Abstract
Considerable interest has been focused on inducing RNA interference (RNAi) in neurons to study gene function and identify new targets for disease intervention. Although small interfering RNAs (siRNAs) have been used to silence genes in neurons, in vivo delivery of RNAi remains a major challenge limiting its applications. We have developed a highly efficient method for in vivo gene silencing in dorsal root ganglia (DRG) using replication-defective herpes simplex viral (HSV-1) vectors. HSV-mediated delivery of short-hairpin RNA (shRNA) targeting reporter genes resulted in highly effective and specific silencing in neuronal and non-neuronal cells in culture and in the DRG of mice in vivo including in a transgenic mouse model. We further establish proof of concept by demonstrating in vivo silencing of the endogenous trpv1 gene. These data are the first to show silencing in DRG neurons in vivo by vector-mediated delivery of shRNA. Our results support the utility of HSV vectors for gene silencing in peripheral neurons and the potential application of this technology to the study of nociceptive processes and in pain gene target validation studies.
INTRODUCTION
RNA interference (RNAi) is an evolutionary conserved, post-transcriptional gene silencing mechanism mediated by two classes of small double-stranded RNA molecules: small interfering RNAs (siRNAs) and microRNAs (miRNAs). MiRNAs are endogenous, regulatory noncoding RNA molecules involved in many developmental and cellular functions (1–3) and have been recently implicated in the pathogenesis of human disease, including neurodegenerative disorders (4). Unlike siRNAs that originate in the cytoplasm, miRNAs are transcribed by RNA pol II as part of a long primary miRNA transcript (pri-miRNA). The pri-miRNA is processed in the nucleus by the enzyme Drosha into a hairpin intermediate, termed precursor miRNA (pre-miRNA), which is subsequently exported to the cytoplasm (5,6). Both siRNAs and miRNAs are generated by Dicer, and increasing evidence suggests that they can act in the same manner to mediate similar effects (7–9).
The recent discovery that RNAi operates in mammalian neurons (10) has generated great excitement, not only with respect to potential applications in functional genomic studies and target validation, but also in harnessing RNAi as a therapeutic strategy to silence disease-causing genes. Although delivery of synthetic siRNAs to the nervous system has achieved silencing of molecular targets in various models of neurological disease including pain (11–13,14), it requires frequent administration and high doses. As a more efficient alternative, targeted delivery of RNAi to neurons can be achieved using viral vectors. Lentiviruses, adenoassociated viruses and more recently, herpes simplex virus have been engineered to deliver short-hairpin RNA (shRNA) to parts of the nervous system (15–20,21). There are no reports, however, of vector-mediated delivery of shRNA to dorsal root ganglion (DRG) neurons in vivo, the main target site for studying nociceptive processes or for the development of new analgesics.
Herpes simplex virus (HSV-1) is a naturally neurotrophic double-stranded DNA virus able to establish life-long latency in neurons. Unlike other viruses that infect neurons, HSV has evolved to be efficiently transported from the nerve terminals innervating the infection site to cell bodies in vivo and has therefore proven particularly efficient at targeting neurons of the DRG following injection into the sciatic nerve. We have previously shown that replication-defective HSV-1 vectors can transduce neurons, and a broad range of non-neuronal cells in culture, with high efficiency, without viral gene expression or toxicity. Deletion of the essential immediate-early (IE) gene ICP4 results in replication-defective viruses. To minimize cytotoxicity, the vectors also contain an inactivating mutation in the gene encoding VP16, which abolishes transactivation of the remaining IE genes (22,23). These vectors are easily produced to high titres using a complementing cell line engineered to express ICP4 and the equine herpes virus homologue of VP16 (24). In vivo, the temporal cascade of viral gene expression is incapable of proceeding past the IE phase resulting in vectors that can establish a persistent state very similar to latency but are unable to reactivate and therefore persist for long periods of time. In addition, by inserting a strong heterologous promoter 1.4 kb downstream of the LAP1 TATA box, a region referred to as LAT P2, we have developed promoter systems that allow prolonged expression of exogenous genes during latency in both the peripheral and central nervous system (22,23).
In the present study, we evaluated the potential of these vectors to deliver RNAi to peripheral neurons using a number of approaches. We show that HSV-mediated expression of shRNA to non-neuronal cells in culture, primary neurons in vitro and DRG neurons in vivo results in effective and specific silencing of targeted genes including the endogenous trpv1 gene, which is involved in nociceptive processing and is therefore a potential target for therapeutic pain relief (25,26).
MATERIALS AND METHODS
Generation of expression cassettes and HSV vectors
The pR19 promoter cassette has been described previously (23). The pR19-Gateway vector consists of the HSV-1 flanking regions (nt 118, 441–120, 219 and nt 120, 413–122, 027) that allow recombination into the LAT region of the HSV genome and the Gateway cassette (Invitrogen) that allows cloning using the recombination properties of bacteriophage lambda. The pR19CMVenh-Gateway vector contains, in addition to the HSV-1 flanking regions, the CMVenhancer element of the CMV IE gene promoter, which was amplified from pR19LacZ (22) using the (forward) 5′-GTTGACATTGATTATTGACTAG-3′ and (reverse) 5′-GGCGAGCTCTGCCAAAACAAACTCCCATTG-3′ primers and cloned upstream of the Gateway cassette. The pR19CMV-Gateway-WCm vector consists of the HSV-1 flanking regions, the Pol II CMV IE gene promoter and a mutated form of the WPRE regulatory element (27) downstream of the Gateway cassette.
The shRNA sequences against the lacZ and gfp genes were designed using online algorithms (Invitrogen). A negative control shRNA sequence that is not predicted to target any known vertebrate gene was supplied by Invitrogen. The shLacZ sense 5′-CACCGCTACACAAATCAGCGATTTCGAAAAATCGCTGATTTGTGTAG-3′ and antisense oligonucleotides and the shGFP sense 5′-CACCGCCACAACGTCTATATCATGGCGAACCATGATATAGACGTTGTGGC-3′ and antisense oligonucleotides were annealed and cloned into pENTR-U6shRNA (Invitrogen) downstream of the U6 promoter between the BamHI-HindIII sites. The U6shLacZ cassette was then inserted into pR19-Gateway and pR19CMVenh-Gateway to generate pR19U6shLacZ and pR19CMVenhU6shLacZ, respectively. The U6shGFP and U6-neg cassettes were inserted into pR19-Gateway to generate pR19U6shGFP and pR19U6-neg. The shRNA sequence against trpv1 (sense 5′-GCGCATCTTCTACTTCAACTTCAAGAGAGTTGAAGTAGAAGATGCGC-3′) was inserted into pENTR-U6shRNA (between the BamHI-HindIII sites). The U6shTRPV1 cassette was then inserted into pR19-Gateway to generate the pR19U6shTrpv1 vector.
The pre-miRNA sequence against lacZ (sense 5′-TGCTGAAATCGCTGATTTGTGTAGTCGTTTTGGCCACTGACTGACGACTACACATCAGCGATTT-3′) and the negative control pre-miRNA sequence (sense 5′-TGCTGAAATGTACTGCGCGTGGAGACGTTTTGGCCACTGACTGACGTCTCCACGCAGTACATTT-3′) were both supplied by Invitrogen cloned into pcDNA™6.2-GW/EmGFP-miR vectors. The pre-miRNA sequence against trpV1 (sense 5′-TGCTGTGTAGTAGCTGTCTGTGTAGCGTTTTGGCCACTGACTGACGCTACACACAGCTACTACA) was cloned into pcDNA™6.2-GW/EmGFP-miR. The miRNA cassettes were transferred into pDONR™221 (Invitrogen) and then into pR19CMV-Gateway-WCm to generate pR19 shuttle vectors.
Replication-defective HSV-1 vectors were generated by calcium phosphate co-transfection (28) of 27/12/M:4 complementing cells with the shuttle plasmids described above and the HSV 1764 4-/27+/RL1+ backbone described previously (23). Genome structures were confirmed by PCR followed by sequencing. Titres were obtained by tenfold serial dilution of the virus in DMEM and addition to complementing cells, which were incubated at 37°C for 30 min, overlaid with growth media containing 1.6% carboxymethyl cellulose (2:1 ratio) and incubated at 37°C for a further 48 h. Viral titres were calculated by counting plaque forming units (pfu) and were typically in the range of 3 × 108 to 2 × 109 pfu/ml. The HSV preparations used in both in vitro and in vivo experiments were generated by growing the virus in Corning 850 cm2 roller bottles. The cell suspension was freeze-thawed to release the virus and centrifuged at 3500 rpm at 4°C for 15 min. The supernatant was filtered through a 5 and 0.45 μm filter. The virus was pelleted by centrifugation at 12 000 rpm at 4°C for 2 h and resuspended in DMEM.
Cell culture, transfection and transduction
The 27/12/M:4 complementing cell line has been described previously (24). BHK-LacZ cells are BHK cells stably transfected with the pR19LacZ construct. All cells were maintained under standard conditions in Dulbecco Modified Eagle Medium (DMEM) supplemented with 200 mM l-glutamine, 10% fetal calf serum, 10% tryptose phosphate broth, 100 units/ml penicillin and 100 μg/ml streptomycin. Continual selection was achieved by maintaining cells in 100 μg/ml of Zeocin and 50 μg/ml of G418 (cell line 27/12/M:4) or 50 μg/ml of G418 (cell line BHK-LacZ). HMBA (3 mM) was included in the growth media to complement the in814 mutation present in the viral backbone (29).
293T cells were transfected using Lipofectamine™ 2000 (Invitrogen) according to manufacturer's instructions in 24-well plates with cells at 80–90% confluency. In co-transfection experiments 100 ng of β-galactosidase- and 1 µg of shRNA-expressing plasmids were used and where indicated also 100 ng of a GFP-expressing plasmid. For transduction with HSV vectors, the virus was diluted in DMEM and the cells were transduced at various concentrations for 30 min, followed by addition of serum-containing media.
Rat primary dorsal root ganglion (DRG) neurons (Cambrex) were thawed, plated at 5 × 103 cells/well on poly-d-lysine coated 24-well plates and grown in neurobasal medium supplemented with B27. Neurons were allowed to mature for 9 days prior to being transduced.
β-Galactosidase activity assay and staining
For determining β-galactosidase protein levels, the High Sensitivity β-Galactosidase Assay kit (Stratagene) was used according to the manufacturer's instructions. Cells were lysed and cell lysates were incubated with a reaction buffer and chlorophenol red-β-d-galactopyranoside (CPRG) substrate. The activity of β-galactosidase was quantified using a microplate reader. For β-galactosidase staining, cells were fixed with 4% paraformaldehyde and incubated overnight with 1× phosphate-buffered saline (PBS) containing 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6O·6H2O, 1 mM MgCl2, and 150 μg of 4-chloro-5-bromo-3-indolyl-β-galactosidase (X-Gal) per milliliter.
Animals and surgery
Experiments were performed using BALB/c mice and Rosa26 transgenic mice (Gt(ROSA)26Sor), which are heterozygous for the ROSA26 retroviral insertion (30) and express β-galactosidase in most tissues of the adult mouse (obtained from the Jackson Laboratory). Mice were injected once directly into the sciatic nerve. Briefly, animals were anaesthetized with halothane or isoflurane, the sciatic nerve was exposed, fine forceps were used to separate the two main branches of the sciatic nerve, the larger branch being the tibial nerve which was injected with 5–7 μl of virus using a 10 μl Hamilton syringe. All animal procedures were carried out in accordance to UK Home Office Regulations.
Histological analysis
DRG were removed, fixed with 4% paraformaldehyde and stained with x-gal overnight. In order to assess expression of both GFP and β-galactosidase, GFP fluorescence was examined prior to x-gal staining, as this otherwise masks fluorescence from GFP. Cell counts were performed in a blinded fashion in whole mount DRG preparations at a magnification of 40×. For sectioning, DRG were isolated, embedded in Tissue Freezing Medium (Triangle Biomedical Sciences, Durham, NC) and immediately frozen in isopentane at −50°C. Total 10 µm cryosections were cut on a Microm HM 500 M Cryostat and placed on Superfrost Plus Gold glass slides (Menzel, Braunschweig, Germany). Slides were subsequently fixed in 70% ethanol. X-gal staining was performed for 2 h. For immunofluorescence, sections were fixed in 4% paraformaldehyde for 15 min, blocked in Image-iT FX Signal Enhancer (Molecular Probes) for 30 min at room temperature, incubated with a mixture of GFP chicken polyclonal (Abcam ab13970, 1/1000) and VR1 rabbit polyclonal (Abcam ab31895, 1/1000) antibodies in 1% BSA for 1 h at room temperature and treated with a mixture of anti-chicken FITC-conjugated (Abcam ab46969, 1/250) and anti-rabbit AlexaFluor 488 (Molecular Probes, 1/200) antibodies for 1 h at room temperature. All photographs were taken using an Axiovert 200M microscope equipped with an Axiocam HRc colour camera (Carl Zeiss, Jena, Germany).
Western blot analysis
DRG were isolated, snap-frozen in liquid nitrogen and immediately homogenized in ice-cold RIPA lysis buffer (Sigma) containing a cocktail of protease inhibitors (Roche, Switzerland). Denatured total cell lysates were run in SDS-PAGE, transferred to nitrocellulose membranes and blocked overnight at 4°C in 5% milk. The blocked membranes were incubated overnight at 4°C with the VR1 polyclonal antibody (Santa Cruz, 1/200), TRPA1 polyclonal antibody (Abcam ab31486, 1/1000), STAT1 polyclonal antibody (Abcam ab31369, 1/1000), or α-tubulin polyclonal antibody (Abcam ab4074, 1/500) followed by 1 h incubation at room temperature with an HRP-conjugated secondary antibody (Abcam ab6721, 1/3000). Immunodetection was performed using the ECL+ reagents (Amersham Biosciences, UK). The blots were scanned and the pixel count and intensity of each band was quantified using the Scion Image software (Scion, Frederick, MD). Signals were normalized against α-tubulin (loading control) and the result was expressed as a percentage of the negative control signal.
Quantitative RT–PCR
Total RNA from cultured HEK293T cells was extracted and DNAseI treated using the RNeasy Mini kit (Qiagen) as per manufacturer's instructions. Three micrograms of total RNA served as template for cDNA synthesis using Oligo dT primers and Superscript III RT in a volume of 20 µl during 1 h at 50°C. This was followed by inactivation of the enzyme at 70°C for 15 min according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Quantitative PCR was performed on an ABI Prism 7900-HT Sequence Detection System (Applied Biosystems) using a qPCR core kit w/o dUTP (Eurogentec). The thermal cycling conditions were 10 min at 95°C, followed by 45 cycles of 15 s at 95°C and 1 min at 60°C. Validated pre-designed Taqman Gene Expression Assays (Applied Biosystems) corresponding to the housekeeping genes ATP5b (Hs00969569_m1), B2m (Hs99999907_m1), GAPDH (Hs99999905_m1), GUSb (Hs99999908_m1) and an interferon response gene ISGF3G (Hs_00196051_m1) were used to generate standard curves on serial dilutions of cDNA. The relative standard curve method was used to calculate the expression values. Values were normalized against the values obtained for the B2m gene.
RNA from whole DRG was extracted using the Rneasy 96 kit (Qiagen) as per manufacturer's instructions. RNA was eluted in 100 µl H2O and vacuum dried to approximately 10 µl. The RNA was subsequently amplified using a modified version of the two-cycle amplification kit from Affymetrix. The reaction was stopped after the first amplification step and a DNAseI digestion was performed. The RNA was cleaned in a 96-well plate, eluted twice in 50 µl H2O and vacuum dried to approximately 20 µl. First strand cDNA synthesis was performed on 1 μg of amplified RNA using random hexamer primers and Superscript II RT. Quantitative PCR was performed using the TaqMan PCR kit (Applied Biosystems). Serial dilutions of cDNA were used to generate standard curves of threshold cycles versus the logarithms of concentration for Sart1 (endogenous control, mouse squamous cell carcinoma antigen recognized by T cells) and Trpv1 or GFP. For Sart1 mRNA levels, the TaqMan Gene Expression Assay (Applied Biosystems) was used. The Trpv1 primers and probe were designed manually: forward primer 5′-AGAATTCAGTGCTGGAGGTGATC-3′, reverse primer 5′-GGCTCCACGAGAAGC ATGTC-3′, and probe sequence carboxyfluorescein-5′-GAGCCAAGGGACCCACCCCATGGAGAGC-3′-tetramethylrhodamine (Eurogentec). The GFP primers and probe were also designed manually: forward primer 5′-AGCAAAGACCCC AACGAGAA-3′, reverse primer 5′-GGCGGCGGTCACGAA-3′, and probe sequence carboxyfluorescein-5′-CGCGATCACATGGTCCTGCTGG-3′-tetramethylrhodamine.
Flow cytometry (FACS)
GFP expression in transfected 293T cells was determined by FACS (fluorescence activated cell sorting) analysis using the FACS Calibur flow cytometer (Becton Dickinson). GFP was identified using a 530 band pass filter. The data was analyzed using CELLQUEST software (Becton Dickinson). A primary gate based on physical parameters (forward and side light scatter, FSC and SSC, respectively) was set to exclude dead cells or debris.
RESULTS
Optimization of shRNA expression
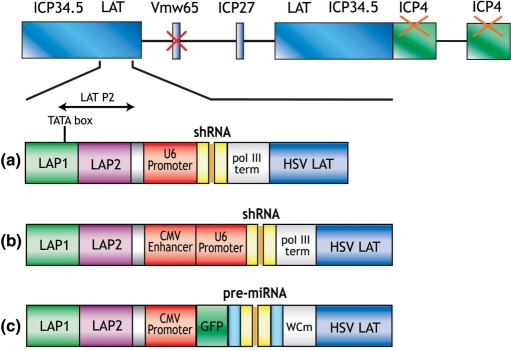
To optimize expression of shRNA from HSV, we generated three expression systems (Figure 1). The pR19U6shRNA system utilizes the U6 Pol III promoter that has been successfully used to silence genes both in vitro and in vivo (Figure 1a). Previous studies have demonstrated that the enhancer element of the Cytomegalovirus IE gene (CMV) Pol II promoter can improve the activity of the U6 or H1 Pol III promoters (31,32). Thus, to test whether shRNA expression could be improved in this manner, we generated pR19CMVenhU6shRNA, where expression of shRNA was placed under the control of a hybrid CMV enhancer-U6 promoter (Figure 1b). Finally, although Pol III promoters have so far been most commonly used for expression of shRNA, there has been an interest in developing systems using Pol II promoters. Our microRNA-like expression system (pR19CMVGFP-miR) takes advantage of the flexibility and variety of Pol II promoters, which unlike pol III promoters, can be inducible and tissue-specific (Figure 1c). When inserted into Pol II mRNA transcripts of irrelevant sequence, pre-miRNA sequences are readily excised to silence expression of target mRNAs with perfect complementarity to the miRNA sequence (33). The sequence of the mature miRNA does not seem to be important for processing and can be altered to target the chosen gene (34). To ensure that the pre-miRNA is properly processed by Drosha, our miRNA cassette contains, in addition to the pre-miRNA sequence, 5′ and 3′ flanking regions derived from the endogenous miRNA-155. A reporter gene encoding green fluorescent protein (GFP) is co-expressed from this system and allows labelling of transduced cells to aid the monitoring of silencing efficiency.
Figure 1.
HSV RNAi expression systems. Schematic representation of the replication-defective HSV-1 genome and the different expression systems used to express shRNA. (a) In pR19U6shRNA, expression of shRNA is driven by the pol III U6 promoter. (b) In pR19CMVenhU6shRNA, expression of shRNA is driven by a hybrid CMVenhancer-U6 promoter. (c) In pR19CMVGFP-miR, the pol II CMV promoter drives expression of both the pre-miRNA sequence and GFP. The shRNA expression cassettes are inserted into the LAT region of the disabled HSV-1 genome. Deletion of ICP4 results in replication-defective vectors. These vectors also contain an inactivating mutation in VP16. Prolonged expression during latency has been achieved by inserting a heterologous promoter after the LAT P2 region.
To compare silencing from these promoter systems, we generated plasmid vectors with a well-characterized hairpin sequence designed to target lacZ, the reporter gene encoding β-galactosidase. Control plasmids were constructed to mediate expression of shRNA against green fluorescent protein (pR19U6shGFP) or a non-target shRNA sequence (pR19U6-neg) under the control of the U6 promoter and a non-target pre-miRNA sequence under the control of the CMV promoter (pR19CMVGFP-miR-neg).
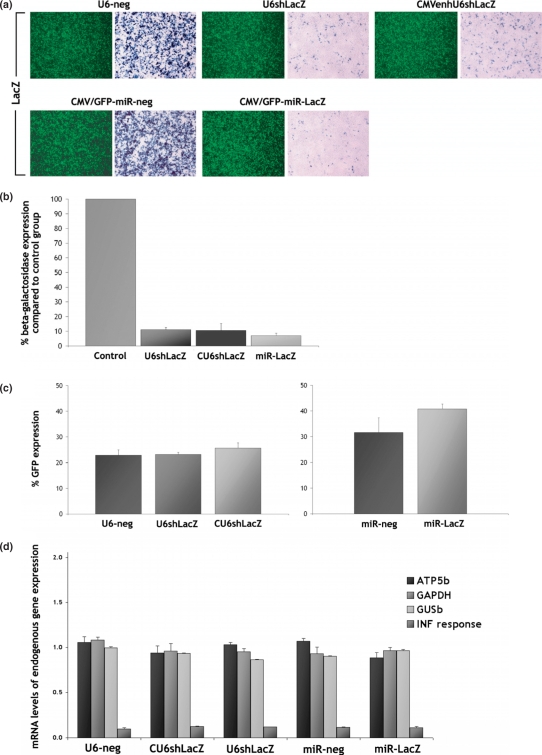
293T cells were co-transfected with a plasmid expressing β-galactosidase and either the pR19U6-neg, pR19U6shLacZ or pR19CMVenhU6shLacZ plasmids. In each experiment, a plasmid expressing GFP was co-transfected to normalize transfection efficiency. Similarly, 293T cells were co-transfected with a β-galactosidase-expressing plasmid and either the pR19CMVGFP-miR-neg or pR19CMVGFP-miR-LacZ plasmids. Transfection efficiency was monitored by the presence of the GFP signal expressed from these constructs. The level of sensitivity achieved with this system is greater than that achieved with endogenous target gene knockdown because the constructs expressing lacZ and shRNA are co-transfected simultaneously and delivery of RNAi to all cells is not required in order to achieve an RNAi response. X-gal staining at 72 h post-transfection demonstrated that each of the RNAi expression systems targeting lacZ substantially reduced β-galactosidase levels (Figure 2a). Analysis of β-galactosidase expression by enzyme activity assay revealed that the pR19U6shLacZ construct silences lacZ by 89.0 ± 1.5% (n = 3, mean ± SD), the pR19CMVenhU6shLacZ construct by 89.4 ± 4.6% and the pR19CMVGFP-miR-LacZ construct by 93.0 ± 1.7% (Figure 2b). Transfection with the negative controls had no effect on β-galactosidase levels indicating that silencing is specific. GFP expression levels were assessed by fluorescent microscopy (Figure 2a) followed by flow cytometry (Figure 2c).
Figure 2.
Silencing evaluation from different promoter systems. (a) 293T cells were co-transfected with a plasmid expressing β-galactosidase and either pR19U6-neg, pR19U6shLacZ or pR19CMVenhU6shLacZ. A plasmid expressing GFP was also co-transfected to monitor transfection efficiency. Similarly, 293T cells were co-transfected with a β-galactosidase-expressing plasmid and either pR19CMVGFP-miR-neg or pR19CMVGFP-miR-LacZ. X-gal staining at 72 h post-transfection revealed that each of the RNAi expression systems substantially reduced β-galactosidase levels compared to the control group (magnification 10×). (b) Enzyme activity assay revealed that pR19U6shLacZ silences lacZ by 89.0 ± 1.5%, pR19CMVenhU6shLacZ by 89.4 ± 4.6% and pR19CMVGFP-miR-LacZ by 93.0 ± 1.7% (n = 3, mean ± SD). (c) GFP expression levels in 293T cells were assessed by flow cytometry at 72 h post-transfection. Transfection with each of the plasmids targeting lacZ had no effect on the levels of GFP. (d) Quantitative RT-PCR at 72 h post-transfection for ATP5b, GAPDH and GUSb revealed that the expression levels of housekeeping gene expression remained essentially unaffected by introduction of shRNA or pre-miRNA into 293T cells. Quantitative RT-PCR for ISGF3G, the interferon-stimulated transcription factor 3 gamma, confirms that silencing is not caused by a non-specific effect associated with induction of the INF response. Values were normalized against the B2m gene.
To investigate RNAi specificity further, we assessed the mRNA levels of four endogenous genes in 293T cells transfected with each of the above constructs. Quantitative RT-PCR for ATP5b, GAPDH and GUSb revealed that there is no significant reduction in the expression levels of any of these housekeeping genes 72 h following introduction of shRNA or pre-miRNA (Figure 2d) and thus, silencing of lacZ cannot be attributed to off-target effects. Moreover, quantitative RT-PCR for ISGF3G, the interferon-stimulated transcription factor 3 gamma, confirms that silencing is not caused by a non-specific effect associated with induction of the interferon (INF) response.
HSV-mediated silencing in non-neuronal cells in culture
Unlike 293T cells, transfection of post-mitotic primary neurons is inefficient and often toxic. The shRNA cassettes were inserted into the LAT region of the replication-defective HSV-1 genome to generate viral vectors (Figure 1).
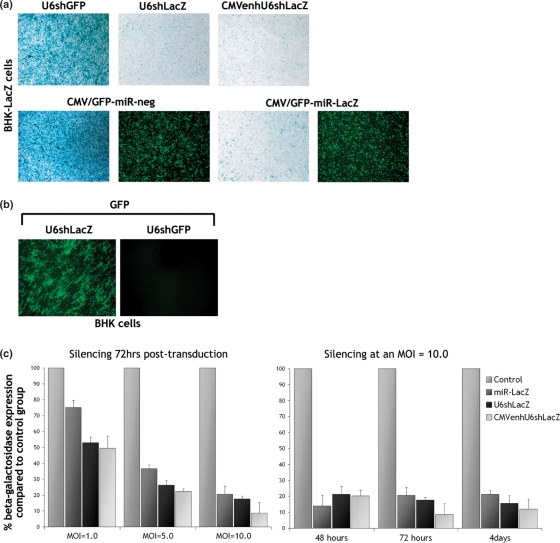
To assess the efficiency of HSV-mediated silencing in vitro, we generated a BHK cell line stably transfected with a construct expressing β-galactosidase (BHK-LacZ). BHK-LacZ cells were transduced with each vector targeting lacZ or the negative control vectors (multiplicity of infection [MOI] = 10.0) (Figure 3a). The expression of functional shRNA from the HSV-U6shGFP negative control (MOI = 5.0) was confirmed by the reduction of GFP levels in BHK cells co-transduced with a HSV vector expressing GFP (MOI = 1.0) (Figure 3b). Expression of miRNA from the HSV-miR-neg control was confirmed by the presence of the GFP signal (Figure 3a). Silencing was assessed by x-gal staining at 72 h post-transduction (Figure 3a). Transduction with the HSV-U6shGFP or HSV-GFP-miR-neg control vectors had no effect on β-galactosidase expression levels. Transduction of BHK-LacZ cells with HSV-U6shLacZ, HSV-CMVenhU6shLacZ or HSV-GFP-miR-LacZ, however, resulted in a significant reduction in β-galactosidase levels. B-galactosidase activity assay performed at various times post-transduction with HSV-U6shLacZ, HSV-CMVenhU6shLacZ or HSV-GFP-miR-LacZ revealed that these reduced protein levels by up to 86.0 ± 7.0% (n = 6, mean ± SD), 91.0 ± 7.0% and 84.0 ± 8.0%, respectively, as compared to controls (Figure 3c). HSV-GFP-miR-LacZ is most effective at 48 h, HSV-CMVenhU6shLacZ at 72 h and HSV-U6shLacZ at 4 days post-transduction, reflecting the different kinetics of expression from the promoters used. β-Galactosidase activity assay performed at 72 h post-transduction (MOI = 1.0, 5.0 and 10.0) revealed that silencing is dose-dependent. Finally, GFP expression in BHK-LacZ cells transduced with HSV-miR vectors was not affected by expression of miR-LacZ as evaluated by fluorescent microscopy (Figure 3a). We can therefore conclude that silencing is not caused by a non-specific effect mediated by the HSV vector backbone. Unlike off-target effects or induction of the INF response, this would most likely be a non-sequence specific effect resulting in silencing of both lacZ and gfp.
Figure 3.
HSV-mediated silencing in non-neuronal cells in culture. (a) BHK-LacZ cells were transduced with each vector targeting lacZ or the negative control vectors (MOI = 10.0). Silencing was assessed by x-gal staining at 72 h post-transduction (magnification 10×). Transduction with the HSV-U6shGFP or HSV-miR-neg control vectors had no effect on β-galactosidase levels, whereas transduction with HSV-U6shLacZ, HSV-CMVenhU6shLacZ or HSV-GFP-miR-LacZ resulted in a significant reduction in β-galactosidase levels. (b) The expression of functional shRNA from HSV-U6shGFP (MOI = 5.0) was confirmed by the reduction of GFP levels in BHK cells co-transduced with a HSV vector expressing GFP (MOI = 1.0) (magnification × 10). (c) Enzyme activity assay 72hrs post-transduction reveals a dose-dependent reduction of β-galactosidase. Enzyme activity assay at various times post-transduction with HSV-U6shLacZ, HSV-CMVenhU6shLacZ or HSV-GFP-miR-LacZ revealed that the vectors reduce protein levels by up to 86.0 ± 7.0%, 91.0 ± 7.0% and 84.0 ± 8.0%, respectively, as compared to the negative control groups (n = 6, mean ± SD).
HSV-mediated silencing in primary neurons
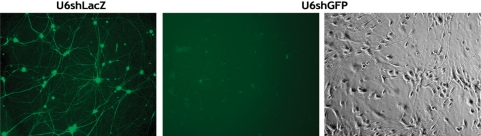
We next sought to determine whether HSV-mediated delivery of shRNA could induce silencing in primary neuronal cells. Rat DRG neuronal cultures were co-transduced with a GFP-expressing vector (MOI = 1.0), and either HSV-U6shLacZ or HSV-U6shGFP (MOI = 5.0). High transduction efficiencies were achieved with almost the entire population of neurons being positive for GFP. Fluorescent microscopy at 48 h post-transduction revealed that transduction with HSV-U6shLacZ had no effect on GFP expression levels, whereas transduction with HSV-U6shGFP resulted in almost complete inhibition of GFP expression (Figure 4). This data indicate highly effective silencing in neurons in vitro induced by HSV-mediated RNAi.
Figure 4.
HSV-mediated silencing in primary neurons. Rat DRG neuronal cultures were transduced with a GFP-expressing vector (MOI = 1.0) and either HSV-U6shGFP or HSV-U6shLacZ (MOI = 5.0). Fluorescent microscopy at 48hrs post-transduction revealed that transduction with HSV-U6shLacZ had no effect on GFP expression levels, whereas transduction with HSV-U6shGFP resulted in a dramatic reduction in the levels of GFP compared to control groups (magnification 10×).
HSV-mediated silencing in vivo
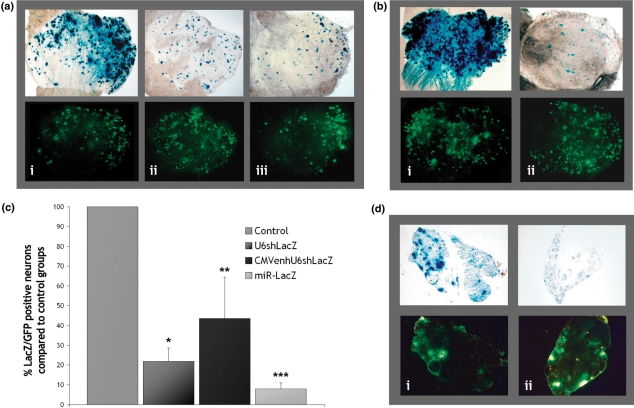
To evaluate the efficiency of HSV-mediated silencing in DRG neurons in vivo, BALB/c mice were injected directly into the sciatic nerve. A single injection (up to 7 μl) was performed to co-deliver a β-galactosidase-expressing vector and each of the vectors targeting lacZ or each of the negative controls. In animals injected with HSV-U6-neg, HSV-U6shLacZ or HSV-CMVenhU6shLacZ (Figure 5a), a GFP-expressing vector was also co-injected to evaluate transduction efficiency and monitor silencing specificity. At day 7, the lumbar DRG (L4/L5) were isolated. Figure 5a and b show representative DRG from injected animals. High transduction efficiencies were achieved in all experiments mainly to the L4 DRG with the majority of neurons being positive for GFP as assessed by fluorescent microscopy (Figure 5a and b). X-gal staining of whole mount DRG preparations revealed a dramatic reduction in β-galactosidase levels in animals injected with HSV-U6shLacZ (Figure 5a-ii), HSV-CMVenhU6shLacZ (Figure 5a-iii) or HSV-GFP-miR-LacZ (Figure 5b-ii) compared to control groups injected with either HSV-U6-neg (Figure 5a-i) or HSV-GFP-miR-neg (Figure 5b-i). In co-injection experiments, effective silencing requires co-transduction with both the β-galactosidase- and RNAi-expressing vectors. In animals injected with the GFP-expressing vector (Figure 5a), expression of GFP from this vector does not allow identification of neurons transduced with the shRNA-expressing vectors. Although a very high degree of LacZ and GFP co-localization is evident when the HSV-U6-neg is co-injected (Figure 5a-i), the high levels of β-galactosidase displayed in some neurons when the HSV-U6shLacZ or HSV-CMVenhU6shLacZ were co-injected may be due to incomplete transduction overlap (Figure 5a-ii and iii). In the DRG of animals injected with HSV-GFP-miR-LacZ (Figure 5b-ii), GFP expression allows identification of transduced neurons and >90% of GFP-positive neurons display markedly reduced or abolished β-galactosidase expression.
Figure 5.
HSV-mediated silencing in DRG neurons in vivo. (a) BALB/c mice were injected once directly into the sciatic nerve with 1 × 106 plaque forming units (pfu) of a HSV β-galactosidase-expressing vector and 5 × 106 pfu of: (i) the HSV-U6-neg control, (ii) HSV-U6shLacZ, or (iii) HSV-CMVenhU6shLacZ. A GFP-expressing vector was also co-injected (1 × 106 pfu) to monitor transduction efficiency and non-specific silencing. (b) 1 × 106 pfu of a β-galactosidase-expressing vector were injected into the sciatic nerve of BALB/c mice with 5 × 106 pfu of: (i) the HSV-GFP-miR-neg control or (ii) HSV-GFP-miR-LacZ. GFP expression from this system allows labelling of transduced cells and monitoring of non-specific silencing. Silencing was assessed by x-gal staining in whole mount L4 DRG preparations at 7 days post-injection (magnification 5×). (c) Silencing was quantified by directly counting the number of GFP-positive versus β-galactosidase-positive neurons in whole mount DRG preparations (at a magnification 40×). HSV-U6shLacZ reduces β-galactosidase protein levels by 78.0 ± 6.8% (*P = 0.001), HSV-CMVenhU6shLacZ by 62.1 ± 20.9% (**P = 0.02) and HSV-GFP-miR-LacZ by 92.0 ± 3.0% (***P = 0.0002) (n = 3, mean ± SD, results analysed for significance using t-test) (d) Rosa26 transgenic mice were injected into the sciatic nerve with 5 × 106 pfu of: (i) HSV-GFP-miR-neg or (ii) HSV-GFP-miR-LacZ. At 7 days post-injection, the L4 DRG were isolated and sectioned. GFP expressed from these vectors allows simultaneous localization of transduced neurons and monitoring of silencing efficacy. Neurons transduced with HSV-GFP-miR-LacZ are GFP positive and display significantly reduced x-gal staining compared to DRG sections taken from control animals (magnification 10×).
We next monitored the level of silencing by directly counting the number of GFP-positive versus LacZ-positive neurons in the whole mount DRG preparations. We found that HSV-U6shLacZ reduces β-galactosidase levels by up to 78.0 ± 6.8% (P = 0.001) (n = 3, mean ± SD), HSV-CMVenhU6shLacZ by up to 62.1 ± 20.9% (P = 0.02) and HSV-GFP-miR-LacZ by up to 92.0 ± 3.0% (P = 0.0002) (Figure 5c). Whereas the number of GFP-positive neurons remained essentially stable at 170 ± 53 cells (n = 15, mean ± SD) in each of the different conditions, the level of LacZ-positive neurons dropped from 464 ± 68 cells (n = 6, mean ± SD) in DRG injected with the negative controls to 122 ± 68, 148 ± 60 and 63 ± 15 cells (n = 3, mean ± SD) in DRG injected with HSV-U6shLacZ, HSV-CMVenhU6shLacZ or HSV-GFP-miR-LacZ respectively. These data confirm robust silencing in peripheral neurons in vivo induced by HSV-mediated RNAi. Injection with the negative controls had no effect on β-galactosidase levels and injection with the vectors targeting lacZ does not seem to have an effect on the levels of GFP indicating silencing specificity. The number of GFP-positive neurons was found to be much lower than the number of LacZ-positive neurons in DRG injected with the negative controls. This is mainly due to the fact that x-gal staining used to visualize LacZ-positive cells is more sensitive than fluorescence microscopy used to visualize GFP-positive cells. Moreover, in the HSV-miRNA vectors, GFP is co-cistronically expressed with the pre-miRNA and this can result in inefficient GFP mRNA processing.
The above results demonstrate silencing in a system where a marker gene has been delivered coincidentally with silencing. We next sought to determine whether lacZ could be effectively silenced in a transgenic mouse model. The HSV-GFP-miR-LacZ vector that was shown to be the most effective at silencing in the co-injection experiments was selected for further testing. Moreover, co-expression of GFP from miRNA vectors allows simultaneous localization of transduced neurons and monitoring of silencing efficacy in transduced cells. Rosa26 mice were injected into the sciatic nerve with the HSV-GFP-miR-neg or the HSV-GFP-miR-LacZ vectors. At day 7, the lumbar DRG from injected mice were isolated and sectioned. Transduction efficiency was high throughout all sections taken from injected animals. Figure 5d shows representative sections from transduced DRG. In all four independent experiments, neurons transduced with HSV-GFP-miR-LacZ (Figure 5d-i), while also GFP-positive, displayed significantly reduced x-gal staining compared to control animals (Figure 5d-ii). β-Galactosidase expression in these sections was also markedly reduced compared to sections taken from the DRG of the non-injected side of the same animals (data not shown).
HSV-mediated silencing of trpv1
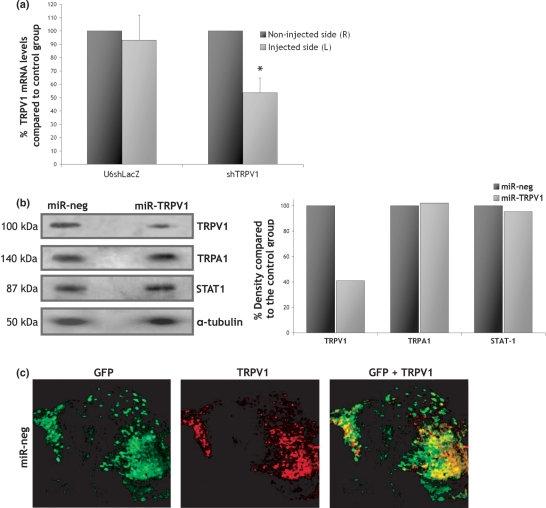
We next investigated whether HSV-mediated RNAi could be used to silence an endogenous neuronally expressed gene. In preliminary experiments, several shRNA constructs expressed under the control of the U6 promoter were evaluated for their ability to knockdown trpv1 by at least 80% in HEK cells (data not shown). The most potent shRNA sequence targeted the region containing nucleotides 1296–1314 of the trpv1 gene. The U6shTrpv1 cassette was inserted into the LAT region of the replication-defective HSV-1 genome and the HSV-U6shTrpv1 vector was injected into the sciatic nerve of BALB/c mice. Mice injected with HSV-U6shLacZ were used as negative controls. A GFP-expressing vector was also co-injected to monitor transduction efficiency. At day 8, the left DRG (injected side) and right DRG (non-injected side) were isolated and analysed for GFP and Trpv1 levels. GFP expression assessed by quantitative RT-PCR confirmed delivery to all injected animals, predominantly to the L4 DRG (data not shown). Quantitative RT-PCR for Trpv1 revealed markedly reduced mRNA levels in the left DRG from all animals injected with HSV-U6shTrpv1 compared to the control group (Figure 6a). Trpv1 mRNA in these animals was reduced by 53.6 ± 11.2% (P = 0.01) (n = 4, mean ± SD). Importantly, HSV-U6shLacZ does not seem to have an effect on the levels of the endogenous trpV1 gene further indicating the specificity of our method.
Figure 6.
HSV-mediated silencing of endogenous trpv1. (A) BALB/c mice were injected once directly into the sciatic nerve with 2.5 × 106 pfu of either HSV-U6shTRPV1 (n = 4) or HSV-U6shLacZ (n = 2). A GFP-expressing vector was also co-injected to assess transduction efficiency (1 × 106 pfu). At day 8, both the left (injected) and right (non-injected) L4 DRG were isolated. Quantitative RT-PCR revealed that TRPV1 mRNA levels in the DRG of animals injected with HSV-U6shTRPV1 were reduced by 53.6 ± 11.2% (mean ± SD) compared to the right DRG. In DRG injected with the HSV-U6shLacZ vector, TRPV1 mRNA levels were essentially unchanged compared to the right DRG. Statistical differences between the two injected groups were determined by one-way t-test, *P = 0.01. (B) BALB/c mice were injected once directly into the sciatic nerve with 5 × 106 pfu of either HSV-GFP-miR-TRPV1 (n = 10) or HSV-GFP-miR-neg (n = 10). At day 8, the L4 DRG isolated from each group were used to extract protein and western blots were performed for TrpV1, TrpA1 and Stat1. Quantification of band density revealed a 59% knockdown of TrpV1 protein in animals injected with HSV-GFP-miR-TRPV1 compared to the negative control, whereas the levels of TrpA1 and Stat1 were unaffected. Values were normalized against α-tubulin. (C) BALB/c mice were injected into the sciatic nerve with 5 × 106 pfu of HSV-GFP-miR-neg (n = 4). Four days post-injection the L4 DRG were sectioned and immunofluorescence for GFP and TrpV1 was performed to reveal the number of TrpV1-expressing neurons transduced by the vector.
We have previously shown that the HSV-GFP-miR is more effective at silencing in vivo than the HSV-U6-shRNA system. Subsequent experiments were therefore performed using a vector expressing miRNA against trpV1. Several pre-miRNA sequences were evaluated (data not shown) and the most potent was used to generate the HSV-GFP-miR-TRPV1 vector. To evaluate silencing at the protein level, 10 BALB/c mice were injected directly into the sciatic nerve with HSV-GFP-miR-neg and 10 mice were injected with HSV-GFP-miR-TRPV1. Eight days post-injection the DRG from injected animals were isolated. Delivery was assessed by fluorescent microscopy for GFP, which confirmed delivery mainly to the L4 DRG. Western blot analysis for TrpV1, TrpA1 and Stat1 was performed on protein extracted from the L4 DRG isolated from each group (Figure 6b). Quantification revealed a 59% reduction in the levels of TrpV1 protein compared to the negative control (average across 10 injections), whilst the levels of TrpA1 and Stat1 remained essentially unchanged indicating that silencing is specific and not caused by off-target effects or induction of the INF response.
The data obtained with both vectors targeting trpV1 is particularly promising as it represents overall Trpv1 levels in both transduced and un-transduced sensory neurons, rather than in transduced neurons only. It should be noted that the 80% knockdown of β-galactosidase expression (Figure 5c) was calculated by counting LacZ-positive versus GFP-positive neurons and therefore reflects the number of neurons out of the population of transduced neurons. The DRG is comprised of a heterologous population of neurons and TrpV1 is mainly expressed in small- and medium- diameter sensory neurons. Thus, we sought to determine whether TrpV1-expressing neurons are efficiently transduced by the vector following injection into the sciatic nerve. BALB/c mice were injected with HSV-GFP-miR-neg and immunofluorescence was performed on L4 DRG sections to investigate the degree of co-localization between GFP expressed by the vector and TrpV1. Figure 6c shows representative sections of DRG isolated 4 days post-injection. Our results confirm that the vector transduces a heterologous population of neurons, including approximately 80% of neurons that express TrpV1. Interestingly, high levels of GFP expression do not coincide with the highest levels of TrpV1 expression. This explains the 53.6% reduction in TrpV1 mRNA using the U6 vector expressing shTRPV1 (Figure 6a) and 59% reduction of TrpV1 protein using the miRNA version (Figure 6b), as compared to the 80% reduction of LacZ.
DISCUSSION
RNAi has become a powerful tool for modulating gene expression. Delivery to neurons however poses specific challenges. Whereas delivery of siRNAs to primary neuronal cultures has been achieved with relatively high efficiency (35), in vivo delivery of RNAi to neurons, and DRG neurons in particular, has been problematic. The development of an efficient method for in vivo delivery of RNAi to peripheral neurons would allow a better understanding of gene function, enable the validation of novel gene targets in drug discovery, and potentially allow the development of new RNAi-mediated approaches to achieving analgesia.
Replication-defective HSV-1 vectors have been extensively used to deliver genes to peripheral neurons, including in animal models of neuropathic and inflammatory pain (36–38) and therefore make ideal candidates for the delivery of RNAi to sensory neurons. However, while a replication-defective HSV-1 vector has been shown to express shRNA in the mouse brain to suppress expression of amyloid precursor protein separately expressed from a lentivirus vector (20), and HSV amplicon vectors have been shown to silence genes in tumour cells in vitro and in vivo (39–41), HSV vectors had not been previously developed to provide silencing in the peripheral nervous system, or of an endogenously expressed gene in neurons in vivo.
Our previously developed replication-defective HSV-1 vectors allow efficient gene delivery to neurons of the central and peripheral nervous system both in vitro and in vivo (22,23) and are particularly efficient at targeting DRG neurons in both mice and rats through retrograde transport following injection into the sciatic nerve. While we have previously identified regulatory elements in the HSV genome that allow efficient expression of exogenous genes from heterologous promoters in a relatively non-promoter specific fashion (23), expression of shRNA from these vectors been had not been previously investigated.
In the present study, we establish that expression of shRNA from these HSV vectors can be achieved using either pol II or pol III promoters. In cell culture, the different RNAi expression systems showed very similar silencing capabilities. The enhancer element of the CMV promoter did not seem to significantly improve silencing from the U6 promoter and this system was found to be the least effective at silencing in vivo. Although the miRNA system was shown to mediate the highest levels of silencing in vivo, a more systematic comparison is required in order to identify whether expressing shRNA from a miRNA-based system is superior to expressing shRNA from pol III promoters. However, expression of GFP together with the shRNA and the potential to use inducible or tissue-specific pol II promoters hold clear advantages for the use of miRNA expression vectors. Furthermore, the miRNA system allows expression of more that one pre-miRNA in tandem and it is therefore possible to simultaneously silence multiple gene targets. Although, as described above, HSV has been previously shown to express shRNA, this is the first demonstration of HSV-mediated expression of shRNA from a pol II miRNA-based system.
We demonstrate that these vectors can induce highly effective silencing of reporter genes in primary neuronal cells and DRG neurons in vivo. Silencing in sensory neurons in vivo was assessed in an overexpression system and more importantly in a transgenic mouse model without any obvious signs of toxicity or inflammation. To our knowledge, this represents the first demonstration of gene silencing in sensory neurons in vivo by vector-mediated delivery of shRNA.
In addition to the silencing of transgenically expressed reporter genes, we also demonstrate in vivo silencing of an endogenous gene. The transient receptor potential vanilloid subtype 1 (Trpv1) is an ideal target for testing the efficacy of HSV-mediated silencing in vivo as it is predominantly expressed in DRG sensory neurons and has been suggested to play a key role in inflammatory pain (25,26,42,43). Our results demonstrate a significant reduction in Trpv1 mRNA levels in sensory neurons from mice injected with the vector expressing shTRPV1 and 59% reduction in TrpV1 protein levels following injection of the vector expressing miR-TRPV1. TrpV1 has been used here as a proof of concept that an endogenous gene can be specifically silenced in DRG using the vector system developed. Since the present aim was to evaluate a means of effective and specific RNAi delivery to DRG neurons in vivo rather that to validate the role of trpV1 itself, the assessment of any behavioural effects is beyond the scope of this study. Previous studies suggest that only a small number of TrpV1-expressing neurons have to be reached by siRNA treatment to evoke an analgesic response and that knockdown of TrpV1 protein by antisense oligonucleotides, despite being too subtle to detect in the DRG or spinal cord, was sufficient to reduce neuropathic pain behaviour (44,45). Thus, 59% knockdown of TrpV1 protein demonstrated by our method would be expected to confer a biological effect. Finally, we explain why complete silencing of trpV1 is not achieved and that the silencing efficacy of our approach with respect to inducing a phenotypic effect will depend upon the efficiency at which the vector transduces the specific types of neurons expressing the target gene.
Specificity has become a major concern in the use of RNAi. Non-specific silencing may result from a non-sequence-specific effect caused by the virus, sequence-specific off-target effects or induction of the INF response. Throughout this study, negative control vectors have had no effect on target gene expression. We also show that the levels of GFP are not affected by expression of shRNA targeting lacZ, and vice versa, thus excluding any potential non-specific effects induced by the HSV vector backbone. Furthermore, we demonstrate that the shRNA and miRNA sequences targeting lacZ have no effect on the expression levels of four endogenous genes, including a gene involved in the INF response, when expressed in non-neuronal cells in culture. More importantly, we show that the shRNA and miRNA sequences targeting trpV1 have no effect on the expression levels of the closely related trpA1 gene or stat1, which is another gene involved in the INF response, when expressed in neurons in vivo.
In conclusion, the present study confirms that replication-defective HSV-1 vectors can be used to efficiently deliver shRNA to peripheral neurons both in vitro and in vivo and induce highly effective and specific silencing of targeted genes. These results highlight the potential of this technology, which can theoretically be directed to target any gene, as a valuable tool for the study of nociceptive processes and the development of new analgesic drugs. Furthermore, these vectors could ultimately be applicable to the development of an HSV vector-based approach to pain control.
ACKNOWLEDGEMENTS
We would like to thank Shalene Singh for helping with the flow cytometry and Ronald De Hoogt for helping with the qRT–PCR. Funding to pay the Open Access publication charges for this article was provided by BioVex Inc.
Conflict of interest statement. This work was funded by BioVex Inc.
REFERENCES
- 1.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 2.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 3.Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- 4.Dostie J, Mourelatos Z, Yang M, Sharma A, Dreyfuss G. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA. 2003;9:180–186. doi: 10.1261/rna.2141503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 6.Lund E, Glutting S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 7.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 8.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 10.Krichevsky AM, Kosik KS. RNAi functions in cultured mammalian neurons. Proc. Natl Acad. Sci. USA. 2002;99:11926–11929. doi: 10.1073/pnas.182272699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorn G, Patel S, Wotherspoon G, Hemmings-Mieszczak M, Barclay J, Natt FJ, Martin P, Bevan S, Fox A, Ganju P, et al. siRNA relieves chronic neuropathic pain. Nucleic Acids Res. 2004;32:e49. doi: 10.1093/nar/gnh044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo MC, Zhang DQ, Ma SW, Huang YY, Shuster SJ, Porreca F, Lai J. An efficient intrathecal delivery of small interfering RNA to the spinal cord and peripheral neurons. Mol. Pain. 2005;28:1–29. doi: 10.1186/1744-8069-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan PH, Yang LC, Shih HC, Lan KC, Cheng IT. Gene knockdown with intrathecal siRNA of NMDA receptor NR2B subunit reduces formalin-induced nociception in the rat. Gene Ther. 2005;12:59–66. doi: 10.1038/sj.gt.3302376. [DOI] [PubMed] [Google Scholar]
- 14.Kumar P, Wu H, McBride JL, Jung KE, Kim MH, Davidson BL, Lee SK, Shankar P, Manjunath N. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 15.Ralph GS, Radcliffe PA, Day DM, Carthy JM, Leroux MA, Lee DC, Wong LF, Bilsland LG, Greensmith L, Kingsman SM, et al. Silencing mutant SOD1 using RNAi protects against neurodegeneration and extends survival in an ALS model. Nat. Med. 2005;11:429–433. doi: 10.1038/nm1205. [DOI] [PubMed] [Google Scholar]
- 16.Raoul C, Abbas-Terki T, Bensadoun JC, Guillot S, Haase G, Szulc J, Henderson CE, Aebischer P. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nat. Med. 2005;11:423–428. doi: 10.1038/nm1207. [DOI] [PubMed] [Google Scholar]
- 17.Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL, Davidson BL. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc. Natl Acad. Sci. USA. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singer O, Marr RA, Rockenstein E, Crews L, Coufal NG, Gage FH, Verma IM, Masliah E. Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat. Neurosci. 2005;8:1343–1349. doi: 10.1038/nn1531. [DOI] [PubMed] [Google Scholar]
- 19.Xia H, Mao Q, Eliason SL, Harper SQ, Martins IH, Orr HT, Paulson HL, Yang L, Kotin RM, Davidson BL. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat. Med. 2004;10:816–20. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- 20.Hong CS, Goins WF, Goss JR, Burton EA, Glorioso JC. Herpes simplex virus RNAi and neprilysin gene transfer vectors reduce accumulation of Alzheimer's disease-related amyloid-beta peptide in vivo. Gene Ther. 2006;13:1068–1079. doi: 10.1038/sj.gt.3302719. [DOI] [PubMed] [Google Scholar]
- 21.Van den Haute C, Eggermont K, Nuttin B, Debyser Z, Baekelandt V. Lentiviral vector-mediated delivery of short hairpin RNA results in persistent knockdown of gene expression in mouse brain. Hum. Gene Ther. 2003;14,:1799–1807. doi: 10.1089/104303403322611809. [DOI] [PubMed] [Google Scholar]
- 22.Lilley CE, Groutsi F, Han Z, Palmer JA, Anderson PN, Latchman DS, Coffin RS. Multiple immediate-early gene-deficient herpes simplex virus vectors allowing efficient gene delivery to neurons in culture and widespread gene delivery to the central nervous system in vivo. J. Virol. 2001;75:4343–4356. doi: 10.1128/JVI.75.9.4343-4356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmer JA, Branston RH, Lilley CE, Robinson MJ, Groutsi F, Smith J, Latchman DS, Coffin RS. Development and optimization of herpes simplex virus vectors for multiple long-term gene delivery to the peripheral nervous system. J. Virol. 2000;74:5604–5618. doi: 10.1128/jvi.74.12.5604-5618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas SK, Lilley CE, Latchman DS, Coffin RS. Equine herpesvirus 1 gene 12 can substitute for vmw65 in the growth of herpes simplex virus (HSV) type 1, allowing the generation of optimized cell lines for the propagation of HSV vectors with multiple immediate-early gene defects. J. Virol. 1999;73:7399–7409. doi: 10.1128/jvi.73.9.7399-7409.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cortright DN, Szallasi A. Biochemical pharmacology of the vanilloid receptor TRPV1. An update. Eur. J. Biochem. 2004;271:1814–1819. doi: 10.1111/j.1432-1033.2004.04082.x. [DOI] [PubMed] [Google Scholar]
- 26.Tominaga M, Tominaga T. Structure and function of TRPV1. Pflugers Arch. 2005;451:143–150. doi: 10.1007/s00424-005-1457-8. [DOI] [PubMed] [Google Scholar]
- 27.Loeb JE, Cordier WS, Harris ME, Weitzman MD, Hope TJ. Enhanced expression of transgenes from adeno-associated virus vectors with the woodchuck hepatitis virus posttranscriptional regulatory element: implications for gene therapy. Hum. Gene Ther. 1999;10:2295–2305. doi: 10.1089/10430349950016942. [DOI] [PubMed] [Google Scholar]
- 28.Stow ND, Wilkie NM. An improved technique for obtaining enhanced infectivity with herpes simplex virus type 1 DNA. J. Gen. Virol. 1976;33:447–458. doi: 10.1099/0022-1317-33-3-447. [DOI] [PubMed] [Google Scholar]
- 29.Ace CI, McKee TA, Ryan JM, Cameron JM, Preston CM. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J. Virol. 1989;63:2260–2269. doi: 10.1128/jvi.63.5.2260-2269.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 31.Xia XG, Zhou H, Ding H, Affar el B, Shi Y, Xu Z. An enhanced U6 promoter for synthesis of short hairpin RNA. Nucleic Acids Res. 2003;31:e100. doi: 10.1093/nar/gng098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ong ST, Li F, Du J, Tan YW, Wang S. Hybrid cytomegalovirus enhancer-h1 promoter-based plasmid and baculovirus vectors mediate effective RNA interference. Hum. Gene Ther. 2005;16:1404–1412. doi: 10.1089/hum.2005.16.1404. [DOI] [PubMed] [Google Scholar]
- 33.Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 34.Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davidson TJ, Harel S, Arboleda VA, Prunell GF, Shelanski ML, Greene LA, Troy CM. Highly efficient small interfering RNA delivery to primary mammalian neurons induces MicroRNA-like effects before mRNA degradation. J. Neurosci. 2004;24:10040–10046. doi: 10.1523/JNEUROSCI.3643-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hao S, Mata M, Goins W, Glorioso JC, Fink DJ. Transgene-mediated enkephalin release enhances the effect of morphine and evades tolerance to produce a sustained antiallodynic effect in neuropathic pain. Pain. 2003;102:135–142. doi: 10.1016/s0304-3959(02)00346-9. [DOI] [PubMed] [Google Scholar]
- 37.Hao S, Mata M, Wolfe D, Huang S, Glorioso JC, Fink DJ. HSV-mediated gene transfer of the glial cell-derived neurotrophic factor provides an antiallodynic effect on neuropathic pain. Mol. Ther. 2003;8:367–375. doi: 10.1016/s1525-0016(03)00185-0. [DOI] [PubMed] [Google Scholar]
- 38.Goss JR, Mata M, Goins WF, Wu HH, Glorioso JC, Fink DJ. Antinociceptive effect of a genomic herpes simplex virus-based vector expressing human proenkephalin in rat dorsal root ganglion. Gene Ther. 2001;8:551–556. doi: 10.1038/sj.gt.3301430. [DOI] [PubMed] [Google Scholar]
- 39.Saydam O, Saydam N, Glauser DL, Pruschy M, Dinh-Van V, Hilbe M, Jacobs AH, Ackermann M, Fraefel C. HSV-1 amplicon-mediated post-transcriptional inhibition of Rad51 sensitizes human glioma cells to ionizing radiation. Gene Ther. 2007;14:1143–1151. doi: 10.1038/sj.gt.3302967. [DOI] [PubMed] [Google Scholar]
- 40.Sabbioni S, Callegari E, Manservigi M, Argnani R, Corallini A, Negrini M, Manservigi R. Use of herpes simplex virus type 1-based amplicon vector for delivery of small interfering RNA. Gene Ther. 2007;14:459–464. doi: 10.1038/sj.gt.3302878. [DOI] [PubMed] [Google Scholar]
- 41.Saydam O, Glauser DL, Heid I, Turkeri G, Hilbe M, Jacobs AH, Ackermann M, Fraefel C. Herpes Simplex Virus 1 Amplicon Vector-Mediated siRNA Targeting Epidermal Growth Factor Receptor Inhibits Growth of Human Glioma Cells in Vivo. Mol. Ther. 2005;12:803–812. doi: 10.1016/j.ymthe.2005.07.534. [DOI] [PubMed] [Google Scholar]
- 42.Caterina MJ, Schumacher MA, Tominaga M, Rosen TA, Levine JD, Julius D. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389:816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- 43.Davis JB, Gray J, Gunthorpe MJ, Hatcher JP, Davey PT, Overend P, Harries MH, Latcham J, Clapham C, Atkinson K, et al. Vanilloid receptor-1 is essential for inflammatory thermal hyperalgesia. Nature. 2000;405:183–187. doi: 10.1038/35012076. [DOI] [PubMed] [Google Scholar]
- 44.Christoph T, Gillen C, Mika J, Grünweller A, Schäfer MK, Schiene K, Frank R, Jostock R, Bahrenberg G, Weihe E, et al. Antinociceptive effect of antisense oligonucleotides against the vanilloid receptor VR1/TRPV1. Neurochem. Int. 2007;50:281–290. doi: 10.1016/j.neuint.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 45.Christoph T, Grünweller A, Mika J, Schäfer MK, Wade EJ, Weihe E, Erdmann VA, Frank R, Gillen C, Kurreck J. Silencing of vanilloid receptor TRPV1 by RNAi reduces neuropathic and visceral pain in vivo. Biochem. Biophys. Res. Commun. 2006;350:238–243. doi: 10.1016/j.bbrc.2006.09.037. [DOI] [PubMed] [Google Scholar]