Abstract
Potent antiretroviral therapy can reduce plasma HIV RNA levels below the threshold of detection for periods of a year or more. The magnitude of HIV RNA reduction in the lymphoid tissue in patients with suppression of HIV RNA levels in plasma beyond 6 months has not been determined. We evaluated levels of HIV RNA and DNA and characterized resistance mutations in blood and inguinal lymph node biopsies obtained from 10 HIV-infected subjects who received 36–52 weeks of indinavir (IDV)/zidovudine (ZDV)/lamivudine (3TC), IDV, or ZDV/3TC. After 1 year of therapy, viral RNA levels in LN of individuals remained detectable but were log10 = 4 lower than in subjects on the triple drug regimen with interruption of therapy or in those treated with ZDV/3TC alone, who had viral loads in their lymph nodes indistinguishable from those expected for untreated patients. In all cases viral DNA remained detectable in lymph nodes and peripheral blood mononuclear cells (PBMC). When plasma virus suppression was incomplete, lymph node and PBMC cultures were positive and drug resistance developed. These studies indicate that pronounced and sustained suppression of plasma viremia by a potent antiretroviral combination is associated with low HIV RNA levels in the lymph nodes 1 year after treatment. Conversely, the persistence of even modest levels of plasma virus after 1 year of treatment reflects ongoing viral replication, the emergence of drug resistance, and the maintenance of high burdens of virus in the lymph nodes.
Reduction in plasma HIV RNA levels is an indicator of antiretroviral activity in the testing of new compounds and a measure of therapeutic efficacy in the clinical management of HIV-infected patients (1). Plasma virus levels become and remain undetectable by HIV RNA quantitative assays in individuals treated with potent combination antiretroviral regimens (2, ††). The corresponding response to antiretroviral therapy in the lymphoid tissues, where the vast majority of virus burden resides (3, 4), has not been characterized beyond 6 months of treatment. Reports correlating reductions in levels of HIV RNA in lymph nodes (LN) and plasma in subjects treated with nucleoside analogs have been contradictory (5–9).
We examined peripheral blood and biopsied LN from 10 subjects receiving potent antiretroviral therapy for up to 1 year for HIV RNA and proviral DNA burden, for infectious virus, and for the development of resistance to antiviral drugs. In eight subjects, in situ hybridization (ISH) in combination with quantitative image analysis (QIA) (10, 11) was performed on LN specimens to corroborate the quantitative data based on analyses of bulk nucleic acid extracts and to localize residual virus burden to specific compartments within LN.§§
METHODS
Patient Recruitment and Initial LN Processing.
Subjects were recruited from the San Diego Cohort of the Merck 035 study 36 to 52 weeks after randomization to therapy with zidovudine (ZDV)/lamivudine (3TC), indinavir (IDV) alone, or ZDV/3TC/IDV, but prior to cross-over to open-label triple drug treatment. With informed consent and the approval of the local institutional review board, open inguinal LN excisions were performed along with simultaneous collection of ≈30 ml of peripheral blood in tubes containing acid/citrate/dextrose. Portions of the LN were immediately snap frozen in liquid nitrogen, and one portion was placed in 3% paraformaldehyde and one in phosphate-buffered saline without CaCl2 and MgCl2 (PBS-A) on ice.
Processing of Blood.
Blood was centrifuged at 258 × g for 12 min. Plasma was stored at −70°C. Peripheral blood mononuclear cells (PBMC) were isolated from the buffy coat by Ficoll/Hypaque density gradient. PBMC were counted and viability was determined by trypan blue exclusion. Cells were divided into aliquots for virus isolation or stored as pellets of 0.5–1 × 106 cells at −70°C.
LN mononuclear cell (LNMC) Suspension.
LNMC were isolated by abrasion of LN fragments on sterile mesh screens with aseptic technique. LNMC were pelleted by centrifugation at 258 × g for 6 min, washed twice in PBS-A, resuspended, and counted. Viability was assessed by trypan blue exclusion. Typically, cell viability exceeded 80%. Cells were divided into aliquots for virus isolation or cryopreserved.
Nucleic Acid Extraction.
RNA was extracted from LN samples with the Qiagen RNeasy total RNA extraction kit (Qiagen, Chatsworth, CA) with the following modifications: LN fragments for RNA extraction were weighed (range of 5–110 mg) then placed in cold lysis buffer (RLT; Qiagen) and homogenized with a tissue grinder. Genomic DNA in the homogenate was fragmented by centrifugation through a Qiashredder column (Qiagen). RNA was eluted in 2 mM Tris/diethyl pyrocarbonate (DEPC)-treated water and stored at −70°C until further processing. Prior to use in quantitation assays, RNA was treated with 10 units of RNase-free DNase I (Stratagene) for 1 hr at 37°C, then reextracted by using the protocol for Amplicor (Roche Molecular Systems, Branchburg, NJ). The standard Amplicor extraction protocol was used for RNA extraction from plasma.
Total DNA was extracted from LN samples and PBMC pellets by using the Qiamp tissue kit (Qiagen) with the following modifications: LN fragments were weighed then minced with sterile scalpels and transferred to individual 1.5-ml Eppendorf tubes. Lysis was with Qiamp lysis buffer and proteinase K at 55°C for 2 hr, then readdition of proteinase K and incubation for another 4 hr at 55°C. Genomic DNA in the lysate was fragmented with Qiashredder columns. PBMC pellets were lysed at 55°C for 1 hr. RNase A was added prior to addition of the buffer AL (Qiagen). DNA was eluted from the Qiamp columns with 2 mM Tris⋅HCl, pH 8.0, at 50°C and stored at −20°C. The concentration of DNA was determined by a semiquantitative ethidium staining procedure (12).
Total HIV RNA Quantitation.
HIV RNA was quantitated with the Amplicor Monitor assay . Plasma samples were processed according to the manufacturer’s specifications. When plasma RNA samples fell below 400 copies per ml, separate aliquots of plasma were assayed by use of the Roche Ultradirect Assay (limit of detection of 20 copies per ml) (13). For HIV RNA levels in LN, total RNA extracted by the Qiagen RNAeasy system was incubated with DNase at 37°C for 1 hr, then reextracted according to the Amplicor protocol. The copies per ml value obtained was transformed to copies per mg of tissue by division by the mass of tissue from which RNA was extracted and processed, followed by a correction factor of 0.2 for the volume of plasma (200 μl) processed to yield the copies per ml value. Two separate LN RNA quantitation assays were performed on RNA extracted from nonadjacent fragments of LN for each patient.
When the initial “copies per ml” values exceeded 106/ml, quantitation was repeated with appropriate dilutions of fresh RNA extracts.
T Cell Antigen Receptor (TCR) Cα mRNA Quantitation.
Aliquots of the DNase-treated, Amplicor reextracted, resuspended RNA were assayed semiquantitatively for TCR Cα mRNA. TCR Cα sequence [codons 143–270 of human TCR Cα gene (14)] were amplified with sense primer TCR 5′-T7 (5′-gtaatacgactcactataggGAACCCTGACCCTGCCGTGTACC- 3′ containing the T7 promoter (lowercase letters), and antisense primer TCR 3′ (5′-AGCCGCAGCGTCATGAGCAGATTAAACCCG-3′) (9), from uninfected PBMC DNA. A complementary RNA quantitation standard was generated by in vitro transcription with T7 RNA polymerase, quantitated spectrophotometrically, diluted, and stored in aliquots at −70°C until use. Samples (10 μl) of resuspended RNA extracts from LN prepared in the Amplicor assay (along with quantitation standards) were heat denatured at 70°C for 3 min, then reverse transcribed at 45°C for 1 hr with 1× Superscript RT buffer, 10 mM dithiothreitol, 0.5 mM each dNTP, 36 units of RNAgard (Pharmacia), 3 units of Superscript II (GIBCO), 50 ng/μl yeast tRNA, 12.5 pmol of TCR 3′ primer, and diethyl pyrocarbonate-treated water to a total volume of 50 μl. A 5-μl sample of each reaction mixture was transferred to thermocycler tubes with Tris⋅HCl, pH 8.3, MgCl2 1.5 mM final, BSA (100 μg/ml), 20 pmol of TCR Cα 5′ primer (5′-GAACCCTGACCCTGCCGTGTACC-3′), 20 pmol of TCR Cα 3′ primer, and mineral oil in a 20-μl reaction volume. Samples were heated to 95°C for 3 min, then transferred to 80°C heat blocks, where 0.5 μl of Taq DNA polymerase (2.5 units) and 1.0 μl of [α-32P]dCTP (specific activity 3,000 mCi/mmol; 1 mCi = 37 MBq) were added to each sample. Perkin–Elmer model 430 Thermocycler settings for the PCR were as follows: denature at 95°C for 30 sec, anneal at 70°C for 30 sec, and extend at 72°C for 1 min for 18 cycles, followed by a final extension at 72°C for 10 min. When these conditions were used, TCR message from as few as 500 PBMC equivalents was detectable. PCR products were resolved by PAGE (6% gel) followed by autoradiography on phosphorscreens (Eastman Kodak). Screens were scanned with a Storm 860 Phosphorimager (Molecular Dynamics) and quantitated with use of the ImageQuant software program (Molecular Dynamics). Logarithms of pixel volumes were extrapolated to copy numbers based on comparative values for the dilution standard of known copy number. Reverse transcriptase (RT) PCRs included a negative control (water), and the efficacy of the DNase treatment was initially established by showing the absence of detectable product in duplicate reactions performed without the addition of RT. In control experiments, in vitro infection of donor PBMC with HIV LAI did not alter TCR Cα message after 3 days at a multiplicity of infection of 0.01 (mean of 25 vs. 24 copies per cell in duplicate assays) and total PBMC and CD8- and monocyte-depleted PBMC yielded comparable levels of TCR Cα message (mean of 24 vs. 19 copies per cell).
HIV DNA Quantitation.
HIV DNA was quantitated by PCR for the HIV gag gene. PCR was performed in duplicate for clinical samples on 100 ng of extracted genomic DNA (≈16,000 cells) along with a dilution standard of spectrophotometrically quantified pNL4-3 HIV plasmid ranging from 600 to 2 copies mixed with 100 ng of genomic DNA from uninfected CEM cells and negative controls. Products were labeled by incorporation of [α-32P]dCTP. Reaction conditions: Total volume of 20 μl, Tris⋅HCl, pH 8.3, 1.5 mM MgCl2, 100 μg/ml BSA, primers SK145 (AGTGGGGGGACATCAAGCAGCCATGCAAAT) and SK431 (GCTATGTCACTTCCCCTTGGTTCTCT) at 20 pmol per reaction (15), and mineral oil overlay. Reaction mixtures were heated to 95°C for 3 min, then transferred to an 80°C heat block, where 1 μl of [α-32P]dCTP and 0.5 μl of Taq DNA polymerase were added. Thermocycler settings were 95°C for 30 sec, 60°C for 30 sec, and 72°C for 1 min for a total of 22 cycles, followed by a 10-min extension at 72°C. Products were resolved by PAGE (8% gel). Autoradiography and copy number extrapolation were as described for the TCR Cα quantitation. When no HIV DNA was detected in a sample, assays were repeated with up to 400 ng of genomic DNA. For samples with positive signals corresponding to <5 copies per 100 ng, values were represented as 1–5 copies per 100 ng. Separately, β-actin (DNA) was quantitated from 100 ng of the same DNA extracts to determine DNA input for PCR. Conditions and methods for this procedure were as above except that primers 317/319 were used (12), the annealing temperature was 62°C, and the PCR was carried out for 18 cycles. The amount of β-actin (DNA) detected in sample extracts was quite consistent (data not shown).
ISH and Quantitative Image Analysis.
The number of copies of HIV-1 RNA in virions bound to follicular dendritic cells (FDCs, the antigen-presenting cells in LN follicles) and in productively infected mononuclear cells (MNCs) was determined by ISH and quantitative image analysis (10). Sections of fixed paraffin-embedded LNs were cut, allowed to adhere to slides, and deparaffinized. After permeabilization the sections were hybridized to a collection of antisense 35S-labeled riboprobes complementary to about 90% of HIV genomic RNA. After hybridization, the sections were coated with nuclear track emulsion, exposed for 1 day, developed, and stained. Silver grains overlying FDCs and their processes or productively infected MNCs were automatically counted in video images of the section with the Metamorph software previously validated (10). Copy numbers were normalized to gram of LN by determining the area of the sections with the Metamorph software program and multiplying by the thickness and density of the tissue (10). To identify cells with <20 copies of HIV-1 RNA per cell, radioautographic exposure times were increased from 1 to 10 days (limit of detection: 1–2 copies of HIV-1 RNA per cell) (16).
Nucleic Acid Sequencing.
RT/nested PCR or nested PCR alone was carried out on nucleic acid extracts from clinical samples using a modification of the GeneChip sequencing protocol using PRT 440s and 440a (Affymetrix, Santa Clara, CA) (H.F.G., J.K.W., C.C.I., D.V.H., and D.D.R., see abstract 577 in §§). The sequence data presented here were from population sequences; however, in one subject (G) sequencing of individual cloned amplicons was carried out (nine clones analyzed). In this case, the data from population sequencing and clonal sequencing were quite comparable. In addition, Applied Biosystems automated sequencing was concurrently performed on the amplicons generated in the Affymetrix procedure, using PRISM dye terminator kits with AmpliTaq DNA polymerase, FS (Perkin–Elmer). The concordance between methods exceeded 99% of all bases sequenced (H.F.G., J.K.W., C.C.I., D.V.H., and D.D.R., unpublished work).
Virus Isolation.
One million LNMC (n = 5) or PBMC (n = 10) were cocultured with 106 2-day prestimulated (phytohemagglutinin, interleukin 2) normal donor PBMC. Cultures were split every 3–4 days and fresh prestimulated donor cells were added weekly. Assays for p24 antigen from culture supernatants were performed weekly. Cultures were maintained for up to 21 days. In four cases where LNMC suspensions were unavailable, 5 to 10 mg of minced LN sample was added to donor PBMC and treated accordingly. Viral recoveries from these two sources of LN material were comparable.
Drug Susceptibility Assays.
Drug susceptibility assays were performed according the the ACTG/DOD consensus PBMC assay (17). For ZDV, a virus isolate was defined as resistant if the IC50 was >0.1 μM; for 3TC, when the IC50 was >10 μM (wild-type and sensitive isolates have an IC50 ≪ 1.0 μM); for IDV, IC50 > 4× that for wild-type LAI virus assayed concurrently.
Statistical Analyses.
Correlation coefficients r, P values, and Wilcoxon rank sum with Student’s t test were determined with use of jmp 3.1.5 (SAS Institute, Cary, NC).
RESULTS
Treatment with the Triple Drug Regimen of ZDV, 3TC, and IDV Can Result in Plasma Virus Suppression to <20 Copies per ml at a Year of Therapy.
All study subjects had comparable starting HIV RNA levels of 4.5–5.5 log10 units/ml of plasma and CD4 cell counts between 70 and 270/mm3 (Table 1). At the time of LN biopsy, the subjects segregated into three response groups based on plasma HIV RNA levels and on change of plasma HIV RNA levels from baseline. Two patients receiving triple therapy (A and B) had undetectable HIV RNA in plasma by the Ultradirect assay (limit of detection of 20 copies per ml) (13). Three individuals (C, D, and J), two on triple therapy and one on IDV alone, had several hundred copies per ml of HIV RNA present. Five subjects, including two on triple therapy and three on ZDV/3TC, had HIV RNA levels >1,000 copies per ml. With the exception of subject I, all subjects experienced a rise in CD4 counts relative to baseline, with a mean increase of 114/mm3 for the 7 subjects receiving an IDV-containing regimen and 44 for the subjects treated with ZDV/3TC alone (P = 0.36). However, the relationship of increases in CD4s for individual subjects over this time period did not correlate with the degree of virus suppression (r = −0.11, P = 0.75).
Table 1.
Clinical and baseline characteristics
| Subject | Prior therapy | Active therapy | CD4 at baseline, cells/mm3 | HIV RNA, log10 copies/ml
|
ΔCD4 count, cells/mm3 | ||
|---|---|---|---|---|---|---|---|
| Plasma baseline | Plasma 1 year | Δ in plasma | |||||
| A | ZDV | ZDV, 3TC, IDV | 243 | 5.2 | <1.3 | −5.2 | 29 |
| B | ZDV, ddC, d4T | ZDV, 3TC, IDV | 274 | 4.7 | <1.3 | −4.7 | 60 |
| C | ZDV | ZDV, 3TC, IDV | 220 | 5.2 | 2.1 | −3.2 | 114 |
| D | ZTDV, ddI, d4T | IDV | 178 | 5.2 | 2.9 | −2.4 | 270 |
| J | ZDV | ZDV, 3TC, IDV | 106 | 5.0 | 2.3 | −2.7 | 192 |
| E | ZDV, ddC | ZDV, 3TC, IDV | 69 | 5.5 | 3.7 | −1.7 | 162 |
| F | ZDV, ddC, ddI | ZDV, 3TC, IDV | 231 | 4.4 | 3.3 | −1.1 | 65 |
| G | ZDV, ddC, ddI | ZDV, 3TC | 199 | 5.0 | 4.3 | −0.7 | 73 |
| H | ZDV | ZDV, 3TC | 119 | 5.2 | 4.8 | −0.4 | 67 |
| I | ZDV | ZDV, 3TC | 188 | 5.1 | 4.3 | −0.8 | −7 |
“Plasma baseline” refers to plasma obtained prior to initiation of the active therapy. “Plasma 1 year” refers to levels in plasma obtained on the day of the LN biopsy. RNA levels in plasma are those determined by quantitative RT-PCR (Amplicor or UltraDirect assay). The threshold of sensitivity for the UltraDirect assay is <1.3. ddC, dideoxycytidine; d4T, didehydrodideoxythymidine; ddI, dideoxyinosine.
Virus RNA Is Greatly Reduced but Remains Detectable in LN after 1 Year of Therapy in Those with Complete Suppression of Plasma Virus.
In the case of four subjects (patients A–D) with either undetectable (n = 2) or very low residual plasma virus (119 and 800 copies per ml) (n = 2), the RNA levels detected in LN were 104 to 105 copies per gram of tissue by the quantitative RT-PCR assay (Amplicor Monitor) applied to bulk extracts of RNA. However, the residual virus levels represented a log10 = 4 reduction compared with untreated or inadequately treated subjects (10, 16). One additional subject (J) with low plasma RNA levels was unevaluable by the quantitative assay on the bulk RNA extracts because of inadequate sampling (Table 2).
Table 2.
HIV RNA and DNA quantitation from bulk nucleic acid extracts and RNA quantitation in LN by ISH and quantitative image analysis
| Subject | Bulk nucleic acid extracts
|
LN ISH and image analysis
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| LN RNA, 1 yr by Amplicor, log10 HIV copies/g | HIV DNA, copies/100 ng
|
HIV RNA by ISH, log10 copies/g
|
Avg HIV RNA copies/MNC, copies/cell | Frequency of MNC, log10 cells/g tissue
|
|||||
| PBMC base | PBMC 1 yr | LN 1 yr | LN FDC | LN MNC | With >20 copies of HIV RNA | With few copies of HIV RNA | |||
| A | 4.1 | 1–5 | 1–5 | 1–5 | ≤4 | — | — | — | 3.1 |
| B | 4.2 | <1 | <1 | 12 | ≤4 | — | — | — | ≤3 |
| C | 4.6 | 42 | 1–5 | 12 | ≤4 | — | — | — | ≤3 |
| D | 4.6 | ND | 1–5 | ND | ≤4 | — | — | — | ≤3 |
| J | ∗ | 1–5 | 1–5 | ∗ | 5.5 | — | — | — | ≤3 |
| E | 7.1 | 21 | 1–5 | 26 | 7.2 | 5.5 | 45 | 3.9 | — |
| F | 8.7 | 9 | 1–5 | 18 | 8.1 | 4.5 | 106 | 4.4 | — |
| G | 8.2 | ND | 12 | 53 | 7.3 | 6.2 | 36 | 4.6 | — |
| H | 8.6 | ND | 6 | 75 | ND | ND | ND | ND | ND |
| I | 7.4 | ND | 1–5 | 48 | ND | ND | ND | ND | ND |
The first data column shows HIV RNA in 1 g (extrapolated) of LN by Amplicor assay (mean of independent determinations on separate samples except for patients A and D, where values are from single assays). The second shows HIV DNA copy numbers in baseline PBMC samples from six subjects and the third shows HIV DNA copy numbers in PBMC at the time of LN biopsy (mean of duplicate assays). For subject B, ≈1 copy was detectable in 400 ng of cellular DNA (64,000 cell equivalents). The fourth column shows HIV DNA copy numbers in 100 ng of LN DNA. ND, not done; ∗, unevaluable. The fifth data column shows HIV RNA copy numbers associated with FDC normalized to a gram of lymphoid tissue. The threshold of detection is 104 copies per gram. The sixth shows RNA copies associated with MNC having >20 copies per cell. The seventh shows average number of RNA copies associated with each positive MNC. The last two columns show respectively the frequency (density) of mononuclear cells having >20 copies of HIV RNA per cell and those with ≪20 copies per cell (typically <5) in 1 g of tissue. The threshold for detection of these cells is 103 cells per gram of tissue. ND, not done; —, not applicable.
Adjacent LN fragments from 8 of 10 subjects were analyzed by ISH with quantitative image analysis (10, 11). The results of these studies correlated well with the quantitation of bulk extracts by using the Amplicor Monitor assay (r = 0.98, P = 0.0001). As previously shown, the majority of HIV RNA is associated with the follicular dendritic network in germinal centers, and levels lower by 1 to 3 log10 units were associated with MNCs. In 4 of the patients with sustained plasma levels of HIV RNA <800 copies per ml (patients A–D), ISH studies failed to detect FDC-associated virus (with a limit of detection of 104 copies per gram) or cell-associated virus with the standard overnight exposure. In one of these patients (A), however, MNCs harboring very low copy numbers of RNA (<20 per cell) were detectable after 10-day exposures. In the case of subject J, a reduced level of approximately log10 = 5 copies per gram of tissue was detectable by ISH studies despite unsuccessful attempts to quantitate from bulk RNA and DNA extracts (Table 2).
In Subjects with Even Modest Plasma Viremia, Very Significant Virus Loads Were Detectable in the Corresponding LNs.
This result was seen in subjects not receiving IDV or in subjects with a history of interruption of IDV therapy. Two subjects (E and F) receiving triple therapy had initial suppression of virus to below the limit of detection but later experienced return of plasma virus at several thousand copies per ml. They had corresponding viral RNA levels in LN of 107 to 108 copies per gram, levels indistinguishable from those found in untreated subjects or in subjects with nucleoside therapy alone (10, 16). In both subjects, treatment histories revealed transient interruption of therapy in the month prior to the LN biopsies (patient E, 6 days and patient F, 3 days), coincident with the rebound of plasma viremia (patient E also had 3 days of treatment interruption 2 months earlier). Serial plasma RNA levels during the course of therapy are shown in Fig. 1. Each of the three subjects receiving only ZDV plus 3TC also had high levels of LN viral RNA. In these cases plasma virus levels reached nadirs of 103 copies per ml but rebounded to 104 copies per ml within several months (data not shown). Again, there was a good correlation between ISH studies and quantitation employing RT-PCR on bulk extracts: the distribution of RNA, the average copy number per MNC harboring detectable HIV RNA, and the density (frequency) of MNCs with >20 copies per cell per gram of tissue were all consistent with what has been defined for untreated and incompletely treated subjects (10) (Table 2).
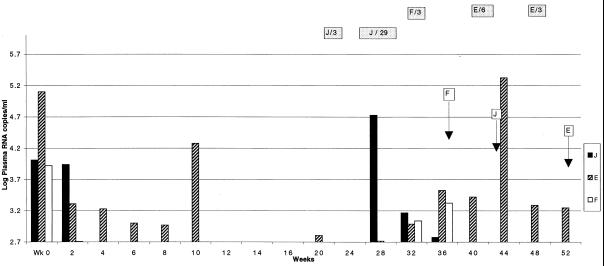
Figure 1.
Serial plasma RNA determinations in three subjects with history of therapy interruptions. Values are presented as log10 copies of HIV RNA per ml by vertical bars, with the lowest values shown being the threshold of detection of the Amplicor assay. Periods of treatment interruptions are shown in shaded horizontal bars with the letter corresponding to the subject followed by the number of days of treatment interruption. Arrows indicate time of LN biopsy for each patient.
HIV DNA Remained Detectable in LN and PBMC from all Patients.
HIV DNA was detectable in LN from all patients evaluable (n = 8), ranging from 1–5 copies to 75 copies per 100 ng of genomic DNA (16,000 cell equivalents). The highest levels were present in the subjects with the highest levels of plasma and LN RNA. Lower levels of viral DNA were detectable in PBMC extracts, ranging from <1 to 12 copies per 100 ng of genomic DNA. In 3 of 6 subjects from whom PBMC at baseline and after 1 year of therapy were available for comparison, a reduction of proviral copy numbers was noted (Table 2).
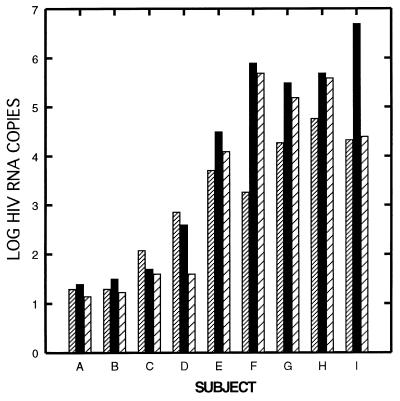
Quantity of HIV RNA and DNA in LN Correlated Well with Plasma HIV RNA After 1 Year of Therapy.
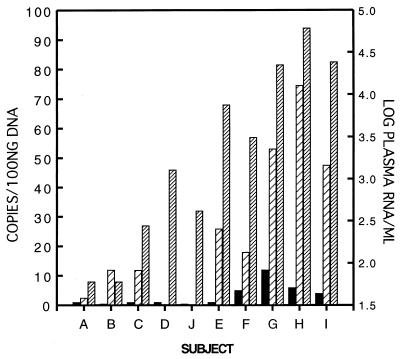
As shown in Fig. 2, this correlation was strong when HIV RNA copy number was normalized either to tissue mass (r = 0.87, P = 0.002) or to TCR Cα mRNA (r = 0.91, P = 0.0006). In one case (J), where no HIV RNA was detectable from assays of extracted LN RNA (performed on two separate LN fragments), no TCR Cα mRNA was detectable (nor was HIV DNA detectable), suggesting an inadequate sampling of LN for the bulk nucleic acid assays. (Approximate limit of detection of TCR Cα mRNA is ≤500 cell equivalents.) Levels of HIV DNA in LN showed a statistically significant correlation with both plasma (r = 0.89, P = 0.027) and LN RNA levels (r = 0.75, P = 0.034). Additionally, plasma viremia correlated with HIV DNA loads in PBMC (r = 0.73, P = 0.026) (Fig. 3).
Figure 2.
Correlation of HIV RNA levels in plasma and LN after 1 year of therapy. Values are represented as log10 copies of HIV RNA for each subject. Plasma was obtained on the same day as the LN biopsy. Densely hatched bars, plasma HIV RNA (log10 copies per ml); black bars, HIV RNA in LN (log10 copies/4 × 106 copies of TCR Cα mRNA); sparsely hatched bars, HIV RNA in LN (log10 copies per mg). Values shown for LN are mean of two assays on different tissue fragments.
Figure 3.
Correlation of HIV RNA levels in plasma and HIV DNA copy numbers in PBMC and lymph node after one year of therapy. Values for HIV RNA in plasma are log10 copies per ml of plasma shown by densely hatched bars (right-hand axis). Values for HIV DNA are in copy numbers per 100 ng of cellular DNA or 16,000 cell equivalents (left-hand axis) shown as black bars for PBMC and sparsely hatched bars for LN. PBMC were obtained on the same day as the LN biopsy. HIV DNA copy numbers represent the mean of duplicate assays performed on the same DNA extract. No LN DNA extract was available for subject D. HIV DNA copy number in PBMC from subject B was approximately 1 in 64,000 cells. Quantitation from LN of patient J was omitted.
Recovery of Virus by Coculture and Detection of Phenotypic and Genotypic Drug Resistance Occurred in All Subjects with Incompletely Suppressed Plasma Virus.
All three subjects receiving ZDV plus 3TC (patients G, H, and I) and the two subjects receiving triple therapy who experienced reemergence of virus after initial suppression (patients E and F) had virus recovered by coculture from LN and/or PBMC. Phenotypic drug assays performed on isolated virus demonstrated emergence of 3TC resistance in four of these five subjects and IDV resistance in one of the two subjects on triple therapy (data not shown). The nucleotide sequence of the virus isolate in the one triple therapy patient from whom phenotypically wild-type virus was obtained (patient E) indicated that this virus was also genotypically wild type (data not shown). However, the sequence of material directly amplified by PCR from the clinical specimens of this patient showed development of IDV resistance mutations as well as the M184V mutation conferring 3TC resistance. In fact, sequencing of material directly amplified by RT-PCR from each of these five subjects demonstrated the acquisition of resistance mutations against one or more of the drugs used (Table 3).
Table 3.
Substitutions conferring drug resistance
| Source | Resistance substitutions
|
||
|---|---|---|---|
| ZDV | 3TC | IDV | |
| J pre | M41L, T215Y/D | L63P | |
| J post | M41L, T215Y/D | L63P | |
| E pre | L63P | ||
| E post | M184V | L63P, V82A (L90W) | |
| F pre | K70R, T215Y | L63P | |
| F post | K70R, T215Y | M184V | L63P, L90M |
| G pre | D67N, K70R, T215Y | K20R, L63P, A71V* | |
| G post | M41L, D67N, K70R, T215Y, K219Q | M184V | K20R, L63P, V82A*† |
| H pre | D67G, K70R, K219Q | L63P | |
| H post | D67G, K70R, K219Q | M184V | L63P |
| I pre | T215F | L63P | |
| I post | M41L, K70R, T215F | M184V | L63P |
Substitutions associated with ZDV, 3TC, or IDV resistance are shown from baseline (pre) and at the time of LN biopsy (post) from each of six subjects (E–J) from whom both “pre” and “post” sequences were obtainable. The baseline data are derived from sequencing of plasma, and the post-treatment data are derived from both plasma and LN samples. There was general agreement of sequences from plasma and LN. Where a discrepancy was noted, a substitution is shown when it occurred in one of the two compartments. Substitutions seen in post-treatment samples not present at baseline are shown in boldface. Numbers in data columns correspond to codon number of either the RT or protease. Amino acid designations follow International Union of Pure and Applied Chemistry single-letter code. A substitution for which a clear association with resistance is unknown is shown in parenthesis.
Occurring in a subject (not on IDV) as a natural polymorphism.
Mutation seen in LN DNA but not in LN RNA or plasma of subject G.
One additional triple therapy patient (J) had virus recoverable by coculture of both PBMC and LNMC despite negligible plasma virus levels. The recovered virus was identical in resistance profile to the virus present at baseline. Review of this patient’s serial RNA levels showed a transient high plasma RNA level corresponding to an interruption of treatment 4 months prior to the LN biopsy. Plasma RNA levels returned to undetectable thereafter (Fig. 1).
DISCUSSION
Therapy with potent combinations of antiviral drugs can rapidly reduce circulating levels of plasma virus to below the level of detection by even the most sensitive assays for HIV RNA for periods of a year or more (2, ††). The ability of these drug regimens to reduce the total body burden of virus has been uncertain because of the large proportion of virus known to be associated with the FDC network in LN (3, 4, 18). In previously published studies, the amount of virus detectable in lymphoid tissue of chronically infected individuals has been uniformly high despite widely varying viral loads in peripheral blood (3, 4). After treatment with regimens of single and combination nucleoside analogues, a correlation between the response in blood and that in LN was seen by some but not by others (5, 6, 8, 9). In the present study, plasma and LN RNA levels were highly correlated. The difference between the observed correlation in this study compared with the earlier ones may be explained by the more potent antiviral drugs used, the availability of relatively large specimens for analysis, and the concurrent quantitation of TCR Cα message, which permitted normalization of the HIV RNA levels to the cellularity of the individual samples studied. In one case (J) this resulted in the identification of an inadequate sample that otherwise would have been considered as a true “undetectable” in the bulk RNA quantitation assays. The separate sample from subject J studied by ISH did have detectable RNA, albeit at low levels, and appeared to have adequate cellularity.
In those individuals achieving plasma RNA levels at or below the limit of detection by the (standard) Amplicor assay (A–D), the extrapolated reduction in LN RNA level of approximately log10 = 4 copies per gram of tissue demonstrated that virus in this tissue compartment can be reduced if viral replication is controlled with potent antiviral drugs. Notably this was achieved in a group of “drug experienced” patients (nearly all with multiple ZDV resistance conferring mutations at baseline). Viral DNA in LN and to a lesser extent in PBMC also correlated with the plasma RNA levels.
In those subjects with incomplete suppression (treated with ZDV plus 3TC: patients G, H, and I) and in those with suppression followed by interruption of therapy (patients E and F), the LN virus loads were indistinguishable from those expected from untreated subjects. In the cases of E and F these are particularly surprising findings. Studies by Cavert et al. have demonstrated that a reduction in the LN virus burden of log10 = 3.5 can be demonstrated following therapy resulting in suppression of plasma virus for 24 weeks (16). Therefore, shortly before therapy interruption, it would have been reasonable to expect a comparable reduction in LN virus levels in these two subjects. However, the levels of HIV RNA in the LNs from subjects E and F were 107 and 108 copies per gram of tissue. Subject J is an additional triple therapy patient who had interrupted therapy. Because his treatment interruption had occurred 4½ months prior to the LN biopsy and virus suppression appears to have been adequate after resumption of therapy, the amount of HIV RNA in the LN detectable by ISH was only modest but was higher than was seen with subjects A–D. To explain these findings, we propose that in some cases, the resumption of virus replication resulting from interruption of therapy can permit a relatively rapid repopulation of the lymphoid tissues to pretreatment levels, effectively “resetting the clock” for the decay of virus in the LNs.
In one individual with very low loads of LN virus, ISH with a 10-day autoradiographic exposure was able to identify rare cells (≈103 per gram of tissue) that harbor few HIV RNA molecules (≈1–5 copy equivalents per cell). These cells did not appear to stain for the macrophage marker CD68 and appeared to be T lymphocytes (data not shown). It is unknown whether these are cells infected with defective virus or represent variant cells infected with replication-competent virus but produce little. This diminished capacity to produce virus and viral antigens may permit these cells to escape immunologic surveillance and to avoid cytopathic or apoptotic death.
A second cellular reservoir (which may or may not be distinct from the first) in these patients that is of interest is those cells (both in PBMC and in LN) that appear to persistently harbor viral DNA. From the patients (n = 6) for whom pretreatment DNA levels were available, it appears that those starting with high PBMC proviral DNA loads (9–42 copies per 100 ng of DNA) experienced declines in PBMC provirus levels, whereas those with low starting PBMC provirus levels showed little measurable change after 1 year of therapy. One possible explanation for this may be that the HIV DNA in patients with a low starting provirus load was more likely to represent DNA in latently infected cells with very long half-lives, whereas the DNA in subjects with higher starting levels was more likely to be in shorter-lived cells (possibly productively infected).
Our attempts to isolate virus by cocultivation using standard protocols were unsuccessful in all but one case when plasma HIV RNA levels were at or below the threshold of detection. Because these subjects also harbored fewer proviral DNA copies, these failures could be based on quantitative rather than on qualitative differences among the subjects. The failure of many of these HIV-infected cells to produce spreading infection in coculture could be explained by infection and persistence of defective viruses or by the existence of a subset of infected cells that are in prolonged quiescence. Both of these identified cellular reservoirs of viral persistence (cells harboring DNA and LNMC with few HIV RNA copy numbers) have potentially important implications for duration of therapy and on the issue of eradication of infection. The successful recovery of virus from one subject (J) who had good suppression of plasma viremia also deserves mention. In this case, the interruption of therapy 4½ months before may not only have “reset the clock” for LN virus decay but also may have allowed reestablishment of a population of latently infected cells capable of activation to a state of viral production. A greater understanding of each of these cell populations is clearly needed.
In every case where levels of plasma RNA were high, multiple mutations associated with drug resistance developed, indicating continued, high levels of virus replication. In triple therapy patients with treatment interruption, there were examples of both the emergence of drug resistance (E and F) and a lack of development of resistance (J). These divergent patterns reflect the stochastic nature of accumulation of drug resistance substitutions for individual drugs (19, 20), which may be magnified in the setting of combination therapy.
In summary, subjects treated with potent antiretroviral drugs who experienced sustained suppression of HIV RNA levels in plasma had virus RNA levels in LN that were 104-fold lower than those expected for untreated subjects. In contrast, incomplete suppression of plasma virus (including cases with interruption of triple therapy) was associated with RNA levels in LN comparable to those from untreated individuals, with positive LN and PBMC virus cultures, and with emergence of multiple drug resistance substitutions in both peripheral blood and LN indicating continued high levels of virus replication. In all cases, virus DNA remained detectable in LN and PBMC. The significance of the persistence of these infected cells is presently unknown. Expanded studies involving more subjects, longer follow-up, and examination of additional tissue compartments will be necessary to further define the durability of responses, the cellular compartments containing persistent HIV, and the potential for eradication of infection.
Acknowledgments
We thank the participating patients; D. Easter, L. Lau, K. Nuffer, G. Dyak, and B. Coon for procurement of LN biopsies; J. Chodakewitz and L. Jonas for helpful comments; N. Keating, S. Albanil, and J. Aufderheide for viral isolation and drug susceptibility assays; and N. Riggs, C. Spina, R. Kornbluth, D. Disharoon, T. Gingeras, and K. Considine for technical advice and assistance. This work has been supported by National Institutes of Health Grants K11 AI 01361 (J.K.W.), AI 27670, AI 38858, AI 36214 (D.D.R.), Center for Aids Research, AI 29164, the Research Center for AIDS and HIV Infection of the San Diego Veterans Affairs Medical Center (D.D.R.), Swiss National Science Foundation Grant 84AD-046176 (H.F.G.), an unrestricted research grant from Merck Research Laboratories, and National Institutes of Health Grant AI 28246 (A.T.H.) for support of the ISH and quantitative image analyses.
ABBREVIATIONS
- LN
lymph node(s)
- ISH
in situ hybridization
- ZDV
zidovudine
- 3TC
lamivudine
- IDV
indinavir
- PBMC
peripheral blood mononuclear cell(s)
- LNMC
LN mononuclear cell(s)
- MNC
mononuclear cell
- FDC
follicular dendritic cell
- TCR
T cell antigen receptor
- RT
reverse transcriptase
Note Added in Proof
Following the submission of this manuscript, virus has been isolated by using a coculture system from subjects A, B, and C after an additional year of undetectable plasma HIV RNA.
Footnotes
Markowitz, M., Cao, Y., Hurley, A., O’Donovan, R., Heath-Chiozzi, M., Leonard, J., Smiley, L., Keiler, A., Johnson, D., Johnson, P. & Ho, D., 11th International Conference on AIDS, 1996, Vancouver.
These studies were presented in part at the 4th Conference on Retroviruses and Opportunistic Infections, Jan. 22–26, 1997, Washington, DC.
References
- 1.Havlir D V, Richman D D. Ann Intern Med. 1996;124:984–994. doi: 10.7326/0003-4819-124-11-199606010-00006. [DOI] [PubMed] [Google Scholar]
- 2.Gulick R, Mellors J, Havlir D, Eron J, Gonzalez C, McMahon D, Richman D, Valentine F, Jonas L, Meibohm A, Chiou R, Deutsch P, Emini E, Chodakewitz J. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 3.Embretson J, Zupancic M, Ribas J L, Burke A, Racz P, Tenner-Racz K, Haase A T. Nature (London) 1993;363:359–362. doi: 10.1038/362359a0. [DOI] [PubMed] [Google Scholar]
- 4.Pantaleo G, Graziosi C, Demarest J F, Butini L, Montroni M, Fox C H, Orenstein J M, Kotler D P, Fauci A S. Nature (London) 1993;362:355–358. doi: 10.1038/362355a0. [DOI] [PubMed] [Google Scholar]
- 5.Lafeuillade A, Tamalet C, Poggi C, Pellegrino P, Tourres C, Izopet J. AIDS. 1997;11:67–72. doi: 10.1097/00002030-199701000-00010. [DOI] [PubMed] [Google Scholar]
- 6.Lafeuillade A, Poggi C, Profizi N, Tamalet C, Costes O. J Infect Dis. 1996;174:404–407. doi: 10.1093/infdis/174.2.404. [DOI] [PubMed] [Google Scholar]
- 7.Meylan P, Burgisser P, Weyrich-Suter C, Spertini F. J Acquired Immune Defic Syndr Human Retrovirol. 1996;13:39–47. doi: 10.1097/00042560-199609000-00007. [DOI] [PubMed] [Google Scholar]
- 8.Cohen O, Pantaleo G, Holodniy M, Fox C, Orenstein J, Schnittman S, Niu M, Graziosi C, Pavlakis G, Lalezari J, Bartlett J, Steigbigel R, Cohn J, Novak R, McMahon D, Bilello J, Fauci A. J Infect Dis. 1996;173:849–856. doi: 10.1093/infdis/173.4.849. [DOI] [PubMed] [Google Scholar]
- 9.Cohen O J, Pantaleo G, Holodniy M, Schnittman S, Niu M, Graziosi C, Pavlakis G N, Lalezari Jacob, Bartlett John A, Steigbigel R T, Cohn J, Novak R, McMahon D, Fauci A S. Proc Natl Acad Sci USA. 1995;92:6017–6021. doi: 10.1073/pnas.92.13.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haase A T, Henry K, Zupancic M, Sedgewick G, Faust R A, Melroe H, Cavert W, Gebhard K, Staskus K, Zhang Z-Q, Dailey P J, Balfour H H, Jr, Erice A, Perelson A S. Science. 1996;274:985–989. doi: 10.1126/science.274.5289.985. [DOI] [PubMed] [Google Scholar]
- 11.Peng H, Reinhart T A, Retzel E F, Staskus K A, Zupancic M, Haase A T. Virology. 1995;206:16–27. doi: 10.1016/s0042-6822(95)80015-8. [DOI] [PubMed] [Google Scholar]
- 12.Spina C A, Guatelli J C, Richman D D. J Virol. 1995;69:2977–2988. doi: 10.1128/jvi.69.5.2977-2988.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulder J, Resnick R, Saget B, Scheibel S, Herman H, Payne H, Harrigan R, Kwok S. J Clin Microbiol. 1997;35:1278–1280. doi: 10.1128/jcm.35.5.1278-1280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rabbitts T H, Lefranc M P, Stinson M A, Sims J E, Schroder J, Steinmetz M, Spurr N L, Solomon E, Goodfellow P N. EMBO J. 1985;4:1461–1465. doi: 10.1002/j.1460-2075.1985.tb03803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynch C E, Madej R, Louie P H, Rodgers G. J AIDS. 1992;5:433–440. [PubMed] [Google Scholar]
- 16.Cavert W, Notermans D W, Staskus K, Wietgrefe S, Zupancic M, Gebhard K, Henry K, Zhang Z Q, Mills R, McDade H, Goudsmit J, Danner S A, Haase A T. Science. 1997;276:960–964. doi: 10.1126/science.276.5314.960. [DOI] [PubMed] [Google Scholar]
- 17.Japour A J, Mayers D L, Johnson V A, Kuritzkes D R, Beckett L A, Arduino J-M, Lane J, Black R J, Reichelderfer P S, D’Aquila R T, Crumpacker C S RV-43 Study Group, AIDS Clinical Trials Group; Virology Committee Resistance Working Group. Antimicrob Agents Chemother. 1993;37:1095–1101. doi: 10.1128/aac.37.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fox C H, Tenner-Racz K, Racz P, Firpo A, Pizzo P, Fauci A S. J Infect Dis. 1991;164:1051–1057. doi: 10.1093/infdis/164.6.1051. [DOI] [PubMed] [Google Scholar]
- 19.Leigh Brown A J, Richman D D. Nat Med. 1997;3:268–271. doi: 10.1038/nm0397-268. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez-Palomino S, Olivares I, Yuste E, Richman D D, Lopez-Galindez C. Antiviral Ther. 1996;1:225–236. [PubMed] [Google Scholar]