]
- 8.Knezetic J, Felsenfeld G. Identification and characterization of a chicken α-globin enhancer. Mol. Cell Biol. 1989;9:893–901. doi: 10.1128/mcb.9.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 10.Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 11.Gribnau J, de Boer E, Trimborn T, Wijgerde M, Milot E, Grosveld F, Fraser P. Chromatin interaction mechanism of transcriptional control in vivo. EMBO J. 1998;17:6020–6027. doi: 10.1093/emboj/17.20.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wijgerde M, Grosveld F, Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature. 1995;377:209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]
- 13.de Laat W, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- 14.Kooren J, Palstra RJ, Klous P, Splinter E, von Lindern M, Grosveld F, de Laat W. Beta-globin active chromatin Hub formation in differentiating erythroid cells and in p45 NF-E2 knock-out mice. J. Biol. Chem. 2007;282:16544–16552. doi: 10.1074/jbc.M701159200. [DOI] [PubMed] [Google Scholar]
- 15.Patrinos GP, de Krom M, de Boer E, Langeveld A, Imam AM, Strouboulis J, de Laat W, Grosveld FG. Multiple interactions between regulatory regions are required to stabilize an active chromatin hub. Genes Dev. 2004;18:1495–1509. doi: 10.1101/gad.289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou GL, Xin L, Song W, Di LJ, Liu G, Wu XS, Liu DP, Liang CC. Active chromatin hub of the mouse alpha-globin locus forms in a transcription factory of clustered housekeeping genes. Mol. Cell Biol. 2006;26:5096–5105. doi: 10.1128/MCB.02454-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klochkov D, Rincon-Arano H, Ioudinkova ES, Valadez-Graham V, Gavrilov A, Recillas-Targa F, Razin SV. A CTCF-dependent silencer located in the differentially methylated area may regulate expression of a housekeeping gene overlapping a tissue-specific gene domain. Mol. Cell Biol. 2006;26:1589–1597. doi: 10.1128/MCB.26.5.1589-1597.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Moura Gallo CV, Vassetzky YS, Huesca M, Scherrer K. A transcription-dependent DNase I-hypersensitive site in a far upstream segment of the chicken α-globin gene domain coincides with a matrix attachment region. Biochem. Biophys. Res. Commun. 1992;184:1181–1189. doi: 10.1016/s0006-291x(05)80013-0. [DOI] [PubMed] [Google Scholar]
- 19.Razin SV, De Moura Gallo CV, Scherrer K. Characterization of the chromatin structure in the upstream area of the chicken α-globin gene domain. Mol. Gen. Genet. 1994;242:649–652. doi: 10.1007/BF00283418. [DOI] [PubMed] [Google Scholar]
- 20.Valadez-Graham V, Razin SV, Recillas-Targa F. CTCF-dependent enhancer blockers at the upstream region of the chicken alpha-globin gene domain. Nucleic Acids Res. 2004;32:1354–1362. doi: 10.1093/nar/gkh301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 22.Liu Z, Garrard WT. Long-range interactions between three transcriptional enhancers, active Vkappa gene promoters, and a 3' boundary sequence spanning 46 kilobases. Mol. Cell Biol. 2005;25:3220–3231. doi: 10.1128/MCB.25.8.3220-3231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beug H, Doederlein G, Freudenstrein C, Graf T. Erythroblast cell lines transformed by a temperature sensitive mutant of avian erythroblastosis virus. A model system to study erythroid differentiation in vitro. J. Cell Physiol. 1979;1:195–207. doi: 10.1002/jcp.1041130427. [DOI] [PubMed] [Google Scholar]
- 24.Beug H, Von Kirchbach A, Doderlin J, Conscience JF, Graf T. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell. 1979;18:375–390. doi: 10.1016/0092-8674(79)90057-6. [DOI] [PubMed] [Google Scholar]
- 25.Nicolas RH, Partington G, Major GN, Smith B, Carne AF, Huskisson N, Goodwin G. Induction of differentiation of avian erythroblastosis virus-transformed erythroblasts by the protein kinase inhibitor H7: analysis of the transcription factor EF1. Cell Growth Differ. 1991;2:129–135. [PubMed] [Google Scholar]
- 26.Rowley PT, Ohlsson-Wilhelm BM, Farley BA. K562 human erythroleukemia cells demonstrate commitment. Blood. 1985;65:862–868. [PubMed] [Google Scholar]
- 27.Splinter E, Grosveld F, de Laat W. 3C technology: analyzing the spatial organization of genomic loci in vivo. Methods Enzymol. 2004;375:493–507. doi: 10.1016/s0076-6879(03)75030-7. [DOI] [PubMed] [Google Scholar]
- 28.Razin SV, Rynditch A, Borunova V, Ioudinkova E, Smalko V, Scherrer K. The 33 kb transcript of the chicken alpha-globin gene domain is part of the nuclear matrix. J. Cell Biochem. 2004;92:445–457. doi: 10.1002/jcb.20066. [DOI] [PubMed] [Google Scholar]
- 29.Weintraub H, Larsen A, Groudine M. Alpha-Globin-gene switching during the development of chicken embryos: expression and chromosome structure. Cell. 1981;24:333–344. doi: 10.1016/0092-8674(81)90323-8. [DOI] [PubMed] [Google Scholar]
- 30.Bruns GA, Ingram VM. Erythropoiesis in the developing chick embryo. Dev. Biol. 1973;30:455–459. doi: 10.1016/0012-1606(73)90102-4. [DOI] [PubMed] [Google Scholar]
- 31.Singal R, vanWert JM, Ferdinand L., Jr Methylation of alpha-type embryonic globin gene alpha pi represses transcription in primary erythroid cells. Blood. 2002;100:4217–4222. doi: 10.1182/blood-2002-02-0457. [DOI] [PubMed] [Google Scholar]
- 32.Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 2007;26:2041–2051. doi: 10.1038/sj.emboj.7601654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat. Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 35.Wurtele H, Chartrand P. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended Chromosome Conformation Capture methodology. Chromosome Res. 2006;14:477–495. doi: 10.1007/s10577-006-1075-0. [DOI] [PubMed] [Google Scholar]
- 36.Razin SV, Kekelidze MG, Lukanidin EM, Scherrer K, Georgiev GP. Replication origins are attached to the nuclear skeleton. Nucleic Acids Res. 1986;14:8189–8207. doi: 10.1093/nar/14.20.8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verbovaia L, Razin SV. Analysis of the replication direction through the domain of a-globin-encoding genes. Gene. 1995;166:255–259. doi: 10.1016/0378-1119(95)00616-8. [DOI] [PubMed] [Google Scholar]
- 38.Tufarelli C, Hardison R, Miller W, Hughes J, Clark K, Ventress N, Frischauf AM, Higgs DR. Comparative analysis of the alpha-like globin clusters in mouse, rat, and human chromosomes indicates a mechanism underlying breaks in conserved synteny. Genome Res. 2004;14:623–630. doi: 10.1101/gr.2143604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hughes JR, Cheng JF, Ventress N, Prabhakar S, Clark K, Anguita E, De Gobbi M, de Jong P, Rubin E, Higgs DR. Annotation of cis-regulatory elements by identification, subclassification, and functional assessment of multispecies conserved sequences. Proc. Natl Acad. Sci. USA. 2005;102:9830–9835. doi: 10.1073/pnas.0503401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown JL, Ingram VM. Structural studies on chick embryonic hemoglobins. J. Biol. Chem. 1974;249:3960–3972. [PubMed] [Google Scholar]
- 41.Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 42.Dodgson JB, Engel JD. The nucleotide sequence of the adult chicken α-globin genes. J. Biol. Chem. 1983;258:4623–4629. [PubMed] [Google Scholar]
- 43.Lewis W, Lee J.-D, Dodgson JB. Adult chicken α-globin gene expression in transfected QT6 quail cells: evidence for a negative regulatory element in the αd globin region. Nucleic Acids Res. 1991;19:5321–5329. doi: 10.1093/nar/19.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.