Abstract
Genome analyses have revealed that members of the Lrp/AsnC family of transcriptional regulators are widely distributed among prokaryotes, including both bacteria and archaea. These regulatory proteins are involved in cellular metabolism in both global and specific manners, depending on the availability of the exogenous amino acid effectors. Here we report the first crystal structure of glutamine receptor protein (Grp) from Sulfolobus tokodaii strain 7, in the ligand-free and glutamine-bound (Grp-Gln) forms. Although the overall structures of both molecules are similar, a significant conformational change was observed at the ligand [l-glutamine (Gln)] binding site in the effector domain, which may be essential for further stabilization of the octameric structure, and in turn for facilitating DNA binding. In addition, we predicted promoter for the grp gene, and these analyses suggested the importance of cooperative binding to the protein. To gain insights into the ligand-induced conformational changes, we mutated all of the ligand-binding residues in Grp, and revealed the importance of Gln binding by biochemical and structural analyses. Further structural analyses showed that Y77 is crucial for ligand binding, and that the residues T132 and T134, which are highly conserved among the Lrp family of proteins, fluctuates between the active and inactive conformations, thus affecting protein oligomerization for DNA binding.
INTRODUCTION
The Lrp/AsnC (also known as feast/famine) family of transcriptional regulators is widely distributed among bacteria and archaea, as an important regulatory system of amino acid metabolism and related processes (1–4). This family of proteins is probably restricted to only prokaryotes, since there are no confirmed Lrp/AsnC homologues within eukaryotic genomes (3). Among the members of the Lrp/AsnC family of proteins, the most-studied one is the Escherichia coli leucine-responsive regulatory protein (Lrp) (1,2). E. coli Lrp is a global regulator of amino acid biosynthesis, transport, protein degradation and intermediary metabolism. It controls a large number of genes and operons, depending on the availability of the effector leucine, which is believed to bind to E. coli Lrp. Recently, a number of Lrp/AsnC family proteins were characterized from archaea (5–7). They are (i) Pyrococcus furiosus LrpA, which inhibits the recruitment of RNA polymerase by blocking the access to the transcription start site (8), (ii) Sulfolobus solfataricus Lrs14, which binds within the promoter region and inhibits the binding of the TATA-box-binding protein (TBP) and transcription factor B (TFB) to the TATA box and TFB-recognition elements (BRE), respectively (9), and (iii) Methanocaldococcus jannaschii Ptr2, which binds upstream of the BRE and the TATA box of the rubredoxin 2 gene, recruits TBP to the TATA box, and enhances transcription in vitro (10).
Members of the Lrp/AsnC family of proteins typically have a molecular mass of around 15 kDa, and exist as multimers in solution. For example, the Lrp/AsnC family proteins from E. coli, Agrobacterium tumefaciens, Pseudomonas aeruginosa and P. furiosus exist as dimers, tetramers, octamers and hexadecamers (11–15), respectively. The target promoters of these proteins often contain a number of binding sites that typically lack obvious inverted repeat elements, and to which binding is usually cooperative.
A 17.4 kDa (pI, 9.58) hypothetical regulator protein, annotated as ST1022 and consisting of 150 amino acid residues, was identified in S. tokadaii strain 7 (16). We named this protein glutamine receptor protein (Grp), and hereafter we refer to it as Grp. A homology search of the Swissprot database using Blast2 revealed that the amino acid sequence of Grp (Q972W6, swissprot id) shares about 68% identity to both the AsnC protein of S. acidocaldarius (Q4J717) (17) and Lrp of S. tokodaii (Q972L7) (16). These two proteins have not been characterized either biochemically or structurally. Furthermore, Grp also displayed moderate identity of about 27–40% to many of the Lrp and AsnC family proteins from various archaeal species.
To date, structural information for the Lrp/AsnC family members is available only for five proteins. They are from Pyrococcus sp. OT3 (PDB codes: 1RI7, 1I1G, 2DBB), E. coli (PDB code, 1CG4) and B. subtilis (PDB code, 2CFX) (18–20). These proteins consist of an N-terminal Helix-Turn-Helix (HTH) DNA-binding domain connected by a linker of about 15 amino acids to the C-terminal α/β fold effector binding domain, where ligands can modulate the protein association, DNA binding and regulatory functions. The functional molecules might be an oligomeric form; in many cases, four dimers are arranged in a non-crystallographic fourfold axis as a disk, making contact through their C-terminal effector domains in the center, with their HTH DNA-binding domains facing out (18–20). Sequence comparisons with the five Lrp/AsnC homologues with known crystal structures revealed that Grp showed 40% identity to the Pyrococcus sp. FL11 protein, which forms a higher order assembly in the presence of l-glutamine (19). In addition to FL11, a number of Lrp/AsnC homologues that modulate protein association by binding to the cognate ligands were described, including E. coli Lrp by l-leucine, E. coli AsnC by l-asparagine, S. solfataricus LysM by l-lysine, Zymomonas mobilis glutamate uptake regulatory protein by l-glutamate, A. tumefaciens PutR by l-proline, P. putida BkdR by l-valine, and P. putida MdeR by methionine (15,20–28). At present, a ligand bound complex is available only for AsnC with its effector l-asparagine (20). However, the structure of AsnC in the absence of the ligand l-asparagine is not available, and thus it is unknown whether a conformational change occurs upon binding to its cognate ligand.
We now report the crystal structures of Grp, in the native (apoprotein) form and the complex with an effector, l-glutamine, refined to 1.82 and 1.80 Å resolution, respectively. As observed in many Lrp/AsnC family proteins, both the native and complex (glutamine-bound, Grp-Gln) structures of Grp form octamers. A comparison of the monomers of the native and complex forms of Grp revealed that their overall conformations are very similar; however, a significant conformational change was observed around the ligand-binding site at the effector domain. To clarify these conformational changes, we solved eight additional mutant crystal structures of Grp. The native, glutamine-bound and mutant structures of Grp provide insights into the possible role of the effector requirement in the binding and wrapping of DNA, and how this widely conserved feature among the Lrp/AsnC family of transcriptional regulators controls gene expression.
MATERIALS AND METHODS
Cloning, expression and purification of Grp
The nucleotide sequence encoding grp (annotated as ST1022) was amplified and cloned into the pET21a(+) expression vector. The resultant plasmid was transformed into the E. coli BL21-CodonPlus (DE3)-RIL-X (Stratagene) strain. The Grp protein was overexpressed at mid-log phase by the addition of Isopropyl-β-D-1 thiogalactopyranoside (IPTG) (final concentration, 1 mM) and was purified as described previously (29).
Dynamic light scattering (DLS)
The oligomeric states of the purified native Grp and its mutants were analyzed by a dynamic light-scattering experiment (DLS), using a DynaPro MS/X instrument (Protein Solutions). The analyses were performed with three protein concentrations (3.0, 1.7 and 0.87 mg/ml) in 20 mM Tris–HCl (pH 8.0) with 0.15 M NaCl. Several measurements were taken at 20°C and were analyzed by the DYNAMICS software, v.3.30 (Protein Solutions).
Crystallization and data collection
Initial crystals of native Grp were grown at 20°C by the sitting drop vapor diffusion method (30), by adding 0.5 μl of protein solution to 0.5 μl of well solution, containing 30% isopropanol, 0.2 M sodium citrate and 0.1 M sodium cacodylate (pH 6.5). Native crystals of Grp grew within a week. The Grp-Gln and T134A-Gln complex crystals were grown under native conditions. The crystals of the other mutants and mutant complexes, including S32A, S32A-Gln, Y77A, Y77A-Gln, T132A, T132A-Gln and T134A, were grown from drops consisting of 1 μl protein and 1 μl of 20% polypropylene glycol P400, 30% isopropanol and 0.07 M sodium citrate, pH 6.2, with the additives 0.01 M FeCl3, 15 μM Cymal-7, 0.2 M 3-(1-methylpiperidinium)-1-propane sulfonate (NDSB-221), 0.01 M taurine, 3% trimethylamine N-oxide, 3% sorbitol, and 0.01 M hexamine cobalt (III) trichloride, respectively. Glutamine (final concentration of 10 mM) was added to the S32A-Gln, Y77A-Gln, T132A-Gln and T134A-Gln drops, whereas 33 mM of glutamine (final concentration) was added to the Grp-Gln drop. The crystals of the native, Grp-Gln and T134A-Gln were soaked in precipitant buffer plus 30% trehalose for cryo-protection, and the data sets were obtained at 100 K using a Jupiter210 CCD detector (RIGAKU MSC Co.) on the RIKEN Structural Genomics Beamline II, BL26B2, at the SPring-8, Hyogo, Japan. For the Y77A-Gln mutant, the data set was collected at the in-house R-axis VII system (RIGAKU MSC). All of the data sets were processed with the HKL 2000 program suite (31). Crystal data statistics are provided in Table 1.
Table 1.
Data collection and refinement statistics of the Grp native, mutants, and their complexes
| Native | Complex | S32A | S32-Gln | Y77A | Y77A-Gln | T132A | T132A-Gln | T134A | T134-Gln | |
|---|---|---|---|---|---|---|---|---|---|---|
| Data collection | ||||||||||
| Cell dimensions (Å) | a = b = 103.771, c = 73.297 | a = b = 103.846, c = 73.992 | a = b = 103.662, c = 74.146 | a = b = 103.663, c = 73.539 | a = b = 103.902, c = 74.483 | a = b = 103.687, c = 74.942 | a = b = 103.697, c = 74.939 | a = b = 103.808, c = 74.464 | a = b = 103.999, c = 74.587 | a = b = 103.946, c = 75.039 |
| Wavelength (Å) | 1.0000 | 1.0000 | 0.9794 | 0.9794 | 0.9794 | 1.54178 | 1.000 | 1.000 | 1.000 | 0.9794 |
| Resolution range (Å) | 50.0–1.82 | 50.0–1.80 | 50.0–1.98 | 50.0–1.44 | 50.0–2.20 | 50.0–2.20 | 50.0–1.90 | 50.0–2.05 | 50.0–2.00 | 50.0–2.15 |
| (1.89–1.82)a | (1.86–1.80) | (2.05–1.98) | (1.49–1.44) | (2.28–2.20) | (2.28–2.20) | (1.97–1.90) | (2.12–2.05) | (2.07–2.20) | (2.23–2.15) | |
| Unique reflections | 18 202 | 19 055 | 14 388 | 35 897 | 10 745 | 10 689 | 16 344 | 12 674 | 14 084 | 11 453 |
| Redundancy | 13.9 (13.6) | 13.6 (11.9) | 14.7 (14.8) | 11.5 (3.5) | 14.6 (14.9) | 9.1 (9.2) | 13.5 (11.5) | 13.1 (11.1) | 7.3 (7.2) | 21.8 (22.1) |
| Completeness (%) | 99.9 (99.9) | 99.9 (100) | 100 (100) | 98.3 (84.5) | 99.9 (100) | 99.6 (100) | 99.4 (96.6) | 97.0 (85.8) | 99.9 (99.9) | 100 (100) |
| Rmerge (%)b | 0.065 (0.484) | 0.063 (0.343) | 0.099 (0.465) | 0.059 (0.490) | 0.092 (0.488) | 0.050 (0.455) | 0.043 (0.453) | 0.058 (0.444) | 0.060 (0.480) | 0.103 (0.475) |
| Refinement statistics | ||||||||||
| Resolution range (Å) | 40.0–1.82 | 40.0–1.80 | 20.0–1.98 | 20.0–1.44 | 20.0–2.40 | 20.0–2.20 | 20.0–1.90 | 20.0–2.05 | 20.0–2.0 | 20.0–2.30 |
| Reflections used in the refinement | 18 201 | 19 044 | 14 174 | 35 802 | 8231 | 10 634 | 16 314 | 12 656 | 14 084 | 9417 |
| Total number of reflections used for working set | 17 097 | 18 044 | 13 468 | 34 020 | 7631 | 10 094 | 15 485 | 11 901 | 13 324 | 8753 |
| R (%)c | 21.2 | 19.60 | 20.90 | 20.70 | 22.90 | 23.20 | 24.2 | 23.8 | 25.1 | 23.20 |
| Total number of reflections used for Rfree | 1104 | 965 | 706 | 1782 | 600 | 540 | 829 | 755 | 760 | 664 |
| Rfree (%)d | 23.0 | 21.38 | 24.50 | 22.30 | 26.40 | 25.80 | 24.4 | 26.5 | 27.7 | 26.70 |
| No. of protein atoms | 1233 | 1233 | 1232 | 1200 | 1214 | 1226 | 1227 | 1203 | 1209 | 1231 |
| No. of ligand atoms | 1 | 2 | 0 | 1 | 0 | 2 | 1 | 1 | 1 | 1 |
| No. of water molecules | 117 | 121 | 115 | 195 | 58 | 43 | 64 | 58 | 99 | 78 |
| Average B factor (Å2) | 31.1 | 26.3 | 25.8 | 21.7 | 54.6 | 51.9 | 50.2 | 55.5 | 43.6 | 37.2 |
| Ramachandran statistics | ||||||||||
| Most favored regions | 91.9 | 90.4 | 89.7 | 92.3 | 88.9 | 92.6 | 90.4 | 90.4 | 89.6 | 89.7 |
| Allowed regions | 8.1 | 9.6 | 10.3 | 7.7 | 11.1 | 7.4 | 9.6 | 9.6 | 10.4 | 10.3 |
| PDB code | 2E7W | 2E7X | 2EFN | 2PN6 | 2EFO | 2EFP | 2PMH | 2YX7 | 2YX4 | 2EFQ |
All of the crystals belong to the space group I422; number of molecules per asu is 1; and the solvent content is about 56%.
All of the refined structures contain one Mg2+ ions, Rmsd in Bond lengths and Bond angles are varied from 0.005 to 0.007 (Å) and 1.1–1.3 (°), respectively.
aValues in parentheses are for the highest resolution shell.
bRmerge = ∑h ∑i | I(h, i) − <I(h)> | /∑h ∑i I(h, i), where I(h, i) is the intensity value of the ith measurement of h and <I(h)> is the corresponding mean value of I(h) for all i measurements.
cR factor = (∑ | |Fobs| − |Fcalc| |/∑|Fobs|)×100, where |Fobs| and |Fcalc| are the observed and calculated structure factor amplitudes, respectively.
dRfree is the same as R factor, but for a 5% subset of all reflections.
Structure determination and refinement
The native Grp structure was determined by the molecular replacement method, using the structure of an archaeal feast/famine regulatory protein, FL11 (PDB code, 1RI7), as a search model. The solution was found by auto-molrep, within the CCP4 program suite, and the refinement was carried out using CNS (32). The protein model was built using the programs Quanta (33) and Coot (34). The final model with 150 residues was refined to a crystallographic R-value of 0.212 (Rfree = 0.230) at 1.82 Å resolution. The structures of the Grp-Gln complex and the S32A, S32A-Gln, Y77A, Y77A-Gln, T132A, T132A-Gln, T134A and T134A-Gln mutants were determined using the native structure of Grp. The final refinement statistics for all of these structures are listed in Table 1. Figures were prepared with the programs Ribbons (35) and Pymol (36).
Site-directed mutagenesis of Grp
Initially, the Grp plasmid was prepared with a Qiagen miniprep kit. A Quickchange site-directed mutagenesis kit (Stratagene) was used to create the Grp mutants (S32A, Y77A, D102A, T132A and T134A), and the plasmids were transformed into JM109 cells. For all of the mutants, N-terminal sequencing was carried out, and they were expressed and purified in a similar manner as the native protein (29).
DNA synthesis, labeling and filter binding assay
Based on a promoter analysis of the grp gene, we prepared four oligos. These oligos (5′-CGTATCTTCCAAAGATTATAAGCGATTTTTTGTAAGTCTTTTTTTATTATAAGTGTTGATTAGATATAA TTCAAAGTAAAGGCTTATATTTTTGTTTATAATATACAGTCT-3′ (111-mer), 5′-CGTATCTTCCAAAGATTATAAGCGATTTTTTGTAAGTCTTTTTTTATTAT-3′ (50-mer), 5′-TAAGTGTTGATTAGATATAATTCAAAGTAAAGGCTTATATTTTTGTTTATAATATACAGTCT-3′ (62-mer) and 5′-ATATTTTTGTTTATAATA-3′ (18-mer)) and their complementary strands were purchased from Fasmac DNA oligos, Japan. The oligos were purified and desalted before annealing to form duplex DNA. The duplex DNAs were used for the binding analysis. We labeled one of strands in each duplex at the 5′ end with [γ-32P]ATP (GE Healthcare, Biosciences) in the presence of T4 polynucleotide kinase (Takara, Japan), and the labeled products were recovered after fractionation by 15% PAGE. To determine the equilibrium dissociation constant (Kd) for these DNAs with Grp, we carried out filter binding assays.
To evaluate the binding activities of the prepared duplex DNAs (111-mer, 50-mer, 62-mer and 18-mer), and to analyze the l-glutamine binding site mutants, a filter binding assay was performed (37,38). Various amounts of the purified Grp protein were incubated in binding buffer (10 mM Tris-HCl pH 8.0, 200 mM NaCl, 20 mM MgCl2, 5 mM β-mercaptoethanol) to give final concentrations of 0–2 μM, and then l-glutamine was added to a final concentration of 10 mM. To this reaction mixture, the DNA duplex was added to a final concentration of 100 nM. The resulting reaction mixture was incubated for 15 min at 27°C, and then was applied to a Millipore nitrocellulose membrane (HAWP filter, 0.45 μM, 13.0 mm diameter), which was previously equilibrated in the binding buffer. The membrane was washed once with 1 ml of binding buffer containing l-glutamine. In the control experiments, the l-glutamine was not included in the reaction mixture, and the washing was omitted. The amount of duplex DNA (radioactivity) retained on the filter was quantitated using a bio image analyzer, BAS 2500 (Fuji Film). The equilibrium dissociation constants (Kd), were determined for different DNA-protein complexes, using the GraphPad Prism 3.0 non-linear regression algorithm (GraphPad Software, USA).
RESULTS AND DISCUSSION
DLS characterization of Grp
To determine whether Grp, like the other members of the Lrp/AsnC family of proteins, forms oligomers in solution, we carried out dynamic light scattering (DLS) experiments. The DLS analysis clearly showed a sharp monomodal fit of the data centered at a 4.6 nm hydrodynamic radius, indicating a homogeneous species that forms an oligomeric structure with an approximate molecular weight of 140 kDa (the calculated molecular mass of the monomer is ∼17 kDa) (data not shown), suggesting that it forms an octameric structure. Several measurements were taken with three protein concentrations (3.0, 1.7 and 0.87 mg/ml), and similar results were obtained (data not shown). The DLS observations thus coincide with the number of molecules in the unit cell of the present crystal structure (8 monomers), which is consistent with the crystal structures of other Lrp/AsnC family proteins.
Overall structure of native Grp
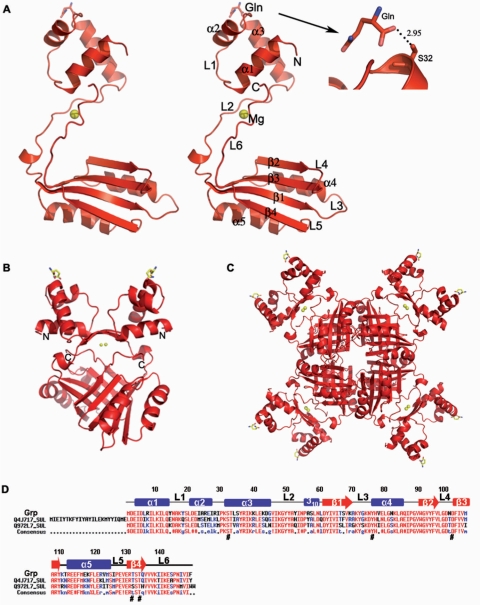
The overall structure of Grp belongs to the α/β family of proteins. It consists of five α-helices and four β-strands, arranged in the order of α1-α2-α3-β1-α4-β2-β3-α5-β4 in the primary structure (Figure 1A–C). The N-terminal α1-3 form a separate helical domain, called the DNA-binding domain, which is composed of ∼45 residues. The β-strands (β1-4) and two other helices (α4 and α5) form another domain, known as the effector-binding domain, which accounts for ∼75 residues. These two domains are well separated, and are connected by a 15 residue loop, L2. Interestingly, the ligand l-glutamine was identified in the DNA-binding domain at helix α3, and we believe that this ligand was derived from the E. coli host. A sequence comparison of Grp with those of the archaeal AsnC from S. acidocaldarius (Q4J717) and the Lrp from S. tokodaii (Q972L7) revealed that the DNA-binding domain residues, especially, within the α2 and α3 regions, are less conserved than the effector-binding domain residues (Figure 1D). However, it is noteworthy to mention that the Ser32 residue of the DNA-binding domain, which interacts with the glutamine ligand, is highly conserved (Figure 1D). The asymmetric subunit contains one molecule, with overall dimensions of 60 × 42 × 30 Å3. In the dimer, the two monomers are related by a crystallographic 2-fold axis, perpendicular to the plane of Figure 1B. The total solvent area buried upon dimerization was ∼2762 Å2 (27%) per monomer. The dimer was stabilized by hydrophobic and hydrogen bonding interactions between the β-sheets and the linker region of the monomers. Four dimers are symmetrically arranged around a crystallographic 4-fold axis and form the quaternary structure, the octameric assembly (Figure 1C). The surface buried by the dimer–dimer association resulted in an accessible surface area loss of ∼2779 Å2 (18%) per dimer. The percentage of solvent accessible surface area buried upon dimerization and oligomerization is comparable to those of E. coli Lrp and B. subtilis AsnC (20). The formation of the octamer is stabilized by the effector domains, especially by the hydrophobic contacts established by the dimer–dimer interface.
Figure 1.
Crystal structure of Grp and amino acid sequence comparison of the closely related proteins. (A) Stereo view of the Grp monomer. Grp is shown in a ribbon model with labels for α-helices, β-strands and loop regions. The bound Mg2+ ion, shown in a cpk model. The N- and C-termini of Grp are indicated by N and C, respectively. (B) Dimer representation of Grp, formed by the crystallographic 2-fold axis. The N- and C-termini are labeled by N and C, respectively. (C) View of the Grp octamer along the 4-fold axis. (D) Sequence alignment of Grp and its closely related proteins, Grp (Q972W6_SUL) from S. tokodaii, AsnC (Q4J717_SUL) from S. acidocaldarius and Lrp (Q972L7) from S. tokodaii. Conserved residues are indicated by red letters. The secondary structural elements in the primary sequences of Grp are indicated as α-helices (bars), β-strands (arrows) and loops (lines).
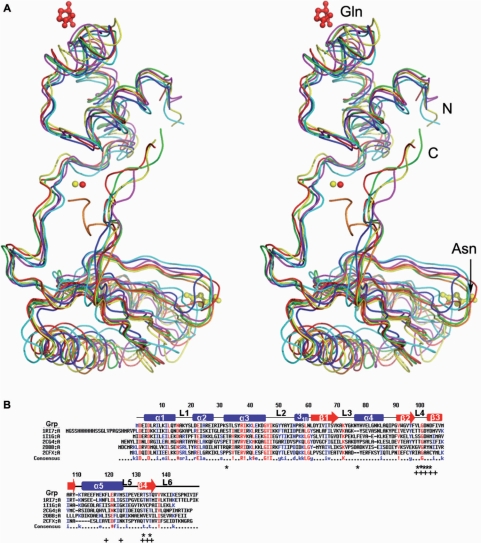
A structural similarity search of the Grp monomer, using DALI (39), readily identified six Lrp/AsnC family proteins, with Z-scores ranging between 11 and 16. These proteins were Pyrococcus sp. FL11 (PDB code, 1RI7), P. furiosus Lrp (PDB code, 1I1G), E. coli AsnC (PDB code, 2CG4), B. subtilis Lrp (PDB code, 2CFX) and P. horikoshii regulators (PDB codes, 2DBB, 2CVI). Superposition of the Grp monomer with these six structures revealed that the overall topology is very similar (Figure 2A). The sequence alignments between Grp and these five proteins showed that FL11 had ∼40% identity, and the identity of all of the other proteins ranged between 23 and 29% (Figure 2B).
Figure 2.
Stereo view of the 3D structures of Grp and sequence comparison of Grp. (A) Superposition of the Grp structure with those of related Lrp/AsnC family proteins. Grp, FL11 (PDB code,1RI7), Lrp (PDB code,1I1G), AsnC (PDB code, 2CG4), Lrp (PDB code, 2CFX) and two putative regulators (PDB codes, 2DBB, 2CVI) are colored red, green, blue, yellow, magenta, cyan and orange, respectively. The bound Mg2+, glutamine, and asparagine are represented by cpk and ball-and-stick models, respectively. (B) Sequence comparison of Grp with other Lrp/AsnC family proteins with known structures. Grp (ST1022), FL11(1RI7), Lrp (I1G), AsnC (2CG4), a putative regulator (2DBB) and Lrp (2CFX) are compared. The secondary structural elements in the primary sequences of Grp are indicated: α-helices (bars), β-strands (arrows) and loops (lines). The glutamine binding residues in Grp (ST1022) and the asparagine binding residues in AsnC (2CG4) are indicated by asterisks and plus signs, respectively.
Crystal structure of Grp complexed with exogenous l-glutamine
The in vitro and genetic analyses of many of the Lrp-like proteins clearly demonstrated that all of these proteins are involved in amino acid metabolism, and that amino acids, such as leucine, asparagine, lysine, glutamine, proline and methionine, serve as ligands (15,20–28). A gel-filtration analysis of the FL11 protein revealed that it forms a higher order assembly structure in the presence of l-glutamine (19). However, the crystal structure of FL11 with glutamine was not available to clarify the mode of ligand binding and its importance for the function. Since FL11 showed ∼40% sequence identity to Grp, it is reasonable to assume that l-glutamine may be a potential effector ligand for Grp. Fortunately, in our native structure, an endogenous l-glutamine molecule was identified at the DNA-binding domain. This clearly suggests that l-glutamine is the probable target ligand for Grp activation. However, the position of the l-glutamine in the native Grp structure is different from the position of the Asn ligand in the structure of E. coli AsnC. In the latter complex structure, the ligand was bound to the effector domain. The lack of l-glutamine binding to the effector domain in the native Grp structure is probably due to the low concentration of endogenous l-glutamine in the protein solution. Based on this presumption, we co-crystallized the Grp with 33 mM of exogenous l-glutamine, to determine whether the ligand could bind to the effector domain. This complex structure (Grp-Gln) was solved by molecular replacement, with the native Grp structure as a model. The data collection and refinement statistics for this complex are shown in Table 1.
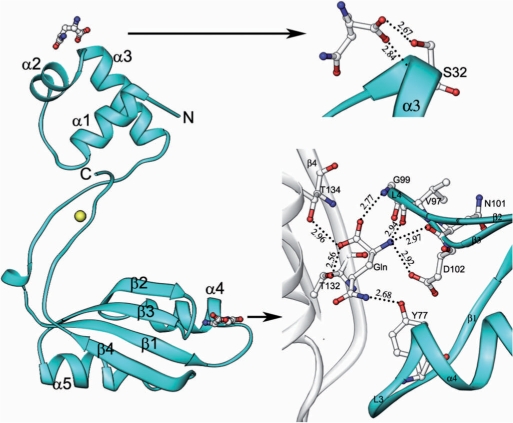
The overall conformation of the complex structure was similar to the native structure, with an r.m.s.d. of 0.83 Å for superposition of 150 Cα atoms (Figure 3). The exceptional quality of the electron density map allowed us to identify two specific glutamine-binding sites unambiguously in the complex structure. One glutamine binds to the N-terminal DNA-binding domain with the α3 helix, as observed in the native protein, and the other one binds to the effector-binding domain at the cleft between the loops L3 (connecting the β1 and α4) and L4 (connecting the β2 and β3) (Figure 3). The latter glutamine has many interactions with the protein molecule, as compared to the former one. At the DNA-binding domain, the C-terminal oxygen atoms OXT and O of the endogenous l-glutamine, form hydrogen bonding interactions with the backbone nitrogen and the side chain oxygen of S32, respectively. At the effector domain, the side chain atom NE2 of l-glutamine hydrogen bonds with the side chain oxygen of Y77. The other side chain atom, OE1, is involved in a water-mediated hydrogen bond with the side chain atom of T132 of the symmetry-related molecule. The backbone nitrogen of the ligand forms a trifurcated hydrogen bond with the side chain atom OD1 of D102 and the backbone oxygens of N101 and V97. The C-terminal oxygen (O) of the ligand forms a bifurcated hydrogen bond with the backbone nitrogens of G99 and the symmetry-related molecule's T134. The other C-terminal oxygen (OXT) also forms a bifurcated hydrogen bond to the side chain OG1 atoms of T132 and T134 of the symmetry-related molecule (Figure 3). These interactions, involving the ligands at both the DNA-binding domain and effector domain, clearly showed that the residues S32, Y77, D102, T132 and T134 are important for the protein-ligand binding. It is interesting to note that these residues are highly conserved within AsnC from S. acidocaldarius and Lrp from S. tokodaii (16,22) (Figure 1D). In the crystal structure of the E. coli AsnC-l-asparagine complex, the ligand occupies the same binding site as observed in the effector-binding site of Grp, although the interaction modes are different (20).
Figure 3.
Crystal structure of the Grp complex bound to exogenous l-glutamine (Gln) and its binding site. On the left side, Grp is shown in a ribbon diagram with labels for α-helices and β-strands, and the bound glutamines are shown as ball-and-stick models colored by atom type (nitrogen, blue; carbon, white; oxygen, red). The neighboring molecule in the dimer, which is involved in interactions with the glutamine, is represented by a white ribbon model. Close-up view of the glutamine-binding site in Grp (right side). Hydrogen bonds are indicated by broken lines. The residues that are involved solely in the interaction with the glutamine are represented by stick models, colored by atom type as mentioned previously.
Conformational change
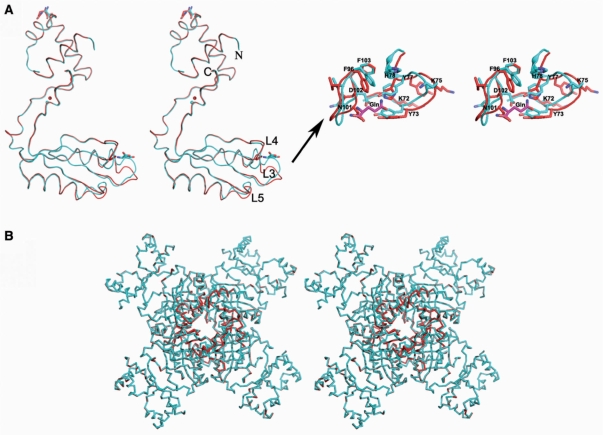
A structural comparison of the native Grp with the complex structure revealed significant conformational changes at two sites in the effector domain-binding site (Figure 4). The first site is the ligand binding region, which is formed by the loops L3 and L4. The second site is the loop L5. A closer inspection of the L4 loop revealed that the accommodation of the glutamine ligand required the backbone and side chain conformations of N101 and D102 to change drastically, such that their Cα positions moved ∼4.5 Å from their original positions. In the native structure, the N101 residue is located inside the ligand binding pocket, whereas D102 is oriented outside (Figure 4). In the presence of exogenous glutamine ligand, a switch-like movement takes place, involving the residues N101 and D102 such that N101 is now located outside the pocket, while D102 has moved inside. In the liganded state, D102 makes a hydrogen bonding interaction with the backbone nitrogen of the ligand. A similar switch-like movement is also observed at the L3 loop, involving the residues Y77 and H78. In the native structure, Y77 is located outside the ligand binding pocket, while H78 is inside. As a result of ligand binding, Y77 and H78 switch their positions. In its new position, Y77 hydrogen bonds with the NE2 atom of the ligand (Figures 3 and 4). A recent analysis of the AsnC protein from Neisseria meningitidis revealed that l-leucine and l-methionine binding increases the thermal stability for octamer formation; however, the crystal structures did not provide any clues about possible conformational changes around the ligand-binding sites (40). Very recently, crystal structure of MtLrp from Mycobacterium tuberculosis H37Rv were complexed with the ligands Phe, His, Tyr, Trp, Leu and Met and the ligand-binding site all the complexes were moved up ∼2 Å as compared with the Gln-binding site (41). The Pyrococcus sp. FFRP, DM1 structures were complexed with Ile and SetMet, the bound ligand occupies the same binding site as observed in the effector-binding site of Grp, although the interaction modes are different (42).
Figure 4.
Conformational changes observed in the Grp complex. (A) Superposition of the native Grp and the exogenous glutamine-bound form (complex) of the Grp protein, represented by a ribbon model (left panel). The native and complex structures are colored red and blue, respectively. The bound glutamines are depicted by stick models. Close-up view of the conformational changes observed in the effector domain are shown in the right panel. (B) Stereo view of the conformational change in the Grp octamer. The free and complex molecules are colored as in Figure 4A.
The solvent accessible surface area buried upon dimerization and oligomerization of the complex increased insignificantly, as compared to the native Grp structure. As we mentioned previously, the octamer formation occurred independent of the ligand; however, the conformational changes observed in the loops and the associated secondary structural elements in the effector domain are evidence of further stabilization of the oligomeric structure (Figure 4B). In addition, the residues T132 and T134 are highly conserved in all of the known Lrp/AsnC family proteins (Figure 2B), and their roles in effector binding suggest that they are important for dimer–dimer interface stabilization in the presence of the ligand, which would further reinforce the quaternary structure. Thus, the glutamine-bound complex increases the stability of the oligomeric structure by forming more hydrogen bonding interactions between the dimers and also through the ligand. The conformational change upon ligand binding seen in the crystal structure of Grp is supported by the previous biochemical studies on E. coli Lrp and P. putida BkdR, which noted a decrease in the intrinsic fluorescence upon the binding of their amino acid effectors to high affinity sites, suggesting a conformational change in both proteins (21,27). In addition, a slight inter-domain rearrangement has been observed upon effector binding in the structurally analogous ACT domain-containing proteins, which are also subject to allosteric regulation by small molecules, typically amino acids (28).
Characterization of the grp gene promoter and modeling of the Grp-DNA complex
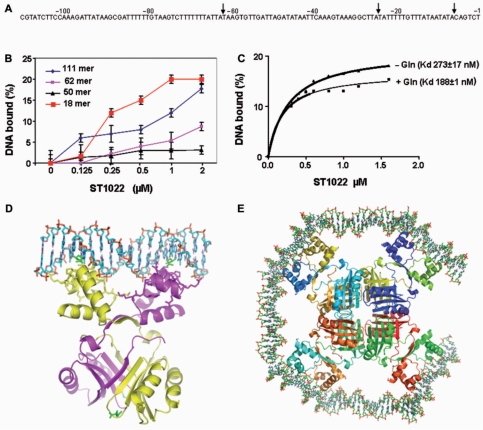
The Lrp/AsnC family proteins bend DNA and form oligomers; therefore, DNA wrapping may play an important role in productively aligning the DNA-bound activator with its targets in the core transcriptional apparatus and, possibly, even in the postrecruitment steps of transcription initiation. DNA bending by the Lrp activators at close proximity to a TATA-box-bound TBP may form the basis of the activation mechanism. The E. coli Lrp protein binds to specific sequences in the promoters of many different genes, including its own (43), and S. solfataricus Lrs14 also specifically binds to multiple sequences in its own promoter (44). In both cases, the binding region overlaps the promoter, suggesting that Lrp and Lrs14 are autoregulated. Therefore, we sought to identify the target sequences of Grp, in order to characterize its DNA-binding site. We used the S. solfataricus Lrs14 promoter sequence as the search model for the Grp protein, and we searched for sequence similarity with a non-coding probe spanning 743 basepairs (bp), located in-between the immediate upstream region of the first ATG of the ST1022 (grp) gene and downstream of the STS117 gene. Interestingly, this analysis suggested that the region spanning from −1 to −111 may be a potential region for Grp recognition (Figure 5A). To test this possibility, a 111-bp duplex oligonucleotide encoding this region was prepared and analyzed for binding with a filter binding assay, using increasing concentrations of Grp. The binding analysis revealed that Grp binds to the 111-bp duplex (Figure 5B). This 111-bp duplex is quite stable, with a free energy of ΔG = −120.4 kcal/mol, calculated using the RNA structure program, version 4.2 (45). Modeling of the DNA duplex with E. coli AsnC suggested that it needs at least 13 bp to cover the DNA-binding site of one monomer, and 18 bp for the spacing between the binding sites for one dimer. Thus, the octameric molecule would require ∼124 bp to cover all four binding sites, which is comparable to our predicted region (−1 to −111) to cover octameric ring of Grp, with a diameter of 127 Å.
Figure 5.
Identification of the probable promoter region, filter binding assay and modeling for Grp. (A) Identification of the probable promoter region for Grp, predicted from the genomic sequences. The fragmented promoter regions (−1 to −62 (62 bp), −62 to −111 (50 bp) and −7 to −25) are highlighted by arrows. (B) A filter binding assay was used to measure the formation of the protein-DNA complexes. Binding reactions were carried out in binding buffer with 100 nM of DNA in the presence of 10 mM l-glutamine. The amount of complex retained on the filter after washing was used to estimate the dissociation constants. (C) Allosteric activation of Grp by l-glutamine. The requirement of glutamine for the DNA binding was analyzed by a filter binding assay, as described in Figure 5B, by using the 18-bp DNA. (D) Possible interaction of the Grp dimer with the 18-bp duplex DNA. The Grp is represented by a ribbon model, and the DNA and glutamines are represented by stick models. (E) A possible arrangement of the DNA binding domains in the octameric molecule. Each monomer in the octamer is represented by a different color. The 14-bp DNA fragments were modeled and placed in-between the docked DNA and the Grp dimer, assuming that the DNA would wrap around the octameric molecule. Since both of the fragments were modeled as B-form DNA and were arranged to cover the octamer ring, the ends of the fragments became close to each other.
To determine the minimum number of base pairs required for Grp binding, we prepared three more DNA fragments containing the regions of −1 to −62 (62-bp), −62 to −111 (50-bp) and −7 to −25 (18-bp) and their complementary strands, and performed a binding analysis with Grp. The 62-bp and 50-bp duplexes showed lower affinity as compared to the 111-bp duplex, and quite interestingly, the 18-bp duplex showed the highest affinity among the four tested duplexes (50 bp < 62 bp < 111 bp < 18 bp) (Figure 5B). This analysis revealed that the 18-bp duplex is the minimum required length for binding to Grp, with affinity comparable to or higher than that of the 111-bp DNA. Further reduction of the length of the duplex may destabilize the protein-DNA complex, since the 18-bp duplex itself had a free energy ΔG = −13.4 kcal/mol. We next analyzed the importance of l-glutamine for Grp oligomerization and DNA binding. The 18-bp minimum DNA duplex was used for the binding studies, and the dissociation constants were calculated. In the presence of l-glutamine, the Kd was 188 ± 1, and in its absence, it was 273 ± 17 nM. This suggests that in both cases, the DNA binds to the protein; however, in the absence of ligand (l-glutamine), the affinity was reduced by onefold (Figure 5C). It is interesting to note that we could not rule out the possibility that endogenous glutamine might have activated DNA binding in the absence of exogenous ligand in the binding assay. In the presence of additional ligand, the effector domain might have become further stabilized for DNA recognition.
Based on our DNA binding analysis, the 18-bp duplex DNA showed affinity comparable to that of the full-length DNA (111 bp), and hence we modeled the 18-bp B-DNA duplex and oriented it to interact with the HtH motif of the Grp protein, which functions in DNA recognition (Figure 5D and E). Although a 13-bp duplex is sufficient to cover the DNA binding surface in the dimer interface, as predicted for other Lrp/AsnC family proteins (19,20), we used 18 bp, based on our analysis and discussed in supplementary discussion-1. Also, a recent structural analysis of the Pyrococcus sp. OT3 FL11-DNA complex revealed that some of our predicted residues, especially, K31, S32. T33 and K39 were interacting with the DNA (46).
Evaluation of the l-glutamine binding site in the effector domain by mutagenic and structural analyses
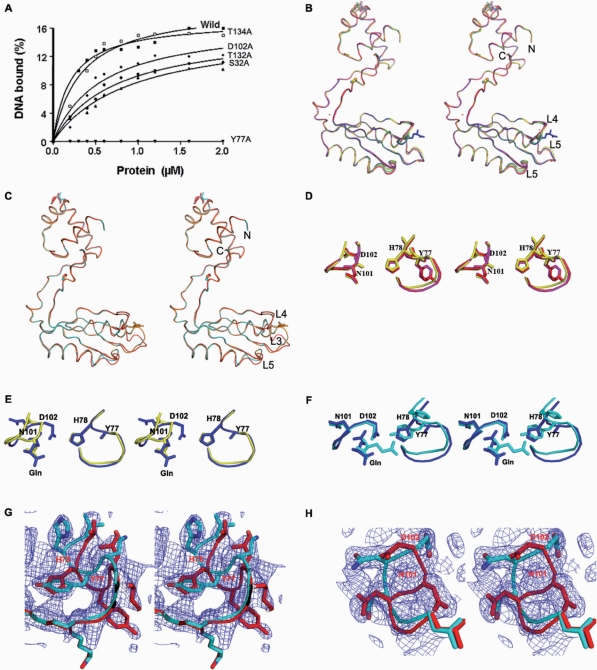
To evaluate the interactions of both exogenous and endogenous l-glutamine with the Grp molecule, we prepared five Grp protein mutants (S32A, Y77A, D102A, T132A and T134A). N-terminal sequencing was performed for all of the purified mutants, and their ability to oligomerize was analyzed by DLS experiments, which yielded results similar to those obtained, one observed for the native Grp. For the DNA-binding studies, we used the 18-bp duplex DNA, since it showed the highest affinity towards Grp, and a filter binding assay was performed (Figure 6A).
Figure 6.
Biochemical and structural snap-shots of the ligand-binding site in the effector domain. (A) DNA binding analyses were carried out to identify the binding ability of the glutamine-binding site mutants, using the 18-bp duplex DNA. The binding reactions were carried out in the presence of 10 mM l-glutamine, as described in Figure 5A. (B) Superposition of Grp mutant structures. The native, S32A, Y77A, Y77A-Gln, T132A, T132A-Gln and T134A structures are colored red, magenta, yellow, blue, orange, wheat and green, respectively. (C) The crystal structures of S32A-Gln and its complex, superimposed on the native Grp protein. The native, complex and S32A-Gln structures are colored red, cyan and orange, respectively. (D–H) Stereo view of structural snap-shots of the effector-binding domain site at the L3 and L4 loops, and mode of glutamine ligand recognition. (D) Superposition of the L3 and L4 loops of the S32A and Y77A mutants on those of the native Grp. (E) Superposition of Y77A and Y77A-Gln. (F) Comparison of the Y77A-Gln and complex structures. (G) Loops L3, showing the two conformations in T132A-Gln and T134A-Gln. (H) Loop L4, showing the two conformations in T134A-Gln.
Among the five mutants, Y77A was completely unable to bind to the DNA. The binding ability of the S32A mutant was reduced by approximately fivefold, and the other mutants, such as D102A, T132A and T134A, showed ∼2- to 4-fold lower binding abilities (Figure 6A), as compared to native Grp. These results provide strong evidence that Y77 is important for DNA recognition, through binding to the l-glutamine. Except for the S32A mutant at the DNA-binding domain, all of the other mutations are at the effector-binding domain. To clarify how these mutants affect the DNA-binding ability of the protein, we crystallized and solved the structures of eight additional mutants and their complexes, including S32A, S32A-Gln, Y77A, Y77A-Gln, T132A, T132A-Gln, T134A and T134A-Gln. All of these structures were solved by molecular replacement, with the native Grp as a search model, and the refinement statistics are provided in Table 1.
The overall structural topology of all of the mutants is similar to the native structure (Figure 6B and C) and they also form an octameric assembly. The S32A, Y77A, T132A, T132A-Gln and T134A structures resemble the native structure, and there was no conformational change observed at the effector binding site (Figure 6D). Interestingly, the endogenous l-glutamine that interacts with S32 of the DNA-binding domain in the native Grp was not found in the S32A mutant structure. Therefore, the S32A mutant structure represents Grp without a ligand at both the DNA binding and effector-binding domains, and it still forms an octamer. This clearly suggests that the ligand is not an absolute requirement for octamer formation, which is also consistent with the previously reported studies of P. furiosus LrpA (19) and E. coli Lrp (11,15).
The overall structure of S32A-Gln resembles the complex structure, and the ligand-binding site at the effector domain also displayed a similar structural rearrangement. This provides further confirmation that the reduced binding activity is due to the lack of one hydrogen bond to the glutamine at the DNA-binding site (Figures 3 and 6C). In Y77A-Gln, a ligand was observed at the effector-binding site, as in the complex; however, the binding modes are significantly different in these two structures. In addition, the mutant (Y77A) complex structure exhibits no conformational change at the L3 loop (residues 72–79), whereas in the L4 loop region (residues 99–102), the conformation has changed, as observed in the complex (Figure 6E and F). This shows that the Y77 residue plays an important role in correcting the conformational change upon ligand binding. In the T132A-Gln structure, the L3 loop adopts two conformations, resembling those observed in the native (inactive) structure and the Grp complex (active) structure. Similarly, both the L3 and L4 loops in the T134A-Gln structure exhibit two conformations (Figure 6G and H). This suggests that the mutation of both the T132 and T134 residues would hamper the ability of the L3 and L4 loops to adopt the active conformation, even in the presence of the ligand. Therefore, we hypothesize that the T132 and T134 residues are important for fixing the proper orientation of the L3 and L4 loops for binding to the ligand, which eventually will generate more hydrogen bonds between the dimers at their interface. These additional hydrogen bonds would further stabilize the octameric structure of Grp. It is important to note that the T132 and T134 residues are highly conserved in all of the known Lrp/AsnC family proteins (Figure 2B)
CONCLUSIONS
The present structural studies revealed the crystal structure of Grp regulator for the first time in the ligand-free and glutamine-bound forms. A comparison of these two structures showed that a significant conformational change occurs at the effector-binding domain upon l-glutamine binding. Closer inspection of this region suggested that this conformational change is responsible for the formation of more hydrogen bonds at the interface of the two dimers. These additional interactions provide more stability to the functional molecule, the octamer, as compared to the native, octameric structure. Further biochemical and mutant structural analyses at the ligand-binding site suggested that the residues interacting with the glutamine are important for the assembly of the octamer, which is required for DNA recognition. In other words, in the presence of l-glutamine, Grp selects one possible conformation of the protein, which is suitable for acting at subsets of the target DNA sites but unsuitable for others, presumably by altering the relative orientation and spacing of the DNA-binding and ligand-binding domains and the associated bound DNA. This proposal is consistent with our present analyses, since comparisons of the native, complex and mutant structures revealed a significant conformational change upon ligand binding at the effector domain, which might support the dimer–dimer and ligand–protein interfaces for further stabilization of octamer formation. We believe a similar mode of ligand activation exists for the other Lrp/AsnC family proteins. Further protein-DNA complex and biochemical analyses are required to obtain more insights into this mode of DNA recognition and bending, and these studies are currently in progress.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors would like to thank C. Kuroishi for cloning, E. Matsunaga and M. Nishio for purification, T. Tanaka for DLS, and K. Gayatri for PDB submission. This work was supported by the RIKEN Structural Genomics/Proteomics Initiative (RSGI), the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science and Technology of Japan. The Open Access publication charges were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Newman EB, Lin R. Leucine-responsive regulatory protein: a global regulator of gene expression in E. coli. Annu. Rev. Microbiol. 1995;49:747–775. doi: 10.1146/annurev.mi.49.100195.003531. [DOI] [PubMed] [Google Scholar]
- 2.Calvo JM, Matthews RG. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinkman AB, Ettema TJ, de Vos WM, van der Oost J. The Lrp family of transcriptional regulators. Mol. Microbiol. 2003;48:287–294. doi: 10.1046/j.1365-2958.2003.03442.x. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki M. Structure and function of the feast/famine regulatory proteins, FFRPs. Proc. Jpn. Acad. 2003;79B:274–289. [Google Scholar]
- 5.Bell SD, Jackson SP. Mechanism and regulation of transcription in archaea. Curr. Opin. Microbiol. 2001;4:208–213. doi: 10.1016/s1369-5274(00)00190-9. [DOI] [PubMed] [Google Scholar]
- 6.Ouhammouch M. Transcriptional regulation in Archaea. Curr. Opin. Genet. Dev. 2004;14:133–138. doi: 10.1016/j.gde.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Geiduschek EP, Ouhammouch M. Archaeal transcription and its regulators. Mol. Microbiol. 2005;56:1397–1407. doi: 10.1111/j.1365-2958.2005.04627.x. [DOI] [PubMed] [Google Scholar]
- 8.Dahlke I, Thomm M. A Pyrococcus homolog of the leucine-responsive regulatory protein, LrpA, inhibits transcription by abrogating RNA polymerase recruitment. Nucleic Acids Res. 2002;30:701–710. doi: 10.1093/nar/30.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiorentino G, Cannio R, Rossi M, Bartolucci S. Transcriptional regulation of the gene encoding an alcohol dehydrogenase in the archaeon Sulfolobus solfataricus involves multiple factors and control elements. J. Bacteriol. 2003;185:3926–3934. doi: 10.1128/JB.185.13.3926-3934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ouhammouch M, Langham GE, Hausner W, Simpson AJ, El-Sayed NM, Geiduschek EP. Promoter architecture and response to a positive regulator of archaeal transcription. Mol. Microbiol. 2005;56:625–637. doi: 10.1111/j.1365-2958.2005.04563.x. [DOI] [PubMed] [Google Scholar]
- 11.Willins DA, Ryan CW, Platko JV, Calvo JM. Characterization of Lrp, and Escherichia coli regulatory protein that mediates a global response to leucine. J. Biol. Chem. 1991;266:10768–10774. [PubMed] [Google Scholar]
- 12.Madhusudhan KT, Huang N, Sokatch JR. Characterization of BkdR-DNA binding in the expression of the bkd operon of Pseudomonas putida. J. Bacteriol. 1995;177:636–641. doi: 10.1128/jb.177.3.636-641.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jafri S, Evoy S, Cho K, Craighead HG, Winans SC. An Lrp-type transcriptional regulator from Agrobacterium tumefaciens condenses more than 100 nucleotides of DNA into globular nucleoprotein complexes. J. Mol. Biol. 1999;288:811–824. doi: 10.1006/jmbi.1999.2715. [DOI] [PubMed] [Google Scholar]
- 14.Brinkman AB, Dahlke I, Tuininga JE, Lammers T, Dumay V, de Heus E, Lebbink JH, Thomm M, de Vos WM, van der Oost J. An Lrp-like transcriptional regulator from the archaeon Pyrococcus furiosus is negatively autoregulated. J. Biol. Chem. 2000;275:38160–38169. doi: 10.1074/jbc.M005916200. [DOI] [PubMed] [Google Scholar]
- 15.Chen S, Rosner MH, Calvo JM. Leucine regulated self association of leucine-responsive regulatory protein (Lrp) from Escherichia coli. J. Mol. Biol. 2001;312:625–635. doi: 10.1006/jmbi.2001.4955. [DOI] [PubMed] [Google Scholar]
- 16.Kawarabayasi Y, Hino Y, Horikawa H, Jin-no K, Takahashi M, Sekine M, Baba S, Ankai A, Kosugi H, Hosoyama A, et al. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain7. DNA Res. 2001;8:123–140. doi: 10.1093/dnares/8.4.123. [DOI] [PubMed] [Google Scholar]
- 17.Chen L, Brügger K, Skovgaard M, Redder P, She Q, Torarinsson E, Greve B, Awayez M, Zibat A, Klenk HP, et al. The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J. Bacteriol. 2005;187:4992–4999. doi: 10.1128/JB.187.14.4992-4999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leonard PM, Smits SH, Sedelnikova SE, Brinkman AB, de Vos WM, van der Oost J, Rice DW, Rafferty JB. Crystal structure of the Lrp-like transcriptional regulator from the archaeon Pyrococcus furiosus. EMBO J. 2001;20:990–997. doi: 10.1093/emboj/20.5.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koike H, Ishijima SA, Clowney L, Suzuki M. The archaeal feast/famine regulatory protein: potential roles of its assembly forms for regulating transcription. Proc. Natl Acad. Sci. USA. 2004;101:2840–2845. doi: 10.1073/pnas.0400109101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thaw P, Sedelnikova SE, Muranova T, Wiese S, Sylvia A, Alonso JC, Brinkman AB, Akerboom J, Oost JV, Rafferty JB. Structural insight into gene transcriptional regulation and effector binding by the Lrp/AsnC family. Nucleic Acids Res. 2006;34:1439–1449. doi: 10.1093/nar/gkl009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen S, Calvo JM. Leucine-induced dissociation of Escherichia coli Lrp hexadecamers to octamers. J. Mol. Biol. 2002;318:1031–1042. doi: 10.1016/S0022-2836(02)00187-0. [DOI] [PubMed] [Google Scholar]
- 22.Chen S, Lannolo M, Calvo JM. Cooperative binding of the leucine-responsive regulatory protein (Lrp) to DNA. J. Mol. Biol. 2005;345:251–264. doi: 10.1016/j.jmb.2004.10.047. [DOI] [PubMed] [Google Scholar]
- 23.Kolling R, Lother H. AsnC: an autogenously regulated activator of asparagine synthetase A transcription in Escherichia coli. J. Bacteriol. 1985;164:310–315. doi: 10.1128/jb.164.1.310-315.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peekhaus N, Tolner B, Poolman B, Kramer R. The glutamate uptake regulatory protein (Grp) of Zymomonas regulator Lrp of Escherichia coli. J. Bacteriol. 1995;177:5140–5147. doi: 10.1128/jb.177.17.5140-5147.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho K, Winans SC. The putA gene of Agrobacterium tumefaciens is transcriptionally activated in response to proline by an Lrp-like protein and is not autoregulated. Mol. Microbiol. 1996;22:1025–1033. doi: 10.1046/j.1365-2958.1996.01524.x. [DOI] [PubMed] [Google Scholar]
- 26.Inoue H, Inagaki K, Eriguchi S, Tamura T, Esaki N, Soda K, Tanaka H. Molecular characterization of the mde operon involved in L-methionine catabolism of Pseudomonas putida. J. Bacteriol. 1997;179:3956–3962. doi: 10.1128/jb.179.12.3956-3962.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madhusudhan KT, Huang N, Braswell EH, Sokatch JR. Binding of L-branched chain amino acids causes a conformational change in BkdR. J. Bacteriol. 1997;179:276–279. doi: 10.1128/jb.179.1.276-279.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ettema TJ, Brinkman AB, Tani TH, Rafferty JB, Van Der Oost J. A novel ligand-binding domain involved in regulation of amino acid metabolism in prokaryotes. J. Biol. Chem. 2002;277:37464–37468. doi: 10.1074/jbc.M206063200. [DOI] [PubMed] [Google Scholar]
- 29.Nakano N, Kumarevel TS, Matsunaga E, Shinkai A, Kuramitsu S, Yokoyama S. Purification, crystallization and preliminary X-ray crystallographic analysis of ST1022, a putative member of Lrp/AsnC family of transcriptional regulators isolated from Sulfolobus tokodaii strain 7. Acta Cryst. 2007;F63:964–966. doi: 10.1107/S1744309107049962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McPherson A. Current approaches to macromolecular crystallization. Eur. J. Biochem. 1990;189:1–23. doi: 10.1111/j.1432-1033.1990.tb15454.x. [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 32.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.Oldfield TJ. A number of real-space torsion-angle refinement techniques for proteins, nucleic acids, ligands and solvent. Acta Crystallogr. 2001;D57:82–94. doi: 10.1107/s0907444900014098. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Carson M. Ribbons. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]
- 36.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- 37.Kumarevel TS, Fujimoto Z, Karthe P, Oda M, Mizuno H, Kumar PKR. Crystal structure of activated HutP: An RNA binding protein that regulates transcription of the hut operon in Bacillus subtilis. Structure. 2004;12:1269–1280. doi: 10.1016/j.str.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 38.Kumarevel TS, Mizuno H, Kumar PKR. Structural basis of HutP-mediated anti-termination and roles of the Mg2+ ion and L-histidine ligand. Nature. 2005;434:183–191. doi: 10.1038/nature03355. [DOI] [PubMed] [Google Scholar]
- 39.Holm L, Sander C. A review of the use of protein structure comparison in protein classification and function identification. Science. 1996;273:595–602. [Google Scholar]
- 40.Ren J, Sainsbury S, Combs SE, Capper RG, Jordan PW, Berrow NS, Stammers DK, Saunders NJ, Owens RJ. The structure and transcriptional analysis of a global regulator from Neisseria meningitidis. J. Biol. Chem. 2007;282:14655–14664. doi: 10.1074/jbc.M701082200. [DOI] [PubMed] [Google Scholar]
- 41.Shrivastava T, Ramachandran R. Mechanistic insights from the crystal structures of a feast/famine regulatory protein from Mycobacterium tuberculosis H37Rv. Nucleic Acids Res. 2007;35:7324–7335. doi: 10.1093/nar/gkm850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okamura H, Yokoyama K, Koike H, Yamada M, Shimowasa A, Kabasawa M, Kawashima T, Suzuki M. A structural code for discriminating between transcription signals revealed by the feast/famine regulatory protein DM1 in complex with ligands. Structure. 2007;15:1325–1338. doi: 10.1016/j.str.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 43.Wang Q, Wu J, Friedberg D, Platko J, Calvo JM. Regulation of the Escherichia coli Lrp gene. J. Bacteriol. 1994;176:1831–1839. doi: 10.1128/jb.176.7.1831-1839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Napoli A, Oost JV, Sensen CW, Charlebois RL, Rossi M, Ciaramella M. An Lrp-like protein of the hyperthermophilic archeaon Sulfolobus solfataricus which binds to its own promoter. J. Bacteriol. 1999;181:1474–1480. doi: 10.1128/jb.181.5.1474-1480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 46.Yokoyama K, Ishijima SA, Koike H, Kurihara C, Shimowasa A, Kabasawa M, Kawashima T, Suzuki M. Feast/Famine regulation by transcription factor FL11 for the survival of the hyperthermophilic archaeon Pyrococcus OT3. Structure. 2007;15:1542–1554. doi: 10.1016/j.str.2007.10.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.