Abstract
Gene targeting refers to the alteration of a specific DNA sequence in an endogenous gene at its original locus in the genome by homologous recombination. Through a gene-targeting procedure with positive–negative selection, we previously reported the generation of fertile transgenic rice plants with a positive marker inserted into the Adh2 gene by using an Agrobacterium-mediated transformation vector containing the positive marker flanked by two 6-kb homologous segments for recombination. We describe here that base changes within the homologous segments in the vector could be efficiently transferred into the corresponding genomic sequences of rice recombinants. Interestingly, a few sequences from the host genome were flanked by the changed sequences derived from the vector in most of the recombinants. Because a single-stranded T-DNA molecule in Agrobacterium-mediated transformation is imported into the plant nucleus and becomes double-stranded, both single-stranded and double-stranded T-DNA intermediates can serve in gene-targeting processes. Several alternative models, including the occurrence of the mismatch correction of heteroduplex molecules formed between the genomic DNA and either a single-stranded or double-stranded T-DNA intermediate, are compared to explain the observation, and implications for the modification of endogenous genes for functional genomic analysis by gene targeting are discussed.
INTRODUCTION
Gene targeting (GT) refers to the alteration of a specific DNA sequence in an endogenous gene at its original locus in the genome by homologous recombination (HR) and, often, to the conversion of the endogenous gene into a designed sequence, i.e. base changes or gene disruptions through gene replacements. Various approaches to GT in flowering plants have been attempted with only limited success (1–8). Plants with the anticipated base changes in the PPO and ALS genes were reproducibly isolated by selecting acquired herbicide resistance (9,10). Transgenic plants with targeted gene disruptions by the insertion of either the selectable hpt gene or the visually screenable GFP gene were repeatedly obtained by their expected phenotypes (11–13). In each of these GT events, Agrobacterium-mediated transformation was used. Two consecutive crossovers are thought to occur at the flanking homologous regions of the selective/screenable sequences on the T-DNA-based vectors and their corresponding genomic regions. While a non-selective base change (silent mutation) was introduced into the PPO and ALS genes adjacent to the specific base changes conferring herbicide resistance in order to distinguish HR from spontaneous mutagenesis events (9,10), no systematic analysis of non-selective base changes at the flanking homologous segments for crossovers in HR has been reported.
We have developed a reproducible GT procedure with positive–negative selection and succeeded in obtaining fertile transgenic rice plants with the Waxy or Adh2 gene disrupted by the insertion of the hpt gene (11,12). In the vectors used for the transformation of embryogenic rice calli by Agrobacterium, the hpt gene was flanked by the Waxy or Adh2 sequences of 6.0–6.8 kb, and two inversely oriented DT-A genes fused with strong and constitutive promoters were placed at both ends of the T-DNA segment adjacent to its border sequences in order to eliminate the overwhelming random integration events (e.g. Adh2; Figure 1). No polymorphic alterations were detected between the Waxy segments in the vector derived from cv. Shimokita and those in the genome of cv. Nipponbare (11). When we used the vector pJHYAd2 (12), in which the homologous Adh2 segments were found to carry multiple base changes, including a newly created KpnI site (Figure 2A), six of seven targeted transgenic plants examined were previously found to carry the KpnI site (12). The observation prompted us to characterize the Adh2 homologous regions in the genome of these targeted transgenic rice plants, and we were able to show the efficient transfer of base changes (point mutations) from the vector to the rice genome. Moreover, a few host genomic sequences were found to be flanked by the point mutations derived from the vector in most of recombinants characterized. A likely explanation for the observation would be a result of the mismatch correction of heteroduplex molecules formed between the genomic and introduced T-DNA sequences. The results offer new insights into the HR processes of GT with positive–negative selection.
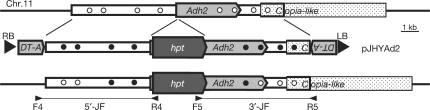
Figure 1.
Detection of the transfer of base changes from a vector to the rice genome in the Adh2 locus. The rice Adh2 locus on chromosome 11 contains a Copia-like retroelement. The vector pJHYAd2 contains the 6.2-kb Adh2 promoter sequence, including a 0.1-kb 5′-untranslated region, the hpt gene for hygromycin B resistance, the 4.0-kb Adh2 region and the 2.0 kb 3′ part of a Copia-like retroelement. The DT-A genes encoding the diphtheria toxin A fragments, which are not drawn to scale, were placed as lethal negative-selection markers at both ends of T-DNA in order to efficiently eliminate transformants carrying entire T-DNA segment(s) (7,12). The large black arrowheads with RB and LB indicate the right and left borders of T-DNA, respectively. The targeted double crossovers at the homologous segments led to the integration of hpt into the endogenous Adh2 sequence. The horizontal lines flanked by small arrowheads under the map represent the 5′- and 3′-junction fragments (5′-JF and 3′-JF) produced only from the disrupted adh2::hpt alleles by PCR amplification. The filled and open circles indicate the base changes in pJHYAd2 and the corresponding genomic sequences, respectively. Homologous crossover sites can be deduced by determining these alternative sequences in the targeted adh2::hpt alleles.
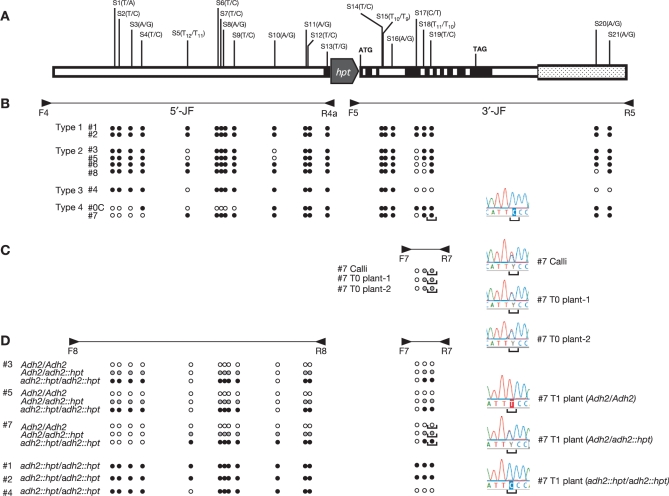
Figure 2.
Base changes transferred from a vector to the rice genome and their transmission into the next generation. (A) Base changes found at the rice Adh2 locus of the recombinants obtained. Twenty-one base-changed sites (S1–S21) were indicated in the Adh2 homologous segments in pJHYAd2. In each site, base changes are indicated as (genome/vector). The black boxes indicate Adh2 exons. Neither hpt nor DT-A is drawn to scale. (B) Base changes of the Adh2 locus found at the targeted adh2::hpt alleles. Base changes, as determined by sequencing the 5′- and 3′-JFs in the heterozygously targeted T0 plants indicated or the #0C callus, are shown under the map. The filled and open circles are as in Figure 1. The sequencing chromatogram shows the sequence containing the base change (indicated by the brackets) at S19 of T0 plant #7 in Type 4. (C) A typical example of the heterozygously targeted callus and T0 plants displaying a mixture of the wild-type and base-changed alleles. The 1.1-kb DNA segments amplified with primers F7 and R7 in #7 recombinants line (Type 4) are a mixture of two fragments that differ from each other at S18 and S19, the latter of which is shown in the sequencing chromatogram. The grey circles indicate a mixture of the wild-type and changed nucleotides. (D) Segregation of the base changes in the selfed progeny of the heterozygously targeted T0 plants. Sequencing of the DNA segments of 6.2 kb amplified with primers F8 and R8 and those of 1.1 kb with primers F7 and R7 from lines #3, #5 and #7 bearing the Adh2/Adh2, Adh2/adh2::hpt and adh2::hpt/adh2::hpt alleles, previously determined by Southern blot analysis (12), revealed that anticipated wild-type or changed nucleotide sequences, indicated by the filled, grey, and open circles as well as the sequencing chromatograms represented by S19. In addition, the base changes in lines #1, #2 and #4 carrying the adh2::hpt/adh2::hpt allele are also shown.
MATERIALS AND METHODS
Sequencing of flanking homologous Adh2 segments
The vector pJHYAd2, the targeted transformed calli, and their transgenic rice plants were described previously (12). Sequencing of the homologous Adh2 segments in pJHYAd2 was performed with the appropriate primers, as described previously (11,12). PCR amplification of the 3′-junction fragment (3′-JF) produced by HR in either targeted transformed calli or targeted transgenic plants (Figure 1) was carried out with primers F5 and R5 (12). Because 5′-JF amplified with primers F4 and R4 (Figure 1) had tended to be insufficient for further analysis, we modified PCR amplification by using PrimeSTAR GXL Taq polymerase (Takara Bio, Ohtsu, Japan) with primers F4 and R4a (5′-TTGGGCCACCTTTTATTACCG-3′) to obtain sufficient 5′-JF (Figure 2B). The cycles of the PCR reactions were as follows: initial denaturation (94°C for 1 min), 15 cycles of denaturation (98°C for 10 s), annealing and extension (66°C for 15 min) and then 19 cycles of denaturation (98°C for 10 s), annealing and extension (66°C for 15 min with the autoextension feature to add 15 s per cycle), and final extension (72°C for 10 min). To avoid detecting PCR artifacts present as a minor fraction in the obtained PCR-amplified fragments, direct sequencing of the PCR-amplified fragments without cloning was performed with the appropriate primers (11,12). For callus #0C, which was accidentally lost during regeneration (12), the genomic sequence was determined in the remaining callus DNA. In addition, the 1.1-kb DNA segment containing S17, S18 and S19 in transformed calli, the primary regenerated T0 plants, and their selfed progeny was amplified using Ex Taq polymerase (Takara Bio) with primers F7 (5′-CCTAACAATCAAATTGATCTAACAAA-3′) and R7 (5′-TCAATTTGTTACTATCATGTTTCATG-3′) for sequencing (Figure 2C): initial denaturation (94°C for 1 min), 30 cycles of denaturation (94°C for 30 s), annealing (60°C for 30 s), extension (72°C for 90 s) and final extension (72°C for 10 min). To confirm that the base changes detected by directly sequencing 5′-JF in T0 plants were in the heterozygous condition and segregated into their selfed progenies, the 6.2-kb segment of the Adh2 promoter region in the T1 segregants was amplified using LA Taq polymerase (Takara Bio) with primers F8 (5′-GAAGTTTATTTACAAAGCCGAGCAAAGAAG-3′) and R8 (5′-ATCCCCCTCTTTTTCAAAGAACAAGAAACC-3′) for sequencing (Figure 2D): initial denaturation (94°C for 1 min), 15 cycles of denaturation (98°C for 10 s), annealing and extension (62°C for 15 min), and then 19 cycles of denaturation (98°C for 10 s), annealing and extension (62°C for 15 min with the autoextension feature to add 15 s per cycle) and final extension (72°C for 10 min).
To gain additional confirmation that the point mutations introduced into the T0-targeted plants were transmitted stably into the next generation, T1 plants with the homozygous adh2::hpt/adh2::hpt alleles were isolated: one plant from the line #1, one plant from the line #2 and two plants from the line #4. Because these homozygous plants would give unambiguous results concerning the adh2::hpt allele, we sequenced the 1.1- and 6.2-kb regions PCR-amplified with the primer sets F7/R7 and F8/R8, respectively.
RESULTS AND DISCUSSION
Base changes on the vector pJHYAd2
The 6.2-kb fragment containing the Adh2 promoter as well as the 6.0-kb fragment containing the Adh2 coding sequence and its adjacent region including the 3′ part of a highly repetitive Copia-like retroelement in pJHYAd2 (Figure 1) was originally prepared from the cv. Nipponbare genome by PCR amplification and was cloned into the backbone vector pINA134 (12). The 6.2-kb Adh2 promoter region in pJHYAd2 was found to contain 13 PCR-induced base changes consisting of 12 single-base substitutions and one single-base deletion (Figure 2A). One of them, the T-to-C transition at S4, generated a new KpnI site. This KpnI site was previously shown to be transferred from pJHYAd2 into the genomes of eight targeted transformants by Southern blot analysis (12). The 6.0-kb Adh2 coding region bore eight base changes with six single-base substitutions and two single-base deletions, and two out of six single-base substitutions resided within the Copia-like retroelement (Figure 2A).
Base changes in the genome of the targeted recombinants
Using the vector pJHYAd2, we previously reported that nine independent transformed calli carrying the anticipated Adh2 disruption (Figure 1) were isolated with a frequency of 1.9 × 10–2 per surviving callus with positive–negative selection and that eight fertile transgenic T0 plants (#1–#8) without ectopic events were subsequently obtained (12). Southern blot analysis revealed all of them to be heterozygotes with a wild-type Adh2 allele and a disrupted adh2 allele by the integration of hpt by HR (Ahd2/adh2::hpt) (12). To determine the sequences of the 6.2- and 6.0-kb homologous segments, we directly sequenced the PCR-amplified junction fragments, 5′- and 3′-JFs, generated by HR. Because these JFs are produced only from the disrupted adh2::hpt alleles, the sequences of the adh2::hpt alleles can be determined even in the heterozygous condition. According to the distribution patterns of the base changes, nine targeted recombinants were classified into four types (Figure 2B). Two recombinants in Type 1 showed that all base changes, together with the selective hpt marker on pJHYAd2, were transferred to the genome, indicating that one crossover occurred to the left of S1 within the 1.4-kb segment and another to the right of S21 only within the 0.4-kb segment in the Copia-like element. Crossovers also occurred at both termini of the homologous region in four recombinants in Type 2. However, not only the base changes from the vector but also the plant genomic sequences were detected, which could not be explained by two simple crossovers. One recombinant #4 in Type 3 must have been generated by one crossover to the left of S1 and another between S16 and S17. The remaining two recombinants in Type 4 must also have been generated through processes similar to those in Type 2, and their right crossovers were likely to be within the 0.4-kb region to the right of S21, whereas the left crossover was between S4 and S5 in recombinant #7 and between S3 and S4 in recombinant #0C. The results clearly show that, in our GT system, point mutations can be efficiently transferred from the vector to the rice genome by HR, a notion also supported by the results of targeted recombinants modified in one of two rice DDM1 genes, OsDDM1a (Os09g0442700) (7, Y. Johzuka-Hisatomi, unpublished data).
Although the junction fragments 3′-JFs were generated only from the disrupted adh2::hpt alleles in either targeted calli or their transgenic T0 plants in the heterozygous condition, such heterozygosity can be seen by sequencing the PCR-amplified fragments produced from the wild-type Adh2 and mutant adh2::hpt alleles together. Figure 2C shows such examples in targeted callus #7 and its two transgenic T0 plants obtained through multiple shoots (11,12). The 1.1-kb PCR-amplified fragments with primers F7 and R7 were anticipated to consist of equal amounts of amplified fragments derived from either the wild-type Adh2 or the mutant adh2::hpt allele. Indeed, direct sequencing of the 1.1-kb PCR-amplified fragments obtained from callus #7 and its T0 plants revealed mixtures of the two alleles indicated by the grey circles at S18 and S19 as well as sequencing chromatograms at S19, whereas that of the 3′-JF from callus #7 or its T0 plants showed only the mutant allele indicated by the filled circles at S18 and S19 as well as a sequencing chromatogram at S19 (Figure 2B and C).
The base changes in the fertile T0 heterogenotes must have been co-segregated with the disrupted adh2::hpt mutation in the selfed T1 progeny. We showed previously that the adh2::hpt mutation in T0 heterozygous lines #3, #5 and #7 was transmitted into the next generation in a Mendellian fashion and that the genomic structures of the segregated T1 plants with the homozygous Adh2/Adh2 alleles, the heterozygous Adh2/adh2::hpt alleles, and the homogygous adh2::hpt/adh2::hpt alleles were confirmed by Southern blot analysis (12). We chose these T1 segregants and examined the base changes in the segregants by direct sequencing of the 6.2-kb PCR-amplified fragment with primers F8 and R8 and the 1.1-kb fragment with primers F7 and R7 (Figure 2D). As expected, plants with the homozygous Adh2/Adh2 alleles and those with the homozygous adh2::hpt/adh2::hpt alleles bore the wild-type sequences and base changes, respectively, as indicated by the respective filled and open circles as well as sequencing chromatograms at S19, whereas those with the heterozygous Adh2/adh2::hpt alleles carried base changes heterozygously, as indicated by the grey circles as well as a sequencing chromatogram at S19. Since the analyzed lines #3 and #5 are the Type 2 recombinants and the line #7 is the Type 4 recombinant, we also examined two homozygous adh2::hpt/adh2::hpt T1 plants #1 and #2 in Type 1 and one such T1 plant #4 in Type 3 (Figure 2D). The results further confirmed that the targeted alleles adh2::hpt in the T0 heterozygous Adh2/adh2::hpt plants were segregated into the homozygous adh2::hpt/adh2::hpt T1 plants. We can thus conclude that the base changes introduced by GT are stably transmitted to the next generation.
T-DNA integrations promoted by non-homologous end-joining and homologous recombination
In Agrobacterium-mediated transformation, T-DNA appears to integrate randomly throughout the plant genome as a single molecule or multiple sequences ligated with each other in various orientations, and T-DNA integration is mediated by non-homologous end-joining (NHEJ) processes (14). It has been well-established that a single-stranded T-DNA molecule is imported into the plant nucleus and becomes double-stranded (15). While the integration of multiple T-DNA sequences is thought to be mediated via double-stranded T-DNA intermediates (16), intermediates for the integration of single-copy T-DNA remain debatable (14). The majority of the randomly integrated single-copy T-DNA molecules are known to contain the entire T-DNA segment with a well-conserved right-border and a left-border sequence that is either conserved or truncated by a few to around 100 bp (17,18). In our gene-targeting procedure with positive–negative selection, the two constitutively expressed DT-A genes at both ends of the T-DNA segment adjacent to its border sequences acted effectively to eliminate transformants carrying entire T-DNA segment(s) integrated randomly and enrich targeted transformants among the surviving calli (7,12). Because the gene carried by T-DNA is expressed rapidly soon after T-DNA becomes double-stranded before integration (19), the transient expression of the double-stranded DT-A gene is thought to kill host plant cells (1,5,20). It is thus assumed that, in the calli that lead to successful GT in our rice system, the complete DT-A gene coding regions with their promoters in the introduced vector must neither become double-stranded nor be transiently expressed before integration (5,20). Thus, either single-stranded or truncated double-stranded T-DNA molecules without the DT-A sequence must serve as intermediates for successful GT (Figures 3 and 4). In the former case, single-stranded T-DNA intermediates are likely to be coated with numerous VirE2 proteins (14,15); it would be interesting to examine whether these VirE2 proteins affect the GT processes. In the latter case, such truncated double-stranded DNA likely can be produced by random priming initiated at a homologous segment between the positive and negative selection markers. The DT-A gene region at the 3′ terminus of the imported single-stranded T-DNA remains single-stranded and is eventually removed by introducing a nick at the homologous single-stranded part. Alternatively, the single-stranded nick is introduced first, and then random priming starts at the remaining homologous segment. Before the DT-A gene at the 5′ terminus of the imported single-stranded T-DNA becomes double-stranded, another nick is introduced to remove the 5′ DT-A gene, which results in the truncated double-stranded T-DNA without the DT-A sequence (5).
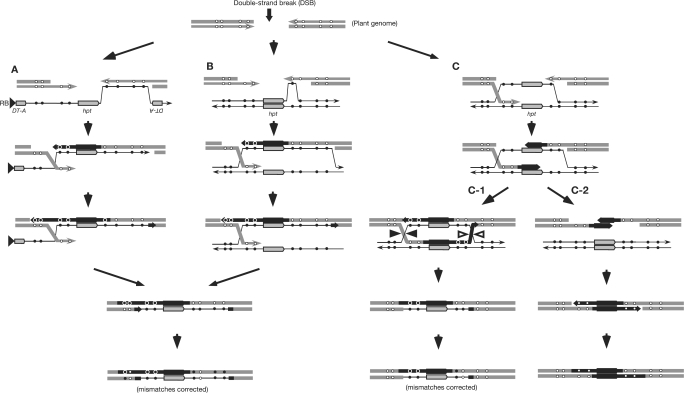
Figure 3.
DSB-induced GT processes with a single-stranded T-DNA intermediate (A) and a truncated double-stranded T-DNA intermediate (B and C). The 5′ ends at the targeted DSB sites in the plant genome are recessed, and one of the resulting 3′-ended single strands can anneal with a homologous single-stranded T-DNA sequence (A) or invade a truncated homologous double-stranded T-DNA region (B) to initiate new DNA synthesis to copy the hpt gene and its flanking region. The heteroduplex region may be expanded by annealing (A) or branch migration (B). The 3′ part of the newly synthesized strand then hybridizes with the homologous region at the opposite side of the break, as postulated by SDSA models (30). The subsequent DNA synthesis in both strands and ligation results in non-crossover TGT recombinants. In successful GT with positive–negative selection, the entire DT-A gene regions with their promoters at the ends of T-DNA in the introduced vector must neither become double-stranded nor express transiently (see text). Alternatively, both 3′-ended single strands from the targeted DSB sites in the plant genome may invade truncated homologous double-stranded T-DNA regions, and the heteroduplex regions may be expanded by branch migration (C). New DNA syntheses are initiated to copy the hpt gene region followed by the formal DSBR model (C-1) or the simple SDSA model (C-2), as described previously (29). Note that only one of two alternative resolutions of the double Holliday structure in the formal DSBR model is illustrated. The horizontal thick grey and thin black lines indicate the single-stranded plant and T-DNA strands, respectively, and their arrowheads represent their 3′ ends. The thick black arrows indicate newly synthesized strands. The filled and open circles represent the base changes in each strand of pJHYAd2 and the corresponding genomic sequences, respectively. The configurations of the DNA strand in (A and B) are drawn in conformity with those in the formal DSBR or simple SDSA models (C). A mismatch correction must occur in the resulting heteroduplex regions.
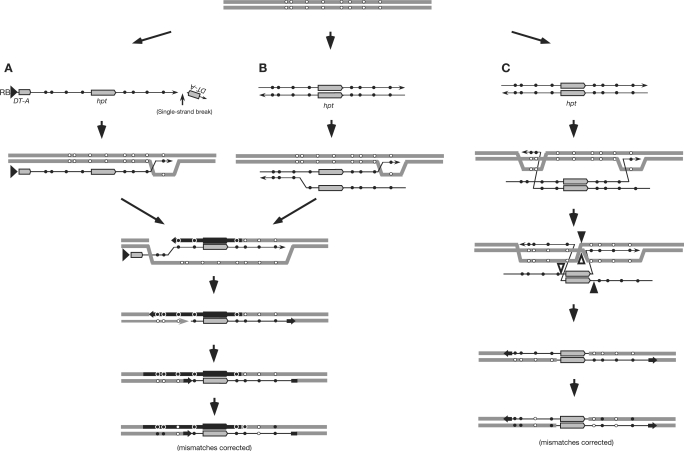
Figure 4.
DSB-independent GT processes with a single-stranded T-DNA intermediate (A) and with a truncated double-stranded T-DNA intermediate (B and C). One-end invading SDSA models (A and B). The broken 3′ end of the single-stranded T-DNA without the DT-A sequence (A) or one of the double strands of the T-DNA intermediate without the DT-A region (B) invades and hybridizes with the homologous plant genomic DNA, and branch migration occurs to form a heteroduplex region. Subsequently, the hybridized plant DNA is nicked and elongated to copy the single-stranded T-DNA template including the hpt gene region, and the synthesized strand anneals with the complementary plant DNA strand. As in Figure 3, mismatch correction occurs in the heteroduplex regions, and non-crossover TGT recombinants are produced. In a two-end invasion model (C), two 3′ ends of the double-stranded DNA may invade the homologous genomic DNA region and the subsequent formation of long heteroduplexes at the flanking homologous regions of the selective hpt gene without DNA synthesis, as postulated by Li et al. (40). Cleavages and ligations at the sites indicated by the open arrowheads as well as those at the sites indicated by the filled arrowheads result in non-crossover TGT recombinants, and a mismatch correction must occur in the heteroduplex regions. Symbols are as in Figure 3.
It is known that base sequence divergence, including base substitutions in the homologous segments of a vector introduced by electroporation into mouse embryonic stem (ES) cells, reduces the GT frequency drastically (21,22). Similarly, base sequence divergence (single base substitutions) between homologous segments led to a significant reduction in the frequency of somatic intrachromosomal recombination in transgenic Arabidopsis plants (23,24). It is thus intriguing whether the introduction of 13 and 8 base changes in the 6.2- and 6.0-kb Adh2 homologous segments of pJHYAd2, respectively (Figure 2A), would cause a significant reduction in the GT frequency, which was reported to be 1.9 × 10−2 per surviving callus (12). Unfortunately, we do not know the GT frequency of Adh2 with no base alterations because all of the available data concerning the GT frequency of Adh2 were obtained in experiments using pJHYAd2. Since a comparable GT frequency of 9.4 × 10–3 per surviving callus was previously obtained in the Waxy locus, in which the vector used carried 6.3- and 6.8-kb Waxy homologous segments without any base changes (11), it is tempting to speculate that the GT frequency of Adh2 with no base alterations may not drastically differ from that with the 21 base changes: 1.9 × 10–2 per surviving callus (12). Because double-stranded DNA molecules appeared to serve as intermediates for both GT in mouse ES cells and intrachromosomal HR in Arabidopsis (21–24), the question of whether the reduction of the HR frequencies caused by base sequence divergence has anything to do with the involvements of double-stranded DNA intermediates could be asked.
Chromosomal double-strand break-initiated gene-targeting processes
The introduction of DSB in the plant genome greatly stimulates the integration of transgenes flanked by homologous sequences on T-DNA introduced into the DSB site, and both anticipated true gene targeting (TGT), generated by double homologous crossovers and one-sided invasion (OSI), resulting from one homologous crossover and another NHEJ at the target locus, were observed (25–28). To account for this observation, DSB-induced synthesis-dependent strand annealing (SDSA) models (29,30) have been proposed: one of the 3′ single-stranded ends of the genomic DNA at the DSB site serves as a primer to initiate DNA synthesis with either single-stranded T-DNA (Figure 3A) (25) or double-stranded T-DNA (Figure 3B) (26) as a template. When the newly synthesized DNA molecule can anneal to the 3′ overhanging homologous sequence on the opposite side of the plant DNA at the DSB site, the resulting recombinant is TGT. If the newly synthesized DNA molecule failed to anneal with another homologous end, however, NHEJ at the second crossover would result in OSI. In the simple SDSA model (29), both of the 3′ single-stranded ends of the genomic DNA at the DSB site can invade homologous regions of the truncated double-stranded T-DNA intermediates to initiate DNA synthesis [Figure 3(C-2)]. Subsequently, newly synthesized DNA molecules are displaced from the template and anneal to each other, and the continuous DNA synthesis and ligation result in TGT. In the formal double-strand break repair (DSBR) model for the generation of non-crossover TGT recombinants [Figure 3(C-1)] (29), the continuous DNA synthesis and ligation result in two Holliday junctions, which can be resolved by cleaving and ligating to yield non-crossover TGT.
We postulated in Figure 2 that apparent double crossovers in the major GT events occur at both ends of the homologous segments; two crossovers occur >10 kb apart, outside of S1 and S21, in Types 1 and 2 recombinants. In each case shown in Figure 2, DNA synthesis of long T-DNA sequences including the hpt gene and subsequent incorporation into the genome are postulated to be involved in producing TGT; therefore, gene conversion events in Type 1 are explainable. For the recombinants in Types 2, 3 and 4, a few sequences from the host genome are flanked by the sequences derived from pJHYAd2, and such an arrangement can be found in both the 5′- and the 3′-JF regions (Figure 2). Generation of the recombinants with these arrangements can be explained by the mismatch correction of heteroduplex molecules formed between the genomic and introduced T-DNA sequences (29–31). If all mismatches in the heteroduplexes formed were corrected into the vector T-DNA sequences, the recombinants in Type 1 could also be produced. The results for recombinants #3, #5, #8 and #0C (Figure 2B) indicate that the formation of long heteroduplexes, rather than the initiation of DNA synthesis, can often occur, and such heteroduplexes must be formed either by hybridization between two single-stranded molecules or by branch migration after invasion of the 3′ single-stranded end into a double-stranded homologous region (Figure 3). Since the hpt gene and its flanking regions in the recombinants must be copied from the T-DNA template, the observation that the hpt-adjacent region (S11–S16) in each recombinant is originated from the vector sequence may be the result of such DNA synthesis. The proposed models in Figure 3 suggest that regions near the crossovers tend to form heteroduplexes, although we cannot determine the actual crossover sites in the individual recombinants obtained in Figure 2B if the occurrence of the heteroduplex formation and subsequent mismatch corrections are considered. It is known that the T:G mismatch that arose through deamination of the 5-methylcytosine residue tended to be corrected to C:G (32). The C:G pair at S17 originated from the genomic sequence was observed in five recombinants (#3, #5, #6, #7 and #8) while its flanking sequences were derived from the vector (Figure 2A). If the T:G mismatch at S17 occurred in the heteroduplex, it would be efficiently corrected to C:G unless the mismatch was resolved by replication. Therefore, the observation at S17 appears to be consistent with the notion that mismatch correction occurred at S17.
As the point mutations carried by a vector were efficiently transferred into the rice genome, gene conversion was observed over the entire region examined (S1–S21), including the hpt gene in Types 1 and 2 and at long tracts of the vector sequence in Type 3 (S1–S16) or in Type 4 (S4/5–S21). This observation appears to be in contrast to the fact that major DSB-induced conversion tracts were reported to be bidirectional and usually shorter than 100–300 bp when double-stranded DNA molecules served in HR with the genomic homologous sequences in yeast (33), Chinese hamster ovary (CHO) cells (34) and mouse ES cells (35). Since DSB-induced targeted integration of a double-stranded transgene with a homologous sequence introduced by particle bombardment in maize was reported very recently (36), it is intriguing to ask whether similar short and bidirectional tracts could be detected in most homologous recombinants obtained by these experiments in plants. It would also be interesting to examine whether such short and bidirectional tracts could be seen when a chromosomal DSB was introduced at a predetermined site in the Agrobacterium-mediated rice gene targeting system with positive–negative selection. According to the models illustrated, no heteroduplex would form in the simple SDSA model involved in the annealing of the newly synthesized DNA strands [Figure 3(C-2)], whereas a long heteroduplex would likely be produced in only one of the two homologous regions flanking the hpt gene in the formal DSBR model [Figure 3(C-1)]. Thus, neither the simple SDSA model nor the formal DSBR model appears to account for the generation of the Type 2 recombinants in Figure 2B.
While we have postulated double crossovers and subsequent heteroduplex formation in Figure 3, the possibility of the occurrence of multiple crossovers without heteroduplex formation could not be completely excluded. For example, template switching during break-induced replication (BIR) initiated by only one-ended DSB was shown to occur in yeast, and such template-switching events were postulated to take place by multiple rounds of strand invasion into the homologous double-stranded DNA region followed by DNA synthesis and dissociation from the template (37). Because it has also been postulated that BIR is suppressed when DSB has two 3′ ends (37) and because the conservative gene conversion processes described in Figure 3 are likely be more frequent events than template switching under the two-ended DSB condition (37), we regarded such a template-switching model to be plausible but less likely to explain the observation in Figure 2A.
Chromosomal double-strand break-independent gene-targeting processes
Several DSB-independent SDSA models for T-DNA-associated HR may account for the efficient transfer of base changes along with the long heteroduplex formation and mismatch correction (Figure 4). One model is a single-stranded T-DNA intermediate model: the 3′ end of the single-stranded T-DNA without the DT-A sequence invades and hybridizes with the homologous double-stranded genomic DNA, and new DNA synthesis is initiated from the nicked genomic DNA to copy the single-stranded T-DNA template (Figure 4A) (20). This model is derived from a microhomology-initiated model for T-DNA integration (17,18), and the 3′ end of the single-stranded T-DNA invades the region with complete homology (instead of microhomology) in the genome. A long heteroduplex can be formed by branch migration rather than DNA synthesis after the invasion of the 3′ terminus of single-stranded T-DNA without the DT-A sequence. Before branch migration stops at the hpt sequence, where heterology begins between the genomic and T-DNA sequences, the hybridized plant DNA is nicked and elongated to copy the T-DNA sequence including the hpt gene region. Before the DT-A sequence near the right border becomes double-stranded and the DT-A gene is expressed transiently, the newly synthesized DNA is displaced from the template T-DNA and annealed to the complementary 3′ overhanging genomic DNA strand in the manner shown in Figure 3A. The processes result in TGT, and two long heteroduplex tracts are formed at both hpt flanking regions.
In yeast, one strand of the transforming double-stranded DNA segment was reported to be assimilated into the homologous genomic sequence to form a long heteroduplex subjected to mismatch correction (38). Although the details of the molecular processes remain obscure, the phenomena appeared to occur only in certain mismatch repair-defective mutants, and the transforming double-stranded DNA molecules used contained no large central heterologous segment, as noted by Longston and Symington (39). Nevertheless, a possible pathway for the assimilation of a single-stranded DNA would be similar to that for the invasion of a single-stranded T-DNA intermediate (Figure 4A and B). In wild-type yeast using a transforming double-stranded DNA molecule with a large heterologous selective marker gene in the middle of the molecule, invasion of two 3′ ends of the double-stranded DNA molecule into the homologous regions of the genomic DNA was found to be the dominant process for GT (Figure 4C), and the assimilation of single-stranded DNA was a minor pathway (39). Comparable results, which have been interpreted as the invasion of two 3′ ends of the double-stranded DNA into the homologous genomic DNA region and the subsequent formation of long heteroduplexes at the flanking homologous regions of a selective marker gene, were also reported in a mouse hybridoma cell line (40), as illustrated in Figure 4C. In this model, no DNA synthesis was postulated to occur during HR processes, and cleavages and ligations near both ends of the heterologous hpt sequence yielded TGT. It remains to be studied whether the presence of two copies of the negative marker gene DT-A affects the alternative pathways in Figure 4B and C.
Because a single-stranded T-DNA molecule is imported into the plant nucleus and becomes double-stranded (15), we have presented and discussed several possible models in which intermediates for successful GT are either single-stranded or double-stranded T-DNA molecules (Figure 3 and 4). While, among the models described here, one of the simplest processes involved in TGT formation appears to be the model for the invasion of a single-stranded T-DNA intermediate (Figure 4A), both single- and double-stranded T-DNA molecules may serve as intermediates in parallel in our gene-targeting system (5). However, we do not yet know which pathway serves as the major pathway for successful gene targeting with positive–negative selection in rice calli.
Implications for rice functional genomic studies
Almost all of the obtained primary transgenic plants with the targeted modifications by our procedure with positive–negative selection were found to carry only one copy of the transgene with the anticipated structure in the heterozygous condition, and no associated ectopic events could be detected (7,8,11,12). We have shown that point mutations carried by the vector were efficiently transferred into the rice genome (Figure 2B). Subsequent elimination of the positive hpt marker by either a site-specific recombinase or a transposase (41,42) could feasibly generate transgenic rice plants containing point mutations without a positive-selection marker. Such ‘clean’ knock-in mutations devoid of selection markers through the elimination of the selection markers using site-specific recombinases have been generated in transgenic mice (43–45), and an important genetic trait conferred by a single-point mutation in mouse was obtained in this way (46). We also found that a few host genomic sequences are flanked by the point mutations derived from the vector in most of the recombinants characterized, which can be explained by the mismatch correction of heteroduplex molecules formed between the genomic and introduced T-DNA sequences. The observation may facilitate the development of localized mutagenesis within only the homologous regions.
ACKNOWLEDGEMENTS
We thank Miwako Matsumoto for technical assistance and Kyoko Takagi for reading the manuscript. This work was supported by grants from the Ministry of Agriculture, Forestry and Fisheries of Japan (Green Technology Project IP1007), the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) from Bio-oriented Technology Research Advancement Institution (BRAIN) in Japan. Y. J.-H. is a recipient of a fellowship from the Japan Society for the Promotion of Science (JSPS) for Young Scientists. Funding to pay the Open Access publication charges for this article was provided by JSPS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Morton R, Hooykaas PJJ. Gene replacement. Mol. Breed. 1995;1:123–132. [Google Scholar]
- 2.Britt AB, May GD. Re-engineering plant gene targeting. Trends Plant Sci. 2003;8:90–95. doi: 10.1016/S1360-1385(03)00002-5. [DOI] [PubMed] [Google Scholar]
- 3.Hanin M, Paszkowski J. Plant genome modification by homologous recombination. Curr. Opin. Plant Biol. 2003;6:157–162. doi: 10.1016/s1369-5266(03)00016-5. [DOI] [PubMed] [Google Scholar]
- 4.Reiss B. Homologous recombination and gene targeting in plant cells. Int. Rev. Cytol. 2003;228:85–139. doi: 10.1016/s0074-7696(03)28003-7. [DOI] [PubMed] [Google Scholar]
- 5.Iida S, Terada R. Modification of endogenous natural genes by gene targeting in rice and other higher plants. Plant Mol. Biol. 2005;59:205–219. doi: 10.1007/s11103-005-2162-x. [DOI] [PubMed] [Google Scholar]
- 6.Tzfira T, White C. Towards targeted mutagenesis and gene replacement in plant. Trends Biotechnol. 2005;23:567–569. doi: 10.1016/j.tibtech.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Iida S, Johzuka-Hisatomi Y, Terada R. Gene targeting by homologous recombination for rice functional genomics. In: Upadhyaya N, editor. Rice Functional Genomics - Challenges, Progress and Prospects. New York, NY: Springer; 2007. pp. 273–289. [Google Scholar]
- 8.Johzuka-Hisatomi Y, Maekawa M, Takagi K, Eun C.-H, Yamauchi T, Shimatani Z, Ahmed N, Urawa H, Tsugane K, Terada R, et al. Homologous recombination-dependent gene targeting and an active DNA transposon nDart-promoted gene tagging for rice functional genomics. In: Hirai A, Sano Y, Hirano H, Sasaki T, editors. Rice Biology in the Genomics Era: Biotechnology in Agriculture and Forestry. Vol. 62. New York, NY: Springer; 2008. pp. 81–94. [Google Scholar]
- 9.Hanin M, Volrath S, Bogucki A, Briker M, Ward E, Paszkowski J. Gene targeting in Arabidopsis. Plant J. 2001;28:671–677. doi: 10.1046/j.1365-313x.2001.01183.x. [DOI] [PubMed] [Google Scholar]
- 10.Endo M, Osakabe K, Ono K, Handa H, Shimizu T, Toki S. Molecular breeding of a novel herbicide-tolerant rice by gene targeting. Plant J. 2007;52:157–166. doi: 10.1111/j.1365-313X.2007.03230.x. [DOI] [PubMed] [Google Scholar]
- 11.Terada R, Urawa H, Inagaki Y, Tsugane K, Iida S. Efficient gene targeting by homologous recombination in rice. Nat. Biotechnol. 2002;20:1030–1034. doi: 10.1038/nbt737. [DOI] [PubMed] [Google Scholar]
- 12.Terada R, Johzuka-Hisatomi Y, Saitoh M, Asao H, Iida S. Gene targeting by homologous recombination as a biotechnological tool for rice functional genomics. Plant Physiol. 2007;144:846–856. doi: 10.1104/pp.107.095992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaked H, Melamed-Bessudo C, Levy AA. High-frequency gene targeting in Arabidopsis plants expressing the yeast RAD54 gene. Proc. Natl Acad. Sci. USA. 2005;102:12265–12269. doi: 10.1073/pnas.0502601102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tzfira T, Li J, Lacroix B, Citovsky V. Agrobacterium T-DNA integration: molecules and models. Trends Genet. 2004;20:375–383. doi: 10.1016/j.tig.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Tzfira T, Citovsky V. Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Curr. Opin. Biotechnol. 2006;17:147–154. doi: 10.1016/j.copbio.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 16.De Buck S, Jacobs A, Van Montagu M, Depicker A. The DNA sequences of T-DNA junctions suggest that complex T-DNA loci are formed by a recombination process resembling T-DNA integration. Plant J. 1999;20:295–304. doi: 10.1046/j.1365-313x.1999.t01-1-00602.x. [DOI] [PubMed] [Google Scholar]
- 17.Tinland B, Hohn B. Recombination between prokaryotic and eukaryotic DNA: integration of Agrobacterium tumefaciens T-DNA into the plant genome. Genet. Eng. 1995;17:209–229. [PubMed] [Google Scholar]
- 18.Brunaud V, Balzergue S, Dubreucq B, Aubourg S, Samson F, Chauvin S, Bechtold N, Cruaud C, DeRose R, Pelletier G, et al. T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertion sites. EMBO Rep. 2002;3:1152–1157. doi: 10.1093/embo-reports/kvf237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narasimhulu SB, Deng X.-B, Sarria R, Gelvin SB. Early transcription of Agrobactertium T-DNA genes in tobacco and maize. Plant Cell. 1996;8:873–886. doi: 10.1105/tpc.8.5.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iida S, Terada R. A tale of two integrations, transgene and T-DNA: gene targeting by homologous recombination in rice. Curr. Opin. Biotechnol. 2004;15:132–138. doi: 10.1016/j.copbio.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 21.Deng C, Capecci MR. Reexamination of gene targeting frequency as a function of extent of homology between the targeting vector and the target locus. Mol. Cell Biol. 1992;12:3365–3371. doi: 10.1128/mcb.12.8.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.te Riele H, Maandag ER, Berns A. Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc. Natl Acad. Sci. USA. 1992;89:5128–5132. doi: 10.1073/pnas.89.11.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Santerre-Ayotte S, Boivin EB, Jean M, Belzile F. A novel reporter for intrachromosomal homoeologous recombination in Arabidopsis thaliana. Plant J. 2004;40:1007–1015. doi: 10.1111/j.1365-313X.2004.02270.x. [DOI] [PubMed] [Google Scholar]
- 24.Opperman R, Emmanuel E, Levy AA. The effect of sequence divergence on recombination between direct repeats in Arabidopsis. Genetics. 2004;168:2207–2215. doi: 10.1534/genetics.104.032896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puchta H, Dujon B, Hohn B. Two different but related mechanisms are used in plants for the repair of genomic double-stranded breaks by homologous recombination. Proc. Natl Acad. Sci USA. 1996;93:5055–5060. doi: 10.1073/pnas.93.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puchta H. Repair of genomic double-strand breaks in somatic plant cell by one-sided invasion of homologous sequences. Plant J. 1998;13:331–339. [Google Scholar]
- 27.Reiss B, Schubert I, Koepchen K, Wendeler E, Schell J, Puchta H. RecA stimulates sister chromatid exchange and the fidelity of double-strand break repair, but not gene targeting, in plants transformed by Agrobacterium. Proc. Natl Acad. Sci. USA. 2000;97:3358–3363. doi: 10.1073/pnas.050582797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puchta H. The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J. Exp. Bot. 2005;56:1–14. doi: 10.1093/jxb/eri025. [DOI] [PubMed] [Google Scholar]
- 29.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haber JE, Ira G, Malkova A, Sugawara N. Repairing a double-strand chromosome break by homologous recombination: revisiting Robin Holliday's model. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003;359:79–86. doi: 10.1098/rstb.2003.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gal S, Pisan B, Hohn T, Grimsley N, Hohn B. Genomic homologous recombination in planta. EMBO J. 1991;10:1571–1578. doi: 10.1002/j.1460-2075.1991.tb07677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu S-Y, Culligan K, Lamers M, Hays J. Dissimilar mispair-recognition spectra of Arabidopsis DNA-mismatch-repair proteins MSH2.MSH6 (MutSα) and MSH2.MSH6 (MutSγ) Nucleic Acids Res. 2003;31:6027–6034. doi: 10.1093/nar/gkg780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sweetser DB, Hough H, Whelden JF, Arbuckle M, Nickoloff JA. Fine-resolution mapping of spontaneous and double-strand break-induced gene conversion in Saccaromyces cerevisiae reveals reversible mitotic conversion polarity. Mol. Cell. Biol. 1994;14:3863–3875. doi: 10.1128/mcb.14.6.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taghian DG, Nickoloff JA. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol. Cell. Biol. 1997;17:6386–6396. doi: 10.1128/mcb.17.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elliott B, Richardson C, Winderbaum J, Nickoloff JA, Jasin M. Gene conversion tracts from double-strand break repair in mammalian cell. Mol. Cell. Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D’Halluin K, Vanderstraeten C, Stals E, Cornelissen M, Ruiter R. Homologous recombination: a basis for targeted genome optimization in crop species such as maize. Plant Biotechnol. J. 2008;6:93–102. doi: 10.1111/j.1467-7652.2007.00305.x. [DOI] [PubMed] [Google Scholar]
- 37.Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- 38.Leung W-Y, Malkova A, Haber JE. Gene targeting by linear duplex DNA frequently occurs by assimilation of a single strand that is subject to preferential mismatch correction. Proc. Natl Acad. Sci. USA. 1997;94:6851–6856. doi: 10.1073/pnas.94.13.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longston LD, Symington LS. Gene targeting in yeast is initiated by two independent strand invasions. Proc. Natl Acad. Sci. USA. 2004;101:15392–15397. doi: 10.1073/pnas.0403748101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, Read LR, Baker MD. The mechanism of mammalian gene replacement is consistent with the formation of long regions of heteroduplex DNA associated with two crossing-over events. Mol. Cell. Biol. 2001;21:501–510. doi: 10.1128/MCB.21.2.501-510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hohn B, Levy AA, Puchta H. Elimination of selection markers from transgenic plants. Curr. Opin. Biotechnol. 2001;12:139–143. doi: 10.1016/s0958-1669(00)00188-9. [DOI] [PubMed] [Google Scholar]
- 42.Gilbertson L. Cre-lox recombination: cre-active tools for plant biotechnology. Trends Biotechnol. 2003;21:550–555. doi: 10.1016/j.tibtech.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 43.Cohen-Tannoudij M, Babinet C. Beyond ‘knock-out’ mice: new perspectives for the programmed modification of the mammalian genome. Mol. Hum. Reprod. 1998;4:929–938. doi: 10.1093/molehr/4.10.929. [DOI] [PubMed] [Google Scholar]
- 44.Müller U. Ten years of gene targeting: targeted mouse mutants, from vector design to phenotype analysis. Mech. Dev. 1999;82:3–21. doi: 10.1016/s0925-4773(99)00021-0. [DOI] [PubMed] [Google Scholar]
- 45.Sorrell DA, Kolb AF. Targeted modification of mammalian genomes. Biotechnol. Adv. 2005;23:431–469. doi: 10.1016/j.biotechadv.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 46.Reichardt HM, Kaestner KH, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P, et al. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]