Abstract
The degradation of the poly(A) tail is crucial for posttranscriptional gene regulation and for quality control of mRNA. Poly(A)-specific ribonuclease (PARN) is one of the major mammalian 3′ specific exo-ribonucleases involved in the degradation of the mRNA poly(A) tail, and it is also involved in the regulation of translation in early embryonic development. The interaction between PARN and the m7GpppG cap of mRNA plays a key role in stimulating the rate of deadenylation. Here we report the solution structures of the cap-binding domain of mouse PARN with and without the m7GpppG cap analog. The structure of the cap-binding domain adopts the RNA recognition motif (RRM) with a characteristic α-helical extension at its C-terminus, which covers the β-sheet surface (hereafter referred to as PARN RRM). In the complex structure of PARN RRM with the cap analog, the base of the N7-methyl guanosine (m7G) of the cap analog stacks with the solvent-exposed aromatic side chain of the distinctive tryptophan residue 468, located at the C-terminal end of the second β-strand. These unique structural features in PARN RRM reveal a novel cap-binding mode, which is distinct from the nucleotide recognition mode of the canonical RRM domains.
INTRODUCTION
Regulation of mRNA turnover is an important process in determining the levels of gene expression and the quality control of mRNA biogenesis (1–3). In eukaryotes, mRNA turnover is controlled by two factors: the length of the poly(A) tail at the 3′ end of the mRNA and the presence of the N7-methyl guanosine (m7GpppG) cap at its 5′ end, which are involved in maturation, transport, translation and degradation of the mRNA (4–7). Cytoplasmic mRNA degradation is initiated by shortening the 3′ poly(A) tail by a variety of deadenylases, leading to the removal of the 5′ cap by a decapping enzyme and the 5′–3′ exonucleolytic degradation of the mRNA (5,8). Several deadenylases have been identified in eukaryotes (4,9). Among them, poly(A)-specific ribonuclease (PARN) is a key enzyme involved in the deadenylation of mRNA in a cap-dependent manner (10–14).
PARN is localized in both the nucleus and cytoplasm in higher eukaryotic cells (12,15). During the meiotic maturation of Xenopus oocytes, PARN participates in the translational silencing of maternal mRNAs (12,16). PARN was recently reported to be a component of the cytoplasmic polyadenylation element binding protein-containing (CPEB-containing) complex, which regulates translation in early embryonic development (17). In the CPEB-containing complex, PARN interacts with the poly(A) polymerase Gld2, inhibits the polyadenylation activity and shortens the poly(A) tail, thus accomplishing translational silencing (17). When the oocytes are induced to mature with progesterone, Aurora A phosphorylates the CPEB-containing complex to expel PARN, resulting in Gld2-catalyzed default polyadenylation and subsequent translation initiation (17).
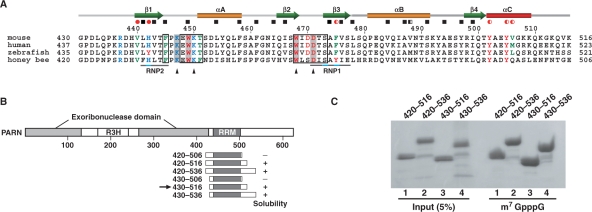
PARN is a divalent metal ion-dependent 3′ exonuclease (10,13,14,18) that specifically catalyzes the 3′–5′ degradation of the single-stranded poly(A) tail of mRNA with a free 3′-hydroxyl group (10,11,13). In addition, PARN interacts with the m7GpppG cap (19,20), which enhances its deadenylation activity (14,19–21). A sequence alignment revealed that PARN contains a region homologous to the DEDDh family of 3′–5′ exonucleases (22), which is interrupted by a long insertion with an R3H domain (23,24). On the other hand, the C-terminal truncated human PARN (23) did not bind to the mRNA cap, suggesting that the putative cap-binding domain resides in the C-terminal region. Thus, PARN contains three domains: the nuclease domain, the R3H domain and the putative cap-binding domain. Recent crystallographic and mutagenesis studies of human PARN revealed that the C-terminal truncated PARN forms a homodimer through its nuclease domain, and that the nuclease domain of each subunit binds to adenosine nucleotides (23). This dimer formation by PARN may be essential for its poly(A)-specific activity. However, the crystal structure lacked structural information on the putative cap-binding domain. As shown in Figure 1, an examination of the amino acid sequences among the PARNs from several species revealed that the cap-binding domain contains sequences homologous to the characteristic RNP1 and RNP2 sequences of the RNA recognition motif (RRM) (16), which are generally responsible for the nucleotide binding activity. Considering the importance of the m7GpppG cap for PARN activity, the cap recognition mechanism should be elucidated.
Figure 1.
(A) Amino acid sequence alignment of the cap-binding domains of PARNs from different species. The positions of secondary structure elements, as observed in the mouse cap-binding domain, are shown at the top of the alignment. The residues involved in hydrophobic core formation, as determined by GETAREA, are indicated by red circles (V440, H442, F475, Y502, Y505 and V506) and black boxes. Completely and partially buried residues are shown by filled and half-filled symbols, respectively. Residues with obvious chemical shift changes (above the mean value + 1 SD) upon cap binding are boxed. Residues directly involved in cap binding are highlighted in gray. The highly conserved ‘RNP1’ and ‘RNP2’ sequences among the canonical RRMs (52) are aligned with the sequences in the cap-binding domain of PARN and are indicated below the sequences. Positions of amino acid substitutions (see text) are denoted by black triangles. (B) Domain structure of PARN and deletion constructs containing the cap-binding domain of PARN with N- and C-terminal flanking sequences of varying lengths. The RRM region was predicted by the secondary structure elements and is shaded dark gray. Plus and minus signs indicate constructs that yielded soluble protein or inclusion bodies, respectively. The arrow identifies the construct that was selected for the solution structure studies. (C) Pull-down assay of the cap-binding domain with m7G(5′)ppp-Sepharose.
In this study, we determined the solution structures of the free and cap-bound forms of the cap-binding domain of mouse PARN by heteronuclear NMR experiments. The 3D structure of the PARN cap-binding domain includes an RRM as predicted by the amino acid sequence analysis. In addition, we performed binding assays with RRM and the m7GpppG cap analog, which identified the important sites for the cap binding. The present results revealed the novel cap-binding mode of PARN RRM, which is unique among the structurally and functionally well-studied RRM family members.
MATERIALS AND METHODS
Cloning
The DNA fragment encoding the cap-binding domain of PARN (residues 430–516) was amplified from the RIKEN FANTOM mouse cDNA clone with the ID 1200003I18 (25), by conventional polymerase chain reaction (PCR), and was subcloned into pCR2.1 (Invitrogen, Carlsbad, CA, USA). The cap-binding domain was expressed with an extra His6-tail, a TEV protease cleavage site, a (Gly–Gly–Ser)2–Gly sequence at the N-terminus and a Ser–Gly–Pro–Ser–Ser–Gly sequence at the C-terminus. Several deletion mutants of PARN that contained the cap-binding domain and its N- and/or C-terminal flanking regions, encoding residues 420–506, 420–516, 420–536, 430–506, 430–516 and 430–536, were amplified by PCR and subcloned into pET15b (Merc, San Diego, CA, USA). Selected point mutations (K447A, K450A, W468L and D471A) were introduced into the cap-binding domain (residues 430–516), using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA).
Expression and purification
The unlabeled and the 15N, 13C-labeled PARN cap-binding domains used for NMR experiments were synthesized by the cell-free protein expression system (26). After the reaction, each protein was isolated by chromatography on a Ni2+ affinity column, and the His6-tag was removed by proteolysis. Subsequent cation-exchange chromatography (HiTrap SP, GE Healthcare, Buckinghamshire, England) yielded the highly purified cap-binding domain. In order to prepare the NMR samples, the protein was transferred into 20 mM sodium phosphate buffer (pH 6.0), 100 mM NaCl, 1 mM 1,4-dl-dithiothreitol-d10 (d-DTT) and 0.02% sodium azide, prepared with either 1H2O/2H2O (9:1) for the 15N, 13C-labeled cap-binding domain or 2H2O for the unlabeled cap-binding domain, by ultrafiltration using a Centricon YM3 filter device (Millipore, Billerica, MA, USA). The protein concentration was increased simultaneously to 0.4–1.0 mM.
The wild-type and mutant forms of the cap-binding domain of PARN were overexpressed in Escherichia coli BL21(DE3) cells (Merc, San Diego, CA, USA) for pull-down assays. The cells were grown in 2× YT medium at 37°C to an OD600 = 0.6, and were induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 20°C. Cells were harvested after ∼16 h by centrifugation, suspended in 20 mM Tris–HCl buffer (pH 7.5) containing 1 M NaCl, 30 mM imidazole, 1 mM 1,4-dl-dithiothreitol (DTT), 0.1 mg/ml lysozyme, DNaseI and protease inhibitor cocktail (Nacalai Tesque, Kyoto, Japan), and lysed with a sonicator. Cell debris and inclusion bodies were removed by centrifugation. The supernatant was loaded on a Ni2+-NTA-agarose column (Qiagen, Inc., Hilden, Germany), which was then washed thoroughly with 20 mM Tris–HCl buffer (pH 7.5), 1 M NaCl, 30 mM imidazole and 1 mM DTT. The proteins were eluted with 20 mM Tris–HCl (pH 7.5), 1 M NaCl and 200 mM imidazole.
The cap analogs (m7GpppG, GpppG and GpppA) and the 10-mer poly(A) oligonucleotide (A10) were purchased from New England Biolabs (Ipswich, MA, USA) and Dharmacon Research, Inc. (Lafayette, CO, USA) respectively. Complexes were prepared by the addition of aliquots of the lyophilized cap analog to the protein NMR samples.
NMR spectroscopy
All NMR spectra of the uniformly 15N, 13C-labeled cap-binding domain in its free form were acquired on Bruker AVANCE 600 and AVANCE 800 spectrometers at a probe temperature of 25°C. Backbone and side chain 1H, 15N and 13C resonances of the free protein were assigned by standard double and triple resonance NMR experiments (27–30). Sequence-specific backbone assignments were achieved by two-dimensional (2D) 1H–15N heteronuclear single quantum correlation (HSQC) and 3D HNCO, HN(CA)CO, CBCA(CO)NH and HNCACB spectra. Assignments of side chain resonances for nonaromatic residues were obtained from 2D 1H-13C HSQC and 3D HBHA(CO)NH, HC(CCO)NH, C(CCO)NH, H(C)CH- correlation spectroscopy (COSY), H(C)CH-total correlated spectroscopy (TOCSY) and (H)CCH-TOCSY, whereas assignments for aromatic residues were performed by H(C)CH-correlation spectroscopy (COSY), aided by 3D 15N- and 13C-separated nuclear Overhauser effect spectroscopy (NOESY)–HSQC spectra, which were recorded with mixing times of 80 ms.
Spectra for the cap-bound state were acquired using an NMR sample containing ∼0.5 mM 15N, 13C-labeled cap-binding domain and ∼2.5 mM unlabeled cap analog, in 90% H2O, 10% 2H2O. 1H, 15N and 13C resonance assignments of the cap-binding domain in complex with the cap were achieved by comparison of the 3D 15N- and 13C-separated NOESY–HSQC spectra between the free and cap-bound states. For the resonance assignment of the m7GpppG cap analog, 1H–13C heteronuclear multiple quantum coherence (HMQC) for the detection of the naturally abundant 13C resonances, 2D homonuclear TOCSY and 2D homonuclear NOESY experiments were performed under the same conditions as in the final stage of the titration experiment (see subsequently). Intermolecular NOEs involving the protons of the cap-binding domain and the cap analog in the complex were obtained from 3D 15N-separated NOESY–HSQC (80, 100 and 200 ms mixing times at 25°C) and 3D 13C-separated NOESY–HSQC (80, 100 and 200 ms at 25°C; 150 ms at 15 and 25°C) spectra. In order to distinguish between the intra- and inter-molecular NOEs, 2D [F1, F2] 13C-filtered NOESY, 2D [F2] 13C-filtered NOESY (31) (150 and 300 ms at 5 and 15°C) and 3D [F1] 13C-filtered [F3] 13C-edited NOESY–HSQC (32) (300 ms at 15 and 25°C) spectra were measured. All NMR data for the structure calculations were processed using the NMRpipe software (33) and were analyzed with KUJIRA, a program suite for interactive NMR analysis (34), used in conjugation with NMRView (35).
Structure calculations
The 3D structures of the cap-binding domain in its free and complex forms were determined with the program CYANA (36–38), which implements automated NOE assignments and structure calculations with torsion angle dynamics steps. Peak lists containing the chemical shifts and the peak volumes of NOEs within the protein were used to obtain lists of intramolecular protein distance restraints. These generated lists were reviewed and combined with a list containing manually assigned NOE distances obtained by resolving ambiguities, such as spectral overlap. In the case of the complex, the lists containing manually assigned intramolecular NOEs within the cap analog and inter-molecular NOEs between the cap and the protein were further added as distance restraints. Secondary structure elements were verified by the NOE patterns: Hαi–HNi+3 and Hαi–Hβi+3 for the α-helical structure, and HNi–HNj, HNi–Hαj+1 and Hαi-1–Hαj+1 for the antiparallel β-sheet. Protein backbone φ, ψ and side chain χ1, χ2 torsion angle restraints were determined by a chemical shift database analysis, using the program TALOS (39), and by inspecting the pattern of intraresidual NOE intensities (40), respectively. Hydrogen bonding restraints for the backbone atoms (rNH–O = 1.7–2.2 Å, rN–O = 2.6–3.3 Å, rNH–C′ = 2.6–3.5 Å, rN–C′ = 3.6–4.6 Å) were also introduced within the secondary structures during the final stage of refinement. Furthermore, stereospecific assignments for the isopropyl methyl and methylene groups were determined with CYANA, by the GLOMSA method (41). The configuration around the glycosidic bond and the sugar puckering of the nucleotide moieties in the cap analog were estimated, based on the intensities of the H8–H2′ and H8–H3′ NOE cross-peaks in the 2D NOESY spectra (42,43). Starting from 100 randomized conformers, final ensembles of 20 and 40 conformers were selected on the basis of the lowest final CYANA target function values for the cap-free and cap-bound forms, respectively. For the cap-binding domain in the cap-bound form, the selected 40 conformers were subsequently energy-minimized, using the SANDER module of AMBER9 (http://amber.scripps.edu) with the generalized Born solvent model (44), and the 20 conformers with the lowest energy were selected for further structure analysis. The qualities of the structures were analyzed using the programs MOLMOL (45) and PROCHECK–NMR (46). The buried residues were defined as those with a relative solvent accessibility of <20%, calculated by GETAREA (http://pauli.utmb.edu/getarea). The coordinates for the ensembles of the 20 conformers of the cap-binding domain in its free and cap-bound forms were deposited in the RCSB Protein Data Bank, under the accession codes 1WHV and 2ROK, respectively.
Pull-down assays with the cap analog
The wild-type and mutant proteins of the cap-binding domain were incubated with m7G(5′)ppp-Sepharose 4B beads (GE Healthcare; Buckinghamshire, England), in a buffer containing 20 mM bis-Tris, pH 5.5, 100 mM NaCl, 1 mM DTT and 1 mM EDTA, for 1 h at 4°C. The beads were washed with the same buffer, and the bound proteins were eluted by adding 2× SDS gel loading buffer and boiling for 5 min. After centrifugation, the supernatant was collected and the proteins were fractionated by 15–25% SDS–PAGE and visualized by staining with Coomassie brilliant blue.
NMR titration experiments
NMR titration experiments of the cap-binding domain with the cap analog, m7GpppG, were performed by recording 2D homonuclear TOCSY (mixing time of 45 ms) and 2D homonuclear NOESY (80, 150 and 300 ms) spectra in 2H2O at 5, 15 and 25°C. Increasing amounts of the unlabeled m7GpppG cap analog were added to 0.4 mM unlabeled cap-binding domain, to achieve molar ratios of 1:0, 1:0.25, 1:0.5, 1:0.75, 1:1, 1:1.25, 1:1.5, 1:5 and 1:12. Titration experiments for 1H–15N resonances were also performed using the 15N, 13C-labeled cap-binding domain with three different cap analogs, m7GpppG, GpppG and GpppA. Two-dimensional heteronuclear 1H–15N HSQC spectra were measured to monitor the titrations. The molar ratios of protein:m7GpppG were 1:0, 1:1 and 1:5; those of protein:GpppG were 1:0, 1:0.1, 1:0.4, 1:0.7, 1:1, 1:1.2, 1:1.4, 1:1.7, 1:2, 1:2.4, 1:3, 1:3.3, 1:3.9 and 1:4.8; and those of protein:GpppA were 1:0 and 1:4.8. These 2D spectra were processed with the XWINNMR (Bruker, Rheinstetten, Germany) and NMRpipe software (33), and were analyzed with Sparky (47). Curve fitting for the determination of the dissociation constant (Kd) was achieved by using the software IGOR Pro (WaveMetrics, Inc., Lake Oswego, OR, USA).
For titration experiments of the cap-binding domain with a 10-mer poly(A) oligonucleotide, A10, the 2D 1H–15N HSQC spectra were acquired at 25°C with 15N, 13C-labeled cap-binding domain in a 90% H2O/10% D2O solution, containing 20 mM sodium phosphate buffer, pH 6.4, 100 mM NaCl and 1 mM d-DTT. Titrations were carried out using 0.2 mM cap-binding domain and various concentrations of unlabeled A10 to achieve a series of different cap-binding domain:A10 molar ratios (1:0, 1:0.1, 1:0.2, 1:0.8, 1:1 and 1:3).
Relaxation measurements
To determine the 15N relaxation rates (R1 and R2) and the steady-state 1H–15N NOE values of the cap-binding domain with and without the cap analog, enhanced-sensitivity experiments were performed using the standard method (48) on a Bruker AVANCE-600 spectrometer equipped with and without a cryoprobe at 25°C. R1 and R2 experiments were measured with relaxation delays of 5, 65, 145, 246, 366, 527, 757 and 1148 ms for R1, and 32, 48, 64, 80, 96, 112, 128 and 144 ms for R2. Duplicate spectra were recorded for delays of 5, 145, 366 and 757 ms (R1), and 32, 64, 96 and 128 ms (R2), to estimate the uncertainties of the peak heights. A recycle delay of 1 s was used for the measurement of the R1 and R2 data. For the steady-state 1H–15N NOE measurement, a relaxation delay of 3 s and a 1H presaturation time of 3 s were used in the NOE experiment, and a 6 s relaxation delay was used in the reference experiment. NMR peak heights determined by Sparky were used for the program ‘sparky2rate’ (http://xbeams.chem.yale.edu/~loria/software.htm). The R1 and R2 values were obtained by fitting the intensities of the peak heights as a function of the relaxation delay to a two-parameter, single exponential curve, using the nonlinear least-squares fitting program, CURVEFIT (A.G. Palmer, Columbia University). The uncertainties of the R1 and R2 values were estimated by means of Monte-Carlo simulations in the CURVEFIT program, based on the peak intensities from duplicated experiments. The steady-state NOE values were obtained from the ratio of the peak intensities in experiments with and without 1H saturation. Errors in NOE values were estimated using the root-mean-square of the background noise of each spectrum (48).
Isothermal titration calorimetry measurements
PARN RRM and the cap analogs (m7GpppG, GpppG and GpppA) were all dissolved in 20 mM sodium phosphate buffer (pH 6.0), 100 mM NaCl and were thoroughly degassed before use. The concentration of PARN RRM in the sample cell was 30 μM, and that of each cap analog was 375 μM.
Isothermal titration calorimetry (ITC) measurements were performed with a Microcal (Amherst, MA, USA) VP–ITC calorimeter. Measurements were made by titration of the cap analogs into the PARN RRM at a temperature of 25°C. For titration experiments, PARN RRM was placed in a 1.4 ml reaction cell. The m7GpppG, GpppG and GpppA were each loaded in the 250 μl injection syringe, and a series of 5 μl injections were made over 16 s, with a spacing of 180 s between injections over 120 min. The reference power was set to 20 μcal/s, and the stirring speed was 300 r.p.m. Parallel control experiments, to correct the heat of mixing, were performed by adding the cap analog to a sample cell containing only buffer, without PARN RRM. The heat generated due to dilution of the titrants was very small and was ignored for the analysis. The thermodynamic data were processed with the Microcal ORIGIN program to extract the enthalpic, entropic and equilibrium constants. Nonlinear least-squares fitting was done using a single site binding model.
RESULTS
Stable and functional cap-binding domain of PARN for structure determination
An amino acid sequence analysis predicted that PARN possesses the putative RNP1 and RNP2 sequences, which led us to assume that the region spanning residues 430–506 (numbering based on mouse PARN) adopts the RRM fold. On the basis of the domain boundary prediction, we designed six protein fragments and produced them by bacterial overexpression: residues 420–506, 420–516, 420–536, 430–506, 430–516 and 430–536 (Figure 1). Each fragment was purified using Ni2+ affinity chromatography (see Materials and methods section). The fragments spanning residues 420–506 and 430–506, which lacked the C-terminal regions, formed inclusion bodies (data not shown).
To select the active fragment for the cap analog, we conducted pull-down assays with the 420–516, 420–536, 430–516 and 430–536 fragments. The four fragments were incubated with m7G(5′)ppp-Sepharose, and the bound proteins were separated by SDS–PAGE (Figure 1C). The protein bands on the gel revealed that all of these fragments possessed binding activity for the cap analog. Therefore, the shortest fragment, spanning residues 430–516, was selected for structural studies of PARN.
Structure determination of the cap-binding domain
Unlabeled or 15N, 13C-labeled proteins corresponding to the cap-binding domain were synthesized by a cell-free protein expression system (26), and the proteins were purified by Ni2+-affinity and cation-exchange chromatography. The NMR experiments for the structure determination of the cap-binding domain were performed using standard techniques (27–29), as described in the Materials and methods section. Secondary structure elements were identified by the patterns of the characteristic backbone NOE connectivities and by the program TALOS (39) (see Materials and methods section). The solution structure was calculated using the CYANA program, based on 1266 NOE-derived inter-proton distance restraints, 14 hydrogen bonds and 143 dihedral angle restraints for the final calculation. Experimental restraints and structural statistics for the 20 lowest energy structures are summarized in Table 1, indicating that residues 440–509 adopted a well-defined structure, with an RMS deviation of 0.33 Å for the backbone atoms.
Table 1.
Structure statistics of free PARN RRM and the RRM:cap analog complex
| Free PARN RRM | RRM: cap complex | |
|---|---|---|
| Restraints for final structure calculations | ||
| Number of NOE distance restraints | 1266 | 1131 |
| Intraresidue | 372 | 348 |
| Sequential (|i−j| = 1) | 367 | 298 |
| Medium–range (1 < |i−j| < 5) | 178 | 154 |
| Long–range (|i−j| ≥ 5) | 349 | 314 |
| Protein–RNA intermolecular | NA | 17 |
| Number of dihedral angle restraintsa | ||
| φ angles | 61 | 42 |
| ψ angles | 58 | 41 |
| χ angles | 24 | 27 |
| Number of hydrogen bond restraintsb | 14 | 18 |
| Structure statistics (20 structures) | ||
| Mean number of NOE violations >0.2 Å | 0 | 0 |
| Mean number of dihedral angle violations >5° | 0 | 1.7 ± 1.5 |
| Average CYANA target function (Å2) | 0.01 | 0.08 |
| Average AMBER energy (kcal mol−1) | NA | −4785 ± 8 |
| RMS deviation to mean coordinatesc | ||
| Backbone heavy atoms (Å) | 0.33 ± 0.07 | 0.52 ± 0.11 |
| All heavy atoms (Å) | 0.74 ± 0.07 | 0.93 ± 0.14 |
| Ramachandran plot analysisd | ||
| Residues in most favored regions (%) | 89.2 | 89.2 |
| Residues in additionally allowed regions (%) | 10.8 | 10.5 |
| Residues in generously allowed regions (%) | 0.0 | 0.3 |
| Residues in disallowed regions (%) | 0.0 | 0.0 |
aφ and ψ angles were derived from the program TALOS. The χ angles contain 18 χ1 and 6 χ2 angle restraints for free PARN RRM and 19 χ1 and 8 χ2 angle restraints for the RRM:cap complex.
bOnly hydrogen bonds supported by NOEs were used.
cResidues 440–509 for free PARN RRM. Residues 440–509 and m7GpppG for the RRM:cap complex.
dFrom PROCHECK-NMR (46) for 20 structures. Residues 440–509 were used.
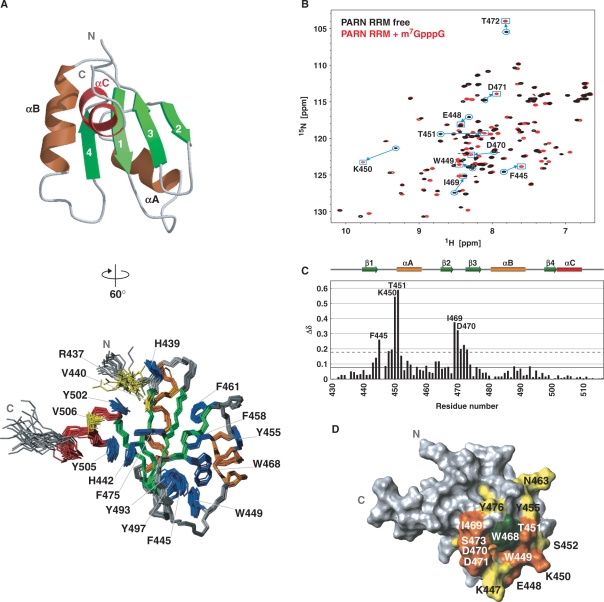
The structure of the cap-binding domain of PARN is composed of four β-strands, β1 (V440–T444), β2 (Q465–W468), β3 (S473–S477) and β4 (R498–T501), and three α-helices, αA (T451–F458), αB (P481–S491) and αC (Y502–K509). It forms a βαββαβα fold, with the first and second α-helices packed against one four-stranded antiparallel β-sheet (Figure 2A). This 3D structure confirmed that the cap-binding domain of PARN contains the RRM fold (the βαββαβ topology). In addition to the RRM fold, the cap-binding domain possesses an extra α-helical structure at its C-terminus (αC, residues 502–509), which we named PARN RRM. The canonical RRM has well-conserved consensus sequences, termed RNP1 and RNP2, which correspond to the β3- and β1-strands, respectively. Both RNP1 and RNP2 include aromatic residues with solvent-exposed side chains that participate in the RNA interaction (49–52). In the case of PARN RRM, however, the αC-helix lies on the RNA recognition surface and covers the β1-strand (Figure 2A). In particular, an aromatic residue within the αC-helix (Y505) is stacked with H442 in RNP2 by a π–π stacking interaction, and Y502 at the N-terminus of the αC-helix participates in a cation–π interaction with R437 from the N-terminal extension of PARN RRM (Figure 2A). Moreover, aliphatic residues [V440 (β1) and V506 (αC)] and aromatic residues [H442 (RNP2), F475 (RNP1), Y502 (αC) and Y505 (αC)] are involved in the hydrophobic interaction between the β-sheet surface and the additional αC-helix, resulting in the formation of the hydrophobic core by the residues on RNP1, RNP2 and the αC-helix (red circles in Figures 1A and 2A). Consequently, the aromatic rings on RNP1 and RNP2, which potentially contribute to the RNA interactions in the case of the canonical RRMs, are completely buried in the hydrophobic core, in the case of PARN RRM. On the other hand, one of the tryptophan residues, W468, which is conserved among the PARNs from several species, is located on the β2-strand. The bulky side chain of this residue is exposed to the solvent (Figure 2A). In addition, the region in the vicinity of W468 is partly stabilized by (i) the side chain of I469, which is not fully buried in the aforementioned hydrophobic core, but interacts with the aromatic rings of its central residues, F475, H442 and Y505; and (ii) a tight turn connecting β2 and β3, which comprises residues D470–S473. These two structural components, as well as the loop connecting β1 and αA, form the rim of a shallow basin. The indole ring of W468 resides on the bottom of the basin.
Figure 2.
3D structure of mouse PARN RRM and titration experiments of PARN RRM with the cap analog. (A) (Left) Ribbon representation of PARN RRM (residues 436–512). The secondary structure elements are labeled and colored green, orange and red for β-strands, αA and αB, and αC, respectively. (Right) Superposition of the 20 conformers with the lowest CYANA target function of PARN RRM (residues 436–512). Aromatic side chains and residues discussed in the text are labeled and colored blue and yellow, respectively. This structure is rotated by 60° around the y-axis in the left panel. (B) 1H–15N HSQC spectra of PARN RRM alone (black) and in the presence of the cap analog at a molar ratio of 1:5 (red). Resonances that shifted significantly are labeled with the residue number and the one-letter amino acid code. (C) Chemical shift changes of PARN RRM upon cap binding. The chemical shift change, ▵δ, was determined asΔ δ = [(Δ δHN)2 + (Δ δN / 6.5)2]1/2 (70), where ▵δHN and ▵δN are the chemical shift differences for HN and 15N, respectively. The mean value and the mean value + 1 SD are shown by continuous and dashed lines, respectively. (D) Mapping of residues with significant chemical shift changes upon cap binding on the PARN RRM structure, which is viewed from the same direction as in A, right panel. Residues with above-average chemical shift changes are colored yellow, and residues with chemical shift changes above the mean value +1 SD are colored orange. The aromatic side chain of W468 is colored green (see also Figure 3).
NMR titration experiments of PARN RRM with the cap analog
PARN reportedly interacts with the cap analog as well as the poly(A) tail (14,19–21). Our pull-down assays (Figure 1C) demonstrated that PARN RRM (residues 430–516 of mouse PARN) itself can interact with the cap analog. In order to identify the cap-binding sites and to investigate whether poly(A) interacts with PARN RRM, NMR titration experiments of PARN RRM were performed using the unlabeled cap analogs (m7GpppG, GpppG and GpppA), and the 10-mer unlabeled poly(A) oligonucleotide (A10). The 1H–15N HSQC spectra of PARN RRM with A10, at molar ratios of 1:0–1:3, revealed that none of the 1H–15N resonances were affected by the addition of A10 (Supplementary Figure S1). This result indicates that A10 does not interact with the PARN RRM fragment, spanning residues 430–516. On the other hand, the 1H–15N HSQC spectra of PARN RRM alone and in the presence of each cap analog at a 1:5 molar ratio revealed that some resonances were shifted by the addition of the cap analogs. The resonances of the PARN RRM residues F445, K450, T451, I469 and D470 underwent large chemical shift changes in the presence of m7GpppG (Figure 2B). The resonances of the same residues showed some chemical shift changes in the presence of GpppG and GpppA as well. However, the magnitudes of their chemical shift changes were much smaller than those for m7GpppG (Supplementary Figure S2). For example, the largest ▵δ value (= [(Δ δHN)2 + (Δ δN / 6.5)2]1/2) was 0.59 p.p.m. for T451 in the presence of m7GpppG, while the values were 0.064 and 0.095 p.p.m. in the presence of GpppG and GpppA, respectively. Even after m7GpppG was added to the solution containing PARN RRM and m7GpppG at a molar ratio of 1:1, to make a ratio of 1:5 in the course of the titration, no further shifting of the resonances of PARN RRM residues was observed. This result indicates that the m7GpppG-bound form of PARN RRM almost completely predominates over the free form after the addition of an equimolar amount of m7GpppG. This was not the case for the GpppG and GpppA titrations. In the latter cases, further shifting was observed upon increasing the protein:cap analog ratio to 1:5, and more shifting was likely to occur by further addition of the cap analogs. These observations are consistent with the thermodynamic parameters acquired by ITC, where we obtained a Kd value of ∼45 μM for the interaction between PARN RRM and m7GpppG. However, the Kd values for the interaction between PARN RRM and either GpppG or GpppA could not be determined under the same experimental conditions, probably due to the 10- to several 100-fold weaker Kd values (Supplementary Figure S3). Therefore, we concluded that the fragment spanning residues 430–516 indeed engages the cap analog, as identified by the pull-down assay (Figure 1C), and that the interaction is highly specific to the N7-methylated cap analog. In addition, our results also indicated that A10 could not bind to the cap-binding site on PARN RRM.
The resonance assignments of the PARN RRM in the complex were achieved by comparing the 3D 15N- and 13C-edited NOESY–HSQC spectra of free and cap-bound PARN RRM. The chemical shift changes in the backbone amide nitrogens and protons between the free and bound forms of PARN RRM were plotted versus the residue number (Figure 2C). Mapping of these shifted residues on the surface of PARN RRM revealed that significant chemical shift changes were observed for the residues in the loop between β1 and αA, and at the C-terminus of β2, which surround the tryptophan residue W468 in the tertiary structure (Figure 2D). This suggests that, as compared with the canonical RRMs, PARN RRM uses a distinct binding surface for the RRM–nucleotide interface. The dissociation constant, Kd, was calculated using the average of the estimated values for the side chain 1H resonances of the representative residues I466 and W468. The Kd value for cap analog binding by PARN RRM was approximately 20 μM with a stoichiometry of 1:1, which is roughly equivalent to the previously reported value (53).
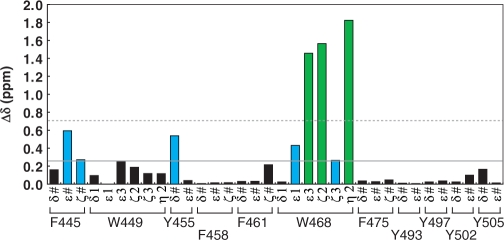
We next focused on aromatic residues, and carried out further NMR titration experiments to monitor the chemical shift changes in the side chain resonances. The 2D TOCSY spectra of the free and cap-bound forms of PARN RRM revealed that the resonances originating from W468 undergo the largest changes upon cap binding among the aromatic residues of PARN RRM (Supplementary Figure S4). The chemical shifts of the aromatic residues F445 and Y455 were also affected by the addition of the cap analog (Figure 3). These residues are located in the vicinity of W468 in PARN RRM, suggesting that W468 of PARN RRM directly participates in the recognition of the m7GpppG cap analog. In contrast, the aromatic residues within the β-sheet surface (F475) and those on the αC-helix (Y502 and Y505), as well as the other conserved tryptophan residue, W449, were scarcely shifted (black bars in Figure 3). As a consequence, PARN RRM uses the exposed tryptophan residue (W468) for the cap recognition, and the recognition is not mediated by the β-sheet surface of the canonical RRMs.
Figure 3.
NMR titration experiments focused on the resonances of aromatic side chains in PARN RRM at 25°C. The chemical shift changes, ▵δ, were obtained by calculating the absolute difference of the 1H chemical shifts in the absence and presence of an excess of the cap analog. Resonances of aromatic side chains with chemical shift changes above the mean value (continuous line) are colored cyan, and those with chemical shift changes above the mean value +1 SD (dashed line) are colored green.
Overall structure of the PARN RRM–cap complex
To solve the complex structure of PARN RRM with the m7GpppG cap analog, the resonances of the cap analog were assigned, using 2D 1H–13C HMQC, 2D TOCSY and 2D NOESY experiments. Cross peaks between the N7-methyl guanosine (m7G) moiety and the second guanine were observed in the NOESY spectrum (Supplementary Figure S5). Subsequently, several 2D and 3D NOESY-type experiments were performed, to assign the inter- and intra-molecular NOEs (see Materials and methods section). The structure of the PARN RRM in complex with the cap analog (hereafter referred to as the RRM:cap complex) was calculated, using the programs CYANA (36–38) and AMBER9 (http://amber.scripps.edu). Structural statistics for the 20 lowest energy structures are summarized in Table 1.
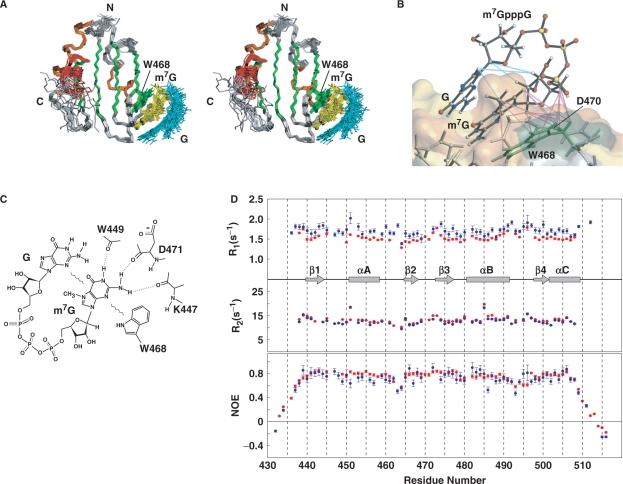
The overall structure of PARN RRM in the cap-bound form turned out to be the same as that of the cap-free form, especially in the region spanning residues 438–507 (Figures 2A and 4A). Consistent with the results of our NMR titration experiments, intermolecular NOEs between the m7G moiety of the cap analog and several amino acid residues of PARN RRM (W468, D470 and D471) were observed in the NOESY spectra (Supplementary Figure S6). The structure of the RRM:cap complex was determined from a total of 1241 NMR-derived restraints, including 1114 intramolecular NOEs, 17 inter-molecular NOEs and 110 dihedral angle restraints (Figure 4B and Table 1). The RRM:cap complex structure revealed that the interaction of PARN RRM with the cap analog is achieved mainly by π–π stacking interactions between the aromatic side chain of W468 of PARN RRM and the m7G base of the cap analog. In addition, intramolecular base-stacking interactions between the m7G and second guanosine bases seem to be present in the complex structure. The aromatic ring of W468, the base of m7G and that of the second guanosine were aligned nearly parallel to each other. In other words, the m7G base was sandwiched between the aromatic ring of W468 on the bottom and the second guanosine base on the top (Figure 4B). The structure of the RRM:cap complex also indicated possible hydrogen bonding interactions between the amino acid residues of PARN RRM in the vicinity of W468 and the bases of the cap analog (Figure 4C): the backbone carbonyl oxygen atoms of K447, W449 and D471 formed NH … O=C hydrogen bonds with N2, N1 and N2 of the m7G base, respectively. Therefore, the 3D architecture of the backbone atoms of these residues is also important for the cap-binding activity of PARN RRM.
Figure 4.
Structure of the PARN RRM:cap complex and NMR dynamics studies for PARN RRM in the presence and absence of the m7GpppG cap analog. (A) Stereo view illustrating a trace of the backbone atoms for the ensemble of the 20 structures of the PARN RRM:cap complex (residues 436–512). Residues in β-strands, αA and αB and αC are colored green, orange and red, respectively. The aromatic side chain of W468 is also depicted in green. The cap analog is colored yellow and cyan for m7Gp and GDP (ppG), respectively. (B) Enlarged view of the interface between PARN RRM and the cap analog. Intra- and inter-molecular NOEs that were observed in NOESY spectra and used as restraints in the structure determination are displayed with cyan and magenta lines, respectively. The surface of the PARN RRM is shown using the same color-code as in Figure 2D. (C) Diagram of the interactions between the m7GpppG cap analog and PARN RRM. Dashed lines indicate hydrogen bonds. Wavy lines symbolize stacking interactions. (D) NMR dynamics studies for PARN RRM with and without the cap analog. 15N R1 (top) and 15N R2 (middle) relaxation rates, and steady-state 1H–15N NOE values (bottom) are shown for free PARN RRM (blue) and the PARN:RRM complex (red). β-strands, helices, loops and turns are indicated schematically.
NMR dynamics studies
To investigate the local mobility of PARN RRM including the αC-helix, in the presence and absence of the cap analog, 2D 1H–15N NMR relaxation experiments were performed using a Bruker 600 MHz spectrometer at a probe temperature of 25°C. The 15N R1 and 15N R2 NMR relaxation rates and the steady-state 1H–15N NOE values for the backbone amides of PARN RRM in its free and complex forms were determined, and are plotted in Figure 4D. Overlapped resonances and those with very poor signal-to-noise ratios were excluded from the relaxation analysis. The average values of the 15N R1 and 15N R2 relaxation rates and the steady-state 1H–15N NOEs were calculated for residues 440–509, excluding the N- and C-terminal flexible regions. The average 15N R1 value was 1.7 s–1 for free PARN RRM. Upon the formation of the RRM:cap complex, the value decreased slightly to 1.5 s–1, as expected from the slight increase in the molecular mass, from 11 to 12 kDa. The 15N R2 values were similar for most of the residues in the free and complex forms (13 s−1) except for T451, which will be described subsequently. The average steady-state 1H–15N NOE values were 0.72 and 0.75 for free PARN RRM and the RRM:cap complex, respectively. The 15N R1 and 15N R2 relaxation rates and the steady-state 1H–15N NOE values of the αC-helix are similar to those of other secondary structure elements in PARN RRM (Figure 4D). Therefore, the entire PARN RRM including the αC-helix (residues 440–509) is stable and well-structured in solution, regardless of the presence of the cap analog.
Concerning the NMR relaxation rates for the amino acid residues located in the proximity of W468, several differences were observed between the free and cap-bound forms. Namely, the 15N R1 and 15N R2 relaxation rates for T451, which is located at the N-terminus of the αA-helix, were higher than the respective average values in the free form (Figure 4D). This indicates internal motions on the micro- to milli-second time scale for T451 in the free form. In the presence of the cap analog, however, the 15N R1 and 15N R2 relaxation rates for T451 were comparable to the average values, suggesting the suppression of internal motions. The 1H–15N resonances originating from residues D470, D471 and S473, which reside within the β2–β3 turn and gather on one side of W468 (Figure 4B), were severely overlapped with other resonances in the case of free PARN RRM. Thus, it was difficult to compare the parameters of the individual dynamics for these residues between the free and cap-bound forms. Inter- and intra-residue NOE connectivities originating from these residues could hardly be detected in the 15N-separated NOESY–HSQC spectrum of the free form. In contrast, in the case of the RRM:cap complex, these resonances were clearly separated in the 1H–15N HSQC spectra, and many inter- and intra-residue NOE connectivities were observed for these residues. Taken together, the NMR relaxation experiments and the NOE connectivities strongly suggest that the structure of the residues within the β2–β3 turn as well as at the N-terminus of the αA-helix, which are all located adjacent to W468, became less mobile upon engaging the cap.
Mutational analysis of PARN RRM
NMR titration experiments indicated that W468 was clearly affected upon cap binding, as shown above (Figure 3). Moreover, the PARN:cap structure suggested that some amino acid residues in the vicinity of W468 are also involved in the cap recognition. To further investigate the functionally important residues of PARN RRM, we constructed four mutant proteins, in which the surface-exposed residues ([0]K447A, K450A, W468L and D471A) were altered, based on our structural study. We then examined the cap-binding activities of these mutant proteins by a pull-down assay. After an incubation with m7G(5′)ppp-Sepharose, the bound proteins were detected by SDS–PAGE (Figure 5). The pull-down assay demonstrated that only the leucine substitution of W468 significantly decreased the interaction between PARN RRM and the cap analog, whereas the other mutations (K447A, K450A and D471A) displayed very little influence on the cap binding (Figure 5). The result indicated that the aromatic ring of W468 is directly responsible for the cap recognition and is a functionally critical residue for the cap-binding activity of PARN RRM. This conclusion coincides with those from our structural and NMR dynamics studies, which showed that W468 is located at the center of the cap-binding interface of PARN RRM. This is also supported by recent data independently obtained for the corresponding tryptophan residue of human PARN RRM (53).
Figure 5.
Pull-down assays of PARN RRM point mutants. The wild-type (WT) and mutant PARN RRM proteins were incubated with m7G(5′)ppp-Sepharose 4B beads, and the bound proteins were eluted by the addition of 2× SDS gel loading buffer. These supernatants were collected, and the proteins were fractionated by 15–25% SDS–PAGE and visualized by staining with Coomassie brilliant blue.
DISCUSSION
The stability of the αC-helix
Analyses of the C-terminal region from mouse PARN, using NMR structures, dynamics studies, and deletion mutagenesis, revealed that PARN RRM has unique structural features, as compared with the canonical RRM family members. First, we showed that the fragment spanning residues 430–516 is properly folded and sufficient to bind to the cap (Figure 1B), while two other fragments (residues 420–506 and 430–506) resulted in inclusion body formation in E. coli cells during protein production. These results led us to propose the important role of the αC-helix (residues 502–509) for the stability and/or solubility of PARN RRM. Actually, our structure of PARN RRM revealed that residues Y502, V506 and Y505 in the αC-helix participated in hydrophobic interactions with the hydrophobic residues on the β-sheet surface (red circles in Figure 1A). As a consequence, the αC-helix completely covers the β1-strand and prevents the canonical RNA binding surface of the RRM from interacting with other molecules (Figure 2A). Furthermore, we demonstrated that the PARN RRM with and without the cap analog is stable and well-structured, and that the position of the αC-helix remains unchanged upon cap analog binding (Figures 2A, 4A and D). NMR dynamics data of PARN RRM in the free and complex forms also revealed that the residues involved in the hydrophobic core formation between the αC-helix and the β-sheet are rigid in both forms. Therefore, we conclude that the hydrophobic residues within the αC-helix exert an important influence on the stability and/or the solubility of the entire PARN RRM, by hydrophobic interactions with the residues within the β-sheet surface.
Many RRM-containing proteins are known to bind with target RNA or protein molecules using the β-sheet surface of RRM and the sequences flanking RRM. One such example is the well-known RNA-binding splicing factor, U1A (54,55). U1A has an α-helical extension termed helix C, which is connected with β4 of the RRM by a short loop (six residues). This structural topology of U1A is similar to that of the PARN RRM, except that the αC-helix of the latter is connected immediately after β4 (Figure 1A). In the case of U1A in the free form, helix C lies across the RNA-binding β-sheet surface and covers a large part of it, to prevent nonspecific RNA from contacting the RNA-binding β-sheet surface. Upon binding to the target RNA, helix C is rearranged to interact with another hydrophobic patch outside the β-sheet surface. This rearrangement allows the amino acid residues on the β-sheet surface, helix C, the β4-helix C loop and the β2–β3 loop to cooperatively recognize the target RNA. Correspondingly, the NMR dynamics data for U1A, 15N T2 value (= 1/R2), of the residues in the two central strands of the β-sheet, within helix C, within the β4-helix C loop and within the β2–β3 loop indicated that these residues undergo conformational exchange processes (56). Importantly, the U1A construct that lacks helix C abolishes the RNA-binding activity, although it is fully soluble (57). Therefore, it is also considered that such a rearrangement of helix C, which regulates the RNA-binding mode of U1A, may also be used in the poly(A) tail recognition by PARN RRM. The interaction between the β-sheet and the αC-helix, however, is very strong in PARN RRM, on the basis of the relaxation experiments. Thus, in the case of PARN RRM, high energy may be needed to displace the αC-helix from the β-sheet surface, and significant stabilization must be achieved upon poly(A) engagement. Actually, our NMR titration experiments indicated that the 10-mer poly(A) oligonucleotide, A10, is not a good substrate to overcome such requirements and to rearrange the αC-helix (Supplementary Figure S1), although the dissociation constant for human PARN RRM and A10 binding was reportedly smaller than ∼50 nM (53). Thus, a longer poly(A) might be required to achieve stronger binding, since a 20-mer poly(A) reportedly binds to PARN RRM more strongly (53). Alternatively, additional poly(A) binding regions outside PARN RRM may be necessary to further stabilize the poly(A) binding.
Comparison of cap recognition between PARN RRM and CBP20
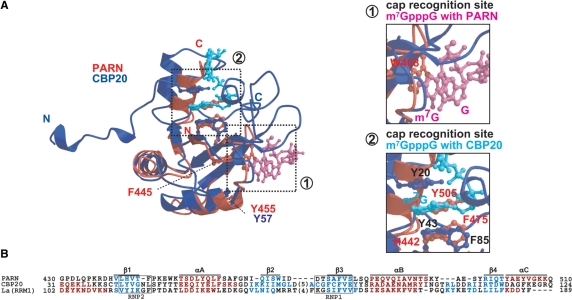
In the mammalian cell nucleus, the nuclear cap-binding complex (CBC), which consists of the CBP20 and CBP80 subunits, binds to the 5′ cap of mRNAs (58,59). Structural and binding studies of CBC with and without the cap analog m7GpppG showed that the CBP20 subunit adopts the classical RRM fold (the βαββαβ topology) and binds directly to the cap in the presence of the CBP80 subunit (59–61). We performed a structural comparison between our PARN RRM (βαββαβα) and CBP20 (βαββαβ) (60,61), to assess the similarity between their βαββαβ regions. As shown in Figure 6, the cap recognition site has also been identified in CBP20 (61), i.e. on the β-sheet surface, which corresponds to the nucleotide binding site of the canonical RRMs (52). In the structure of CBP20, the m7G base is sandwiched between two tyrosine residues, Y43 on the β-sheet surface and Y20 in the N-terminal extension (60,61). Surprisingly, PARN RRM has a distinctive cap recognition site, with respect to the βαββαβ topology (Figures 4A and 6A). On the β-sheet surface of PARN RRM, the aromatic ring of H442 (corresponding to Y43 in CBP20) stacks with the phenol ring of Y505 on the characteristic αC-helix by π–π stacking interactions (Figures 2A and 6A), as described in the Results. A comparison between the two cap-bound structures of PARN RRM and CBP20 revealed that the aromatic ring of Y505 on the αC-helix in PARN RRM occupies the position corresponding to that of the m7G base in the CBP20-cap complex, even in the presence of the cap analog (Figure 6A). Interestingly, the Y505 residue is highly conserved among several species of PARN RRMs (Figure 1A). This suggests that the stacking between Y505 and H442 is a common feature of PARN RRMs. This stacking prevents the aromatic residues on the β-sheet surface of PARN RRM from interacting with the cap.
Figure 6.
Structural comparison and sequence alignment between RRMs. (A) A superposition of the 3D structures of PARN RRM (red) with the m7GpppG cap analog (magenta), and the CBP20 complex (blue) with the m7GpppG cap analog (cyan) (PDB ID 1H2T). Side chains discussed in the text are labeled. The right panel shows an enlarged view of the boxed area in the left panel. (B) Structure-based sequence alignment between the RRMs of CBP20, La protein and PARN. The well-conserved consensus sequences, RNP1 and RNP2, are boxed. Residues involved in β-strands and α-helices are highlighted in cyan and red, respectively. The secondary structure elements observed in PARN RRM are shown at the top of the alignment.
The cap-binding specificity of PARN RRM
The specific binding of the cap to W468 of PARN RRM was confirmed by the mutation W468L, which indicated that this tryptophan residue plays a central role in the cap recognition (Figure 5). The other mutant proteins, K447A, K450A and D471A, displayed scarcely any influence on the interaction [0]with the cap, despite the fact that these mutated residues are located in the vicinity of W468 (Figure 5). Thus, changing the functional groups of the side chains of these residues did not affect the cap binding by PARN RRM. These results for the mutational analysis, combined with the description of possible hydrogen bonding interactions in the PARN RRM:cap complex structure (Figure 4C), clearly explained how the cap recognition is achieved. Indeed, K447 in the β1–αA loop and D471 in the β2–β3 turn are directly involved in the recognition of the cap analog in the complex structure. However, they utilized their backbone carbonyl oxygen atoms for the recognition, instead of the functional groups of their side chains. Therefore, the mutation of these two residues displayed no detectable effect on the cap-binding activities. The backbone and side chain of K450 lacked specific interactions with the cap. In addition to the cases of K447 in the β1–αA loop and D471 in the β2–β3 turn, the backbone atoms of W449 in the β1–αA loop also participate in the cap binding (Figure 4C). Our NMR dynamics data revealed that the internal motions of the amino acid residues in the β2–β3 turn as well as those at the N-terminus of the αA-helix, which are all located adjacent to W468, were suppressed by the cap binding (Figure 4D). This observation indicates that the residues in the β1–αA loop, including the two residues K447 and W449 mentioned above, play an auxiliary role in the cap binding. Therefore, given that the 3D architecture of the backbone atoms in the β1–αA loop and the β2–β3 turn is formed by these residues, we conclude that the favorable 3D geometries of the loop and the turn are also important for the specific recognition of the cap by PARN RRM. Notably, the lengths of the β1–αA loop and the β2–β3 turn of PARN RRM are significantly shorter than those of the canonical RRMs (Figure 6B). This structural feature probably ensures the backbone stability by reducing the amplitude of the backbone motions, thereby facilitating the specific interaction of PARN RRM with the cap analog.
Full-length PARN reportedly deadenylates the m7GpppG-capped RNA better than the nonmethylated GpppG- or GpppA-capped RNAs (19–21). Our NMR titration and ITC data showed that PARN RRM has stronger affinity for the m7GpppG than for the GpppG and GpppA cap analogs. Therefore, we concluded that the specific recognition of m7G by PARN is indeed attributable to PARN RRM. These data revealed the importance of the methylation. Interestingly, the intermolecular hydrogen bond network between PARN RRM and the m7G, which were determined by our structural study, could be formed by nonmethylated guanine as well. Therefore, how could the m7GpppG be discriminated from the others? One would expect that the methyl group is surrounded by many atoms and is involved in van der Waals interactions with them. However, our NOE data revealed that only W468 is in the proximity of m7G (Supplementary Figure S6), and the structure showed that there is a π–π stacking interaction between W468 and m7G (Figure 4B). It is known that the methylation of guanine results in electron deficiency of the guanine base π-orbitals, which favorably stack with the electron-rich π-orbitals of the tryptophan indole (62). Thus, the methylation of the guanine base is required to reinforce the π–π stacking interaction. Taken together, our results suggest that the stabilization of the π–π stacking interaction is the main driving force of m7GpppG recognition.
The Kd values between m7GpppG and PARN RRM, determined by our NMR titration and ITC experiments, were in the micromolar to several tens of micromolar range. These values are coincident with those obtained by previous fluorescence titration experiments (53). The m7GpppG cap structure is substantially recognized by CBC in the nucleus and by eIF4E in the cytoplasm. These Kd values are ∼10 nM for CBC and 200 nM for eIF4E (60). Thus, the order of the affinities for m7GpppG is: CBC > eIF4E > PARN RRM. As described in the previous section and in Figure 6, CBP20, a subunit of CBC, adopts a similar RRM fold to that of PARN RRM. However, CBP20 uses a canonical RNA-binding surface for strong binding to the m7GpppG cap. Intriguingly, considering the regulatory roles of PARN, it is assumed that PARN RRM accommodates the m7GpppG cap on the unique binding surface on the RRM fold in order to weaken the affinity for the cap, as compared with the binding by CBP20 and eIF4E.
Unique structural features of RRMs
Our NMR studies of PARN RRM revealed that the aromatic side chain of W468, which forms the cap-binding center, protrudes from the β2 strand into the space between the β2-strand and the αA-helix (Figure 2A). On the other hand, the canonical RRM structures, such as CBP20 (61), U1A (63,64), sex-lethal protein (65), hnRNP D0 (66), Musashi1 (67) and PABP (68), do not have a tryptophan residue at the position equivalent to W468 (Figure 6B). The PARN RRM contains highly conserved, bulky aromatic residues (F445 and W449) (Figure 1A), and their side chains form the hydrophobic core of the protein in the immediate vicinity of W468 (Figure 2A). In most RRMs, however, the positions corresponding to F445 and W449 in PARN RRM are occupied by conserved aliphatic residues, which also contribute to the formation of the hydrophobic core, e.g. a leucine residue at the position of F445 (52) [the exception is the RRM1 of La protein, which has phenylalanine at the corresponding position (69)]. Consequently, since the side chains of F445 and W449 of PARN RRM are bulkier than those of aliphatic residues, these aromatic side chains might invade the space where W468 would be located. Therefore, W468 protrudes into the solvent, leading to the unique interaction with the cap analog in PARN RRM.
We have determined a vast number of RRM structures (over 90 structures), which have been deposited in the Protein Data Bank, as part of the structural genomics project at RIKEN. Among them, the structure of an RRM from the methenyltetrahydrofolate synthetase domain-containing protein (MTHFSD, PDB ID 2E5J), which adopts the canonical RRM fold, contains a tryptophan residue at the C-terminal end of the β2-strand, the position equivalent to that of W468 in PARN RRM (Supplementary Figure S7). This tryptophan residue of MTHFSD RRM, however, was unable to engage a nucleotide by a π–π stacking interaction, as in the case of PARN RRM, since the indole ring of the tryptophan is completely covered by a hydrophobic residue located at the N-terminus of the αA-helix, and therefore is buried in the hydrophobic core. This indicates that the existence of a tryptophan residue at the sequence position equivalent to that of W468 in PARN RRM does not necessarily mean that it is the site for nucleotide engagement. It seems that the use of the indole ring of the corresponding tryptophan residue for cap recognition is unique to the PARN RRM, and that a tryptophan at the equivalent position in the canonical RRMs would not be used for nucleotide-binding, but for hydrophobic core formation.
The N-terminal region of PARN, consisting of the exonuclease and R3H domains, is known to form a homodimer that is directly involved in poly(A) engagement (14,23). Our findings provide new information regarding the cap recognition by PARN RRM. At the moment, however, the functional significance of the duplicated cap-binding abilities and the exonuclease activities remains unclear. It will be important to study the full-length PARN protein to clarify its mechanisms in the quality control and the translational regulation of mRNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank Mr Masato Aoshima, Mr Toshihiko Ogawa, Mr Takeshi Nagira, Ms. Yasuko Tomo, Dr Satoru Watanabe and Mr Takushi Harada for preparing the PARN RRM proteins. This work was supported by the RIKEN Structural Genomics/Proteomics Initiative (RSGI), the National Project on Protein Structural and Functional Analysis, Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 2.Maquat LE, Carmichael GG. Quality control of mRNA function. Cell. 2001;104:173–176. doi: 10.1016/s0092-8674(01)00202-1. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell P, Tollervey D. mRNA turnover. Curr. Opin. Cell Biol. 2001;13:320–325. doi: 10.1016/s0955-0674(00)00214-3. [DOI] [PubMed] [Google Scholar]
- 4.Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- 5.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 6.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 7.von der Haar T, Gross JD, Wagner G, McCarthy JE. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat. Struct. Mol. Biol. 2004;11:503–511. doi: 10.1038/nsmb779. [DOI] [PubMed] [Google Scholar]
- 8.Coller J, Parker R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell P, Tollervey D. mRNA stability in eukaryotes. Curr. Opin. Genet. Dev. 2000;10:193–198. doi: 10.1016/s0959-437x(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 10.Åström J, Åström A, Virtanen A. In vitro deadenylation of mammalian mRNA by a HeLa cell 3′ exonuclease. EMBO J. 1991;10:3067–3071. doi: 10.1002/j.1460-2075.1991.tb07858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Åström J, Åström A, Virtanen A. Properties of a HeLa cell 3′ exonuclease specific for degrading poly(A) tails of mammalian mRNA. J. Biol. Chem. 1992;267:18154–18159. [PubMed] [Google Scholar]
- 12.Körner CG, Wormington M, Muckenthaler M, Schneider S, Dehlin E, Wahle E. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J. 1998;17:5427–5437. doi: 10.1093/emboj/17.18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Körner CG, Wahle E. Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J. Biol. Chem. 1997;272:10448–10456. doi: 10.1074/jbc.272.16.10448. [DOI] [PubMed] [Google Scholar]
- 14.Martínez J, Ren YG, Thuresson AC, Hellman U, Åström J, Virtanen A. A 54-kDa fragment of the Poly(A)-specific ribonuclease is an oligomeric, processive, and cap-interacting Poly(A)-specific 3′ exonuclease. J. Biol. Chem. 2000;275:24222–24230. doi: 10.1074/jbc.M001705200. [DOI] [PubMed] [Google Scholar]
- 15.Chiba Y, Johnson MA, Lidder P, Vogel JT, van Erp H, Green PJ. AtPARN is an essential poly(A) ribonuclease in Arabidopsis. Gene. 2004;328:95–102. doi: 10.1016/j.gene.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 16.Copeland PR, Wormington M. The mechanism and regulation of deadenylation: identification and characterization of Xenopus PARN. RNA. 2001;7:875–886. doi: 10.1017/s1355838201010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JH, Richter JD. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell. 2006;24:173–183. doi: 10.1016/j.molcel.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 18.Ren YG, Kirsebom LA, Virtanen A. Coordination of divalent metal ions in the active site of poly(A)-specific ribonuclease. J. Biol. Chem. 2004;279:48702–48706. doi: 10.1074/jbc.M403858200. [DOI] [PubMed] [Google Scholar]
- 19.Dehlin E, Wormington M, Körner CG, Wahle E. Cap-dependent deadenylation of mRNA. EMBO J. 2000;19:1079–1086. doi: 10.1093/emboj/19.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao M, Fritz DT, Ford LP, Wilusz J. Interaction between a poly(A)-specific ribonuclease and the 5′ cap influences mRNA deadenylation rates in vitro. Mol. Cell. 2000;5:479–488. doi: 10.1016/s1097-2765(00)80442-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez J, Ren YG, Nilsson P, Ehrenberg M, Virtanen A. The mRNA cap structure stimulates rate of poly(A) removal and amplifies processivity of degradation. J. Biol. Chem. 2001;276:27923–27929. doi: 10.1074/jbc.M102270200. [DOI] [PubMed] [Google Scholar]
- 22.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu M, Reuter M, Lilie H, Liu Y, Wahle E, Song H. Structural insight into poly(A) binding and catalytic mechanism of human PARN. EMBO J. 2005;24:4082–4093. doi: 10.1038/sj.emboj.7600869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liepinsh E, Leonchiks A, Sharipo A, Guignard L, Otting G. Solution structure of the R3H domain from human Sμbp-2. J. Mol. Biol. 2003;326:217–223. doi: 10.1016/s0022-2836(02)01381-5. [DOI] [PubMed] [Google Scholar]
- 25.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 26.Kigawa T, Yabuki T, Matsuda N, Matsuda T, Nakajima R, Tanaka A, Yokoyama S. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J. Struct. Funct. Genomics. 2004;5:63–68. doi: 10.1023/B:JSFG.0000029204.57846.7d. [DOI] [PubMed] [Google Scholar]
- 27.Clore GM, Gronenborn AM. Determining the structures of large proteins and protein complexes by NMR. Trends Biotechnol. 1998;16:22–34. doi: 10.1016/S0167-7799(97)01135-9. [DOI] [PubMed] [Google Scholar]
- 28.Bax A. Multidimensional nuclear magnetic resonance methods for protein studies. Curr. Opin. Struct. Biol. 1994;4:738–744. [Google Scholar]
- 29.Cavanaph J, Fairbrother WJ, Palmer AGI, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. San Diego, CA, USA: Academic Press; 1996. [Google Scholar]
- 30.Clowes RT, Crawford A, Raine AR, Smith BO, Laue ED. Improved methods for structural studies of proteins using nuclear magnetic resonance spectroscopy. Curr. Opin. Biotechnol. 1995;6:81–88. doi: 10.1016/0958-1669(95)80013-1. [DOI] [PubMed] [Google Scholar]
- 31.Iwahara J, Wojciak JM, Clubb RT. Improved NMR spectra of a protein-DNA complex through rational mutagenesis and the application of a sensitivity optimized isotope-filtered NOESY experiment. J. Biomol. NMR. 2001;19:231–241. doi: 10.1023/a:1011296112710. [DOI] [PubMed] [Google Scholar]
- 32.Zwahlen C, Legault P, Vincent SJF, Greenblatt J, Konrat R, Kay LE. Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: Application to a bacteriophage l N-peptide/boxB RNA complex. J. Am. Chem. Soc. 1997;119:6711–6721. [Google Scholar]
- 33.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi N, Iwahara J, Koshiba S, Tomizawa T, Tochio N, Güntert P, Kigawa T, Yokoyama S. KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high-throughput NMR structure studies. J. Biomol. NMR. 2007;39:31–52. doi: 10.1007/s10858-007-9175-5. [DOI] [PubMed] [Google Scholar]
- 35.Johnson BA, Blevins RA. NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 36.Güntert P. Automated NMR structure calculation with CYANA. Meth. Mol. Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 37.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 38.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 39.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 40.Powers R, Garrett DS, March CJ, Frieden EA, Gronenborn AM, Clore GM. The high-resolution, three-dimensional solution structure of human interleukin-4 determined by multidimensional heteronuclear magnetic resonance spectroscopy. Biochemistry. 1993;32:6744–6762. doi: 10.1021/bi00077a030. [DOI] [PubMed] [Google Scholar]
- 41.Güntert P, Braun W, Wüthrich K. Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. J. Mol. Biol. 1991;217:517–530. doi: 10.1016/0022-2836(91)90754-t. [DOI] [PubMed] [Google Scholar]
- 42.Machida M, Yokoyama S, Matsuzawa H, Miyazawa T, Ohta T. Allosteric effect of fructose 1,6-bisphosphate on the conformation of NAD+ as bound to L-lactate dehydrogenase from Thermus caldophilus GK24. J. Biol. Chem. 1985;260:16143–16147. [PubMed] [Google Scholar]
- 43.Koide S, Yokoyama S, Matsuzawa H, Miyazawa T, Ohta T. Conformation of NAD+ bound to allosteric L-lactate dehydrogenase activated by chemical modification. J. Biol. Chem. 1989;264:8676–8679. [PubMed] [Google Scholar]
- 44.Xia B, Tsui V, Case DA, Dyson HJ, Wright PE. Comparison of protein solution structures refined by molecular dynamics simulation in vacuum, with a generalized Born model, and with explicit water. J. Biomol. NMR. 2002;22:317–331. doi: 10.1023/a:1014929925008. [DOI] [PubMed] [Google Scholar]
- 45.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 46.Laskowski RA, Rullmann JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 47.Goddard TD, Kneller DG. SPARKY 3. San Francisco: University of California; 2006. [Google Scholar]
- 48.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 49.Dreyfuss G, Swanson MS, Pinol-Roma S. Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation. Trends Biochem. Sci. 1988;13:86–91. doi: 10.1016/0968-0004(88)90046-1. [DOI] [PubMed] [Google Scholar]
- 50.Adam SA, Nakagawa T, Swanson MS, Woodruff TK, Dreyfuss G. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol. Cell Biol. 1986;6:2932–2943. doi: 10.1128/mcb.6.8.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swanson MS, Nakagawa TY, LeVan K, Dreyfuss G. Primary structure of human nuclear ribonucleoprotein particle C proteins: conservation of sequence and domain structures in heterogeneous nuclear RNA, mRNA, and pre-rRNA-binding proteins. Mol. Cell Biol. 1987;7:1731–1739. doi: 10.1128/mcb.7.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 53.Nilsson P, Henriksson N, Niedzwiecka A, Balatsos NA, Kokkoris K, Eriksson J, Virtanen A. A multifunctional RNA recognition motif (RRM) in poly(A)-specific ribonuclease (PARN) with cap and poly(A) binding properties. J. Biol. Chem. 2007;282:32902–32911. doi: 10.1074/jbc.M702375200. [DOI] [PubMed] [Google Scholar]
- 54.Avis J, Allain F.H.-T, Howe PWA, Varani G, Neuhaus D, Nagai K. Solution structure of the N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding. J. Mol. Biol. 1996;257:398–411. doi: 10.1006/jmbi.1996.0171. [DOI] [PubMed] [Google Scholar]
- 55.Allain F-HT, Gubser CC, Howe PWA, Nagai K, Neuhaus D, Varani G. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formation. Nature. 1996;380:646–650. doi: 10.1038/380646a0. [DOI] [PubMed] [Google Scholar]
- 56.Mittermaier A, Kay LE, Forman-Kay JD. Analysis of deuterium relaxation-derived methyl axis order parameters and correlation with local structure. J. Biomol. NMR. 1999;13:181–185. doi: 10.1023/A:1008387715167. [DOI] [PubMed] [Google Scholar]
- 57.Scherly D, Kambach C, Boelens W, van Venrooij WJ, Mattaj IW. Conserved amino acid residues within and outside of the N-terminal ribonucleoprotein involved in U1 RNA binding. J. Mol. Biol. 1991;219:577–584. doi: 10.1016/0022-2836(91)90651-l. [DOI] [PubMed] [Google Scholar]
- 58.Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 59.Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj IW. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 60.Calero G, Wilson KF, Ly T, Rios-Steiner JL, Clardy JC, Cerione RA. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat. Struct. Biol. 2002;9:912–917. doi: 10.1038/nsb874. [DOI] [PubMed] [Google Scholar]
- 61.Mazza C, Segref A, Mattaj IW, Cusack S. Large-scale induced fit recognition of an m7GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 2002;21:5548–5557. doi: 10.1093/emboj/cdf538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ishida T, Doi M, Ueda H, Inoue M, Sheldrick GM. Specific ring stacking interaction on the tryptophan-7-methylguanine system: comparative crystallographic studies of indole derivatives-7-methylguanine base, nucleoside, nucleotide complexes. J. Am. Chem. Soc. 1988;110:2286–2294. [Google Scholar]
- 63.Allain FH, Howe PW, Neuhaus D, Varani G. Structural basis of the RNA-binding specificity of human U1A protein. EMBO J. 1997;16:5764–5772. doi: 10.1093/emboj/16.18.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagai K, Oubridge C, Jessen TH, Li J, Evans PR. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990;348:515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- 65.Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999;398:579–585. doi: 10.1038/19242. [DOI] [PubMed] [Google Scholar]
- 66.Nagata T, Kurihara Y, Matsuda G, Saeki J, Kohno T, Yanagida Y, Ishikawa F, Uesugi S, Katahira M. Structure and interactions with RNA of the N-terminal UUAG-specific RNA-binding domain of hnRNP D0. J. Mol. Biol. 1999;287:221–237. doi: 10.1006/jmbi.1999.2616. [DOI] [PubMed] [Google Scholar]
- 67.Nagata T, Kanno R, Kurihara Y, Uesugi S, Imai T, Sakakibara S, Okano H, Katahira M. Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1. J. Mol. Biol. 1999;287:315–330. doi: 10.1006/jmbi.1999.2596. [DOI] [PubMed] [Google Scholar]
- 68.Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 69.Teplova M, Yuan YR, Phan AT, Malinina L, Ilin S, Teplov A, Patel DJ. Structural basis for recognition and sequestration of UUUOH 3′ termini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol. Cell. 2006;21:75–85. doi: 10.1016/j.molcel.2005.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mulder FA, Schipper D, Bott R, Boelens R. Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J. Mol. Biol. 1999;292:111–123. doi: 10.1006/jmbi.1999.3034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.