Abstract
Foxa2 (HNF3β) is a one of three, closely related transcription factors that are critical to the development and function of the mouse liver. We have used chromatin immunoprecipitation and massively parallel Illumina 1G sequencing (ChIP–Seq) to create a genome-wide profile of in vivo Foxa2-binding sites in the adult liver. More than 65% of the ∼11.5 k genomic sites associated with Foxa2 binding, mapped to extended gene regions of annotated genes, while more than 30% of intragenic sites were located within first introns. 20.5% of all sites were further than 50 kb from any annotated gene, suggesting an association with novel gene regions. QPCR analysis demonstrated a strong positive correlation between peak height and fold enrichment for Foxa2-binding sites. We measured the relationship between Foxa2 and liver gene expression by overlapping Foxa2-binding sites with a SAGE transcriptome profile, and found that 43.5% of genes expressed in the liver were also associated with Foxa2 binding. We also identified potential Foxa2-interacting transcription factors whose motifs were enriched near Foxa2-binding sites. Our comprehensive results for in vivo Foxa2-binding sites in the mouse liver will contribute to resolving transcriptional regulatory networks that are important for adult liver function.
INTRODUCTION
Forkhead box A2 (Foxa2, HNF3β) is one of three closely related transcription factors, whose expression is an initiating factor in the earliest stages of liver specification during embryonic development and normal liver homeostasis in the adult liver (1–3). Foxa1 (HNF3α), Foxa2 and Foxa3 (HNF3γ) were initially identified in liver cells by their ability to interact with the promoters of two important liver-expressed genes—transthyretin (Ttr) and alpha-1-antitrypsin (A1AT) (4). The three members of this group contain a highly conserved winged-helix DNA-binding domain and are critically important in mouse liver development and maintenance. While mutational analysis has shown that the three FoxAs are functionally redundant in the mouse liver, with all three able to bind to the same sequence (5), Foxa1 or Foxa2 are most critical for the specification of the liver (6). Foxa2 is the first of the three family members to be expressed in the embryo prior to gastrulation, with Foxa1 and Foxa3 expression following at the onset of gastrulation (1). All three are expressed in the embryonic endoderm cells that constitute the precursor cells for all gut organs, and all three remain present and active in the adult liver. In the adult, Foxa2 plays a major role in glucose and lipid metabolism, targeting genes such as glucose-6-phosphatase, catalytic (G6pc) and tyrosine aminotransferase (Tat) (3,7). In addition to its role as an important transcription factor regulating gene expression in liver development and function, Foxa2 can also open compacted chromatin, allowing for the activation of transcription from silenced genes, and so can act as a pioneer transcription factor in liver signaling cascades (8).
In the adult, the liver is involved in the maintenance of general homeostasis. It is comprised primarily of hepatocytes and has many varied roles including regulation of metabolic activity, cholesterol production and secretion, production of plasma proteins, chemical detoxification, drug and alcohol elimination and, during embryonic development, site of red blood cell production. The cellular functions of the normal liver are governed through the dynamic interaction of a central group of transcription factors that regulate liver-specific gene expression. This group includes HNF4, Foxa1, Foxa2, Foxa3, Tcf1/2 (HNF1α/β), Onecut1 (HNF6) and C/EBPα. These transcription factors interact in highly complex ways that are continually regulated throughout prenatal and postnatal development (9).
Until recently, the transcriptional regulatory functions of FoxA proteins were studied on a gene-by-gene basis. However, the advent of chromatin immunoprecipitation coupled with microarray technology (ChIP-chip) has allowed the identification of in vivo binding locations on a larger scale. Several groups have used ChIP-chip to investigate the in vivo binding sites of important liver transcription factors, including Foxa2 (9–17). Most microarray studies have targeted promoter regions that are closely associated with transcriptional start sites (TSS) of genes; however, it has been increasingly recognized that transcription factors have the ability to regulate gene activity from greater distances. A study in the hepatoma cell line HepG2, using ChIP combined with a microarray that covered the ENCODE regions representing 1% of the human genome, demonstrated that in vivo transcription factor-binding sites (TFBS) often occur outside of traditional promoter regions (14). Furthermore, recent genome-wide ChIP-sequencing experiments have highlighted the benefits of binding site analyses of transcription factors that are not restricted to specific genomic regions or by the need for annotated genes (18–21). Newer approaches to massively parallel sequencing (22–24) have the ability to more accurately detect transcription factor binding in any genomic region at much lower cost than earlier global sequencing methods. Since Foxa2 acts not only as an important transcriptional regulator in liver development and maintenance but also as competence factor in accessibility of DNA by other transcription factors, understanding its genomic wide binding locations is imperative to understanding of gene regulation in the liver.
This study presents the first genome-wide investigation of in vivo Foxa2 binding target regions in the adult mouse liver. We combined the power of a second-generation DNA sequencer with ChIP and quantitative PCR (qPCR) validation to show not only high coverage of well-characterized Foxa2 target sites and genes, but also novel aspects of Foxa2 interaction with the mouse genome. We show that the majority of Foxa2-binding sites are within 10 kb upstream of TSS and within the first introns of genes. In addition, we provide evidence that Foxa2 binds a large number of targets in unannotated genomic regions. We also show that Foxa2's apparent influence on gene expression remains extensive in the adult liver, with close to half of abundantly liver-expressed genes being associated with at least one Foxa2-binding site. Finally, we have demonstrated the presence of additional transcription factor motifs within Foxa2-binding site sequences that represent potential Foxa2-interacting partners. We present analyses of the expression of each transcription factor in the liver and function of the group of genes with which each motif is associated. We have identified a large number of novel Foxa2 in vivo binding sites, which may be involved in specific gene regulation or permissive gene regulation through the opening of chromatin. We have also confirmed Foxa2 binding to many novel sites using ChIP–qPCR indicating the high quality of our dataset. By identifying Foxa2 binding locations in combination with gene expression and bioinformatics analysis of potential cointeracting transcription factors, we provide novel insights into Foxa2 transcriptional networks in the adult mouse liver.
METHODS
ChIP
Adult female C57Bl/6J mouse livers were homogenized in 1% formaldehyde and incubated at room temperature (RT) for 10 min, prior to the addition of glycine to 0.125 M followed by incubation at RT for 5 min. Cells were pelleted, washed and resuspended in 5 volumes ChIP cell lysis buffer (10 mM Tris–Cl, pH 8.0, 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40) containing protease inhibitor cocktail tablets (Roche, Laval, QC, Canada). Cells were then rehomogenized, incubated on ice for 5 min and repelleted. The pellet was resuspended in 3 volumes ChIP nuclear lysis buffer (1% SDS, 5 mM EDTA, 50 mM Tris–Cl, pH 8.1) supplemented with protease inhibitor cocktail tablets (Roche). Cells were sonicated on ice-water (Sonicator 3000, Misonix, Farmingdale, NY, USA) for 20 cycles of 30 s on, 40 s off and 60 μg of chromatin was precleared with Protein G beads (100 μl, Active Motif), Protein Inhibitor Cocktail (0.5 μl, Active Motif) and ChIP dilution buffer (to 250 μl, 0.01% SDS, 1.1% Triton X-100, 167 mM NaCl, 16.7 mM Tris–Cl, pH 8.1). Beads were precipitated and 3 μg of Foxa2 [HNF-3β (M-20): sc-6554, Santa Cruz] antibody or normal rabbit IgG (sc-2027, Santa Cruz, Santa Cruz, CA, USA) was added to supernatants. Fresh Protein G beads were also blocked with 1 mg/mg BSA and 0.1 mg/mg herring sperm DNA in ChIP dilution buffer. Following overnight incubation, rocking at 4°C, the samples were incubated with the blocked beads for 4 h rocking at 4°C. The beads were then precipitated and washed in low salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–Cl, pH 8.1, 150 mM NaCl), high salt buffer (low salt buffer with 500 mM NaCl), lithium chloride buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris–Cl, pH 8.1) and twice with TE buffer. Bead complexes were eluted twice by addition of 125 μl elution buffer (1% SDS, 0.1 M NHCO3) and rotation for 15 min at RT. NaCl was added to 0.192 M to reverse crosslink and samples were incubated overnight at 65°C. Samples were then incubated with Proteinase K (Invitrogen, Carlsbad, CA, USA) and RNaseA (Sigma-Aldrich, Oakville, ON, Canada) for 2 h at 50°C. DNA was purified by two rounds of phenol–chloroform extraction and ethanol precipitation and resuspended in 50 μl dH2O.
Sequencing
Foxa2-bound DNA (4.7 ng) was purified by SDS–PAGE to obtain 100–300 bp fragments and sequenced on an Illumina 1G sequencer according to Robertson et al. (24). Briefly, size fractionated DNA was extracted and a single adenosine was added using Klenow exo– (3′ and 5′ exo minus; Illumina). Illumina adaptors were then added and DNA was subjected to 20 cycles of PCR according to manufacturer's instructions. We then purified DNA and performed cluster generation and 27 cycles of sequencing on the Illumina cluster station and 1G analyzer following the manufacturer's instructions.
Sequencing analysis and peak production
Resulting sequences were mapped to the NCBI Build 36 (mm8) reference mouse genome to produce 13 984 706 mapped reads that were extended to 200 bp length XSETs and overlapped to create peaks according to Robertson et al. (24). To further define the peak dataset, each group of XSETs that represented DNA fragments with identical fragment start coordinates was ‘collapsed’ to a single XSET. Peaks generated from the resulting filtered reads were thresholded at a peak height of 10, based on the stabilization of the FoxA-like binding sequences enrichment curve (Supplementary Figure 1), creating a high confidence set of Foxa2-binding sites with an estimated false discovery rate of 0.05. The dataset containing all 11 475 peaks can be downloaded from www.bcgsc.ca/data/chipseq.
Foxa2 enrichment analysis
A set of 42 DNA sequences known to bind FoxA proteins in mouse, human or rat was extracted from the literature. The sites, with supporting evidence, are available at the ORegAnno database (www.oreganno.org) (25). Because sequence lengths ranged from 7 to 22 bp, we created a Foxa2 model by adding flanking sequences. We aligned the flanked sequences by scanning them with the Foxa2 position weight matrix (PWM) from the JASPAR database (jaspar.cgb.ki.se) (26). For each flanked sequence, we retained the top scoring sub-sequence that contained at least 5 bp from the binding site originally reported in the literature. The logo for the resulting 42 aligned sequences, 39 of which were unique, is shown in Supplementary Figure 2. To assess the extent to which ChIP–Seq peaks were enriched in Foxa2-like binding sequences, we used a custom Java algorithm to scan peak sequences with a 10-mer representation of each known functional site. We permitted either no mismatches or up to one mismatch at positions that had an information content below 0.6 bits [Supplementary Figures 1 and 7 in Robertson et al. (24)]. We also scanned peak sequences with a PWM constructed from the aligned site sequences, using the above algorithm (Figure 1B). The score assigned to a tested DNA sequence was the ratios of the probabilities of observing a given nucleotide from the PWM model, and from the background model, for all positions in the matrix. We assumed the background probabilities to be 0.25 for all four nucleotides.
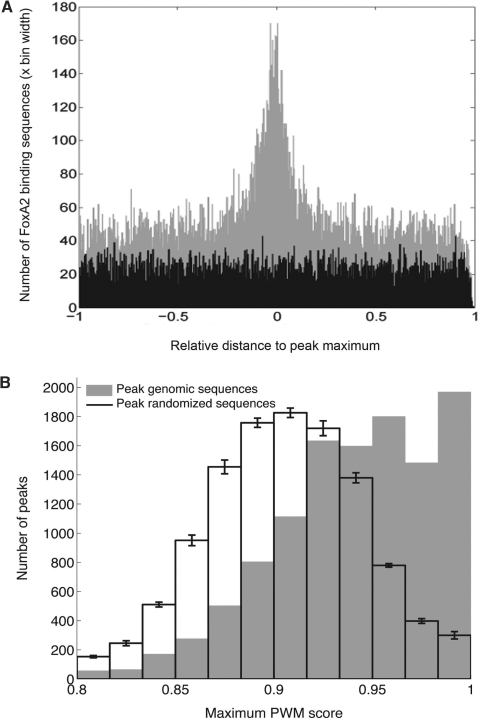
Figure 1.
(A) Distances from sites similar to Foxa2-binding sequences to the peak maximum, normalized to peak width. This graph shows that Foxa2-binding sequence sites tended to cluster around peak maxima. (B) Peak enrichment in Foxa2-like sequences, as assessed with PWM scan scores. The Foxa2 PWM used was created from the aligned sequences used to generate the sequence logo shown in Supplementary Figure 2 (see Methods section). The graph indicates that peak genomic sequences contained higher scoring Foxa2-binding sequences than randomized peak sequences; that is, peaks were enriched relative to a random expectation.
Quantitative real-time PCR
Quantitative real-time PCR (qPCR) was performed on an ABI 7900HT Fast Real-Time PCR System using 2x SYBR® Green Master PCR Mix according to the manufacturer's instructions (both from Applied Biosystems, Foster City, CA, USA). The fold enrichment of each target site was calculated as 2 to the power of the cycle threshold (cT) difference between an IgG immunoprecipitated sample and a Foxa2 immunoprecipitated sample. We selected primer sets for six negative regions, all of which showed low enrichment between 0.8 and 1.2. Forty-two other validation sites were chosen over a range of subgroups as defined in the results (41 positive and 1 negative). All primers are listed in Supplementary Table 3.
SAGE
The adult mouse liver long Serial Analysis of Gene Expression (longSAGE) library (SM100) was produced as previously described (27). Briefly, a LongSAGE library was created from 4.7 μg of DNase-treated total RNA using the I-SAGE Long kit and protocol (Invitrogen) and sequenced on capillary DNA sequencers (Applied Biosystems). Tag sequences were mapped to the genome using NCBI Build 36 mouse assembly. SAGE tag sequences for this library are publicly available at www.mouseatlas.org.
SiRNA analysis
SiRNA experiments were performed in the Hepa1-6 hepatocyte cell line. Cells were grown and maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% FBS. Control and mouse Foxa2 siRNA pools were obtained from Santa Cruz and cells were treated with 100 nM siRNA overnight in serum-free DMEM. Media was replaced the following day with DMEM with 10% FBS and cells were cultured for 72 h, prior to total RNA isolation by using Trizol according to the manufacturer's instructions. RNA was reverse transcribed using Superscript III (Invitrogen) to obtain cDNA for use in QPCR reactions. Results presented are an average of three independent experiments.
Transcription factor comotif analysis
The MATCH v2.6 algorithm was used with a profile filter that specified low false positives and good quality models to scan sequences derived from the 3016 peaks that contained at least one exact 10-mer match to one of the 39 known functional sequences, as well as sequences from the 2494 peaks that did not contain such a match (28). A total of 363 vertebrate TRANSFAC Professional v9.3 PWMs were assessed. Scanned sequences extended ±250 bp from each putative Foxa2-binding site under a peak, or for peaks without exact match hits, from the location of the peak maximum. The statistical significance of the number of scan hits per kilobytes for each model was estimated by generating and scanning 100 third-order Markov random sequences from a mouse genomic model outside of open reading frames (rsat.ulb.ac.be/rsat), each of which had a length equal to the combined length of all scanned peak sequences (29). For each TFBS model, we calculated an empirical P-value and z-score from peak versus random scan results, and used the P-value, and then the z-score, to rank the 363 models.
RESULTS
Identification and characterization of Foxa2-binding sites in vivo
To determine locations of in vivo Foxa2 binding in the mouse adult liver, we sequenced Foxa2-bound DNA fragments by combining chromatin immunoprecipitation and massively parallel Illumina 1G sequencing (ChIP–Seq) (22–24). The sequences were then mapped to the mouse genome (NCBI Build 36, UCSC mm8) (see Methods section). Foxa2-bound regions are represented by peaks consisting of overlapping mapped sequences. To determine a high confidence subset of peaks at a minimal peak height, we determined the number of peaks containing a Foxa2-binding sequence. We extracted and aligned a set of 42 sequences (39 of which are unique) that are known from the literature to bind FoxA proteins in mouse, human or rat (see Methods section). This resulted in a 10-mer binding sequence model (Supplementary Figure 1, position 2–11). We scanned all peaks, allowing a maximum of 1 bp mismatch, and assessed the relationship between peak height and the fraction of peaks containing Foxa2-binding sequences (Supplementary Figure 3A). As anticipated, with increasing peak heights, the percentage of peaks that contained a Foxa2-binding site sharply increased until a peak height of 10, where the curve of enrichment began to level out. Similar comparisons of randomized peak sequences did not show the same increase in binding sequences with respect to peak height. The level of plateau of peaks occurred at peak height 10, resulting in a dataset of 11 475 high confidence peaks of height 10 or above, of which, 8981 (78%) contained at least one Foxa2-binding sequence (FDR = 0.05). Overall, we identified 22 758 individual binding sequences, with an average of 2.5 binding sequences per peak (Supplementary Figure 3B). The peaks had an average width of 728 bp and an average height of 16 (Table 1). Foxa2-binding site were preferentially found close to the peak maximum (Figure 1A) and most peaks had a Gaussian-like shape, suggesting that the location of the Foxa2-binding site responsible for a peak can be inferred from the shape of the peak and the location of the peak height maximum. Peaks were distributed across the mouse genome with approximately equal numbers of peaks per chromosome when normalized for chromosome length (Supplementary Figure 2A). To further confirm that our peaks were enriched in potential Foxa2 binding sequences, we created a PWM based on our aligned literature-derived Foxa-binding sequences and scored each peak (see Methods section). Significantly, the number of peaks containing a high PWM score was highly enriched in our dataset over randomized peak sequences (P = 0) (Figure 1B).
Table 1.
Global peak properties and peak locations relative to UCSC mm8 known genes
| Minimum peak height | 10 |
| # Peaks | 11 475 |
| # Individual DNA reads under peaks | 285 176 |
| Average (median) peak height | 16 (13) |
| Average (median) peak length (bp) | 727.5 (654) |
| # Peaks within ±1 kb TSSa | 694 (6.0%) |
| # Peaks in 10 kb upstream TSS regiona | 1800 (15.7%) |
| # Intragenic peaksa | 5575 (48.6%) |
| # Peaks in 1st introna | 1852 (16.1%) (33.2% of intragenic) |
| # Peaks within extended gene region of annotated genesa | 7808 (68.0%) |
| # Peaks >50 kb from a gene | 2356 (20.5%) |
| # Genes (extended gene region) with peaks | 5060 |
| # Genes (extended gene region) with peak and SAGE tag | 2781 (43.5% of genes with tag) |
aPeak maximum falls within these regions.
While relatively few peaks (6%) had a maximum within 1 kb of a TSS, a higher than expected percentage of peaks (33.2%) had a maximum within a first intron. A peak was associated with a gene if the peak's maximum was within a region that extended from 10 kb upstream from the gene's TSS to 1 kb downstream of its transcription termination site. Genes with a peak and SAGE tag refers to peaks with maxima within the extended region of a gene that also had SAGE tag(s).
We compared the locations of the high-confidence peaks to UCSC mm8 annotated genes, repetitive regions and conserved genomic elements and found that 68% of peaks were located within an extended gene region encompassing 10 kb upstream of the TSS to 1 kb downstream of the 3′UTR. Interestingly, only 6.0% of the peaks were located within 1 kb upstream or downstream of the TSS of annotated genes, while 15.7% of the peaks were located within 10 kb upstream of the TSS and 16.1% were localized to a first intron (Table 1). We also observed that 20.5% of all peaks were more than 50 kb from any annotated gene (Table 1). Of these peaks, 40% overlapped conserved genomic regions, suggesting that either Foxa2 can act at remote distances from genes or that a number of Foxa2-regulated genes have yet to be annotated. Only 5% of these remote peaks overlapped microsatellite repeats (data not shown). This level of overlap is consistent with only 4% of peaks in the total dataset overlapping microsatellite repeats (data not shown), indicating a low occurrence of peaks over sequence repeat regions. Thus, the Foxa2-binding sites we observed are not preferentially located close to an annotated TSS but are observed at a range of locations across the mouse genome.
Peaks were associated with known Foxa2-target genes and binding sites
To validate our high-confidence peak set in silico, we looked for peaks that overlapped known, characterized Foxa2-binding sites in the mouse liver. Of a set of 16 binding sites described in vitro for the mouse liver (representing 10 target genes) (30–42), our dataset demonstrated peaks overlapping nine sites that were associated with six genes. To confirm that those previously published sites over which we did not see a peak were in fact negative for Foxa2 binding in the adult liver, we investigated the Hhex and PTG Foxa2-binding sites by qPCR on replicate ChIP experiments. Neither of these sites showed enrichment by qPCR (Hhex = 0.7 average enrichment, PTG = 2.2 average enrichment), indicating little or no Foxa2 binding. This suggests that some Foxa2 sites characterized in vitro are not always occupied in the adult mouse liver in vivo. Of those sites associated with a peak, the location of the site corresponded closely to the location of the peak maximum. From a set of 50 genes known to be regulated by FoxA proteins (where the exact binding site may or may not be characterized) (3,4,9,34,42–67), we observed peaks in the extended gene regions of 36 genes (72%) (Supplementary Table 1). Overall, our success at identifying known Foxa2 targets indicates that our dataset has high coverage of bona fide in vivo Foxa2-binding sites across the genome.
Of the nine well-characterized sites over which we saw peaks, we tested the fold enrichment of five by ChIP–qPCR. The sites chosen were located in the Foxa2, transthyretin (Ttr) and cytosolic phosphoenolpyruvate carboxykinase 1 (Pck1) promoters, the albumin (Alb) enhancer and the hepatocyte nuclear factor 4 alpha (HNF4α) enhancer (Figure 2A). Enrichment was observed for all sites, with the greatest enrichment seen at the HNF4α enhancer (52.9-fold) (Figure 2A). Interestingly, the HNF4α promoter site, which has also been reported to be bound by Foxa2, was not overlapped by a peak of height 10 or above and showed enrichment of only 2.5 by qPCR, confirming data shown by Hatzis et al. (36) that the HNF4α promoter site is likely not occupied by Foxa2 in the adult mouse liver in vivo. We also tested additional, previously uncharacterized, sites in the promoter/enhancer regions of Foxa2 regulated genes over which peaks of height 10 and above were observed. These included a site in the Alb gene, 5′ to the known enhancer and promoter sites (Figure 3A), as well as peaks in the alpha fetoprotein (Afp), vitronectin (Vtn) and tyrosine aminotransferase (Tat) genes. All four showed enrichment by qPCR, ranging from 14- to 30-fold (Figure 3B), indicating that we were able to identify additional binding sites associated with established Foxa2-target genes.
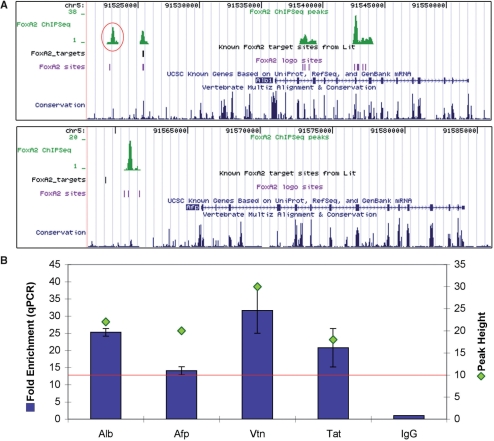
Figure 2.
Characterized target Foxa2-binding sites. (A) Table of six characterized target sites in five target genes showing peak presence or absence, peak height and fold enrichment by qPCR. All three sites that contained a peak also showed enrichment above 2-fold. (B) UCSC genome browser mm8 screenshots of four target sites at two of the target genes. Of the HNF4α enhancer and promoter sites, only the enhancer was covered by a peak in our adult liver dataset. The Ttr gene has two closely related promoter sites from different publications, both of which were overlapped by a single peak in our dataset. An additional track displaying the Foxa2-like binding sequence sites from scanning our 10-mer model is also shown (‘Foxa2 logo sites’).
Figure 3.
Characterization of novel target sites associated with known Foxa2-target genes. (A) UCSC genome browser screenshots of peaks associated with the Alb and Afp genes. The Alb novel site is an additional location to the characterized target site; both sites were overlapped by peaks, indicating binding by Foxa2 in the liver. The Afp novel site is an alternate location 5′ to the site characterized in embryonic liver. (B) QCPR fold enrichment for four novel target sites in characterized Foxa2-target genes averaged over three biological replicate Foxa2 ChIP experiments. Both the fold enrichment and the peak height are shown; the red line indicates the minimum peak height threshold for our dataset (i.e. 10 XSETs). All enrichments were calculated using IgG enrichment as a control in all graphs showing qPCR results. All four novel target sites had fold enrichments above 13.
Many genes expressed in the liver contained Foxa2-binding sites
To correlate Foxa2 binding with gene expression, we determined the number of genes that are both transcribed in the liver and potentially regulated by Foxa2. We generated a transcriptional profile of active genes by constructing a longSAGE library from adult mouse liver, using the protocol described previously (27,68). We sequenced this library to a depth sufficient to detect abundant and moderately abundant transcripts (66 272 high quality tag counts). At this depth, many transcripts are represented by only one tag (singletons). However, these have previously been shown to represent bona fide transcripts (27). These tag counts represented 14 026 tag sequence types that mapped to 6396 annotated genes (NCBI Build 36, UCSC mm8). We then overlapped data from this library to our ChIP–Seq dataset to identify genes that were both expressed and associated with in vivo Foxa2 binding (Supplementary Figure 4). We found that 2781 of these genes had one or more Foxa2-binding site(s), suggesting that 43.5% of genes expressed in the adult liver can be associated with an active Foxa2-binding site.
We used Gene Ontology (GO) analysis (FatiGO, fatigo.bioinfo.cipf.es) to identify shared functional properties for the group of expressed genes that were associated with at least one Foxa2 peak (Supplementary Table 2) (69). Six of the top 10, hierarchical level 5, Biological Process categories were associated with metabolism, indicating that these genes were primarily involved in the main biological function of the adult liver. However, we also noted that ‘transcription’ ranked fourth in the top ten Biological Process categories (16.8% of genes), and that ‘transcription factor activity’ ranked sixth in the top ten, hierarchical level 5, Molecular Function categories (6.5% of genes). This suggests that those liver-expressed genes potentially regulated by Foxa2 also involved in transcriptional processes and supports the role of Foxa2 as a major transcription factor regulating gene expression in the liver.
We further characterized ChIP–Seq's ability to identify novel Foxa2-binding sites using ChIP–qPCR on biological triplicates. We tested peaks that ranged in heights and locations, including peaks both with and without a Foxa2-binding sequence. While all peaks chosen were associated with genes that our SAGE results indicated were transcribed in the liver, we divided the genes into those that had published evidence for expression or function in the liver and those that did not (70–77). Of the 16 sites tested, 14 showed an enrichment of >3-fold by qPCR (Figure 4A and B). Of peaks associated with genes with characterized roles in the liver, the greatest enrichment of 30-fold was seen for ApoA2, which is a member of the apolipoprotein family, involved in metabolism. Among the genes lacking characterized roles in the liver, the greatest enrichments of 22.7- and 14.8-fold were seen for the undefined Gtl3 and AI132487 genes, respectively. Of the 16 sites characterized by qPCR, only two did not show enrichment above 3-fold: Cask (does not contain a Foxa2-binding sequence) and Dock8 (contains a Foxa2-binding sequence) (1.2- and 1.5-fold enrichment, respectively). Interestingly, 10 out of the 11 peaks that did not contain Foxa2-binding sequences did show enrichment by qPCR, indicating that the presence of such a sequence within DNA is not essential Foxa2 binding. This observation suggests that there are unidentified Foxa2-binding sequences located within these peaks, or that Foxa2 is localized to these regions through protein–protein interactions.
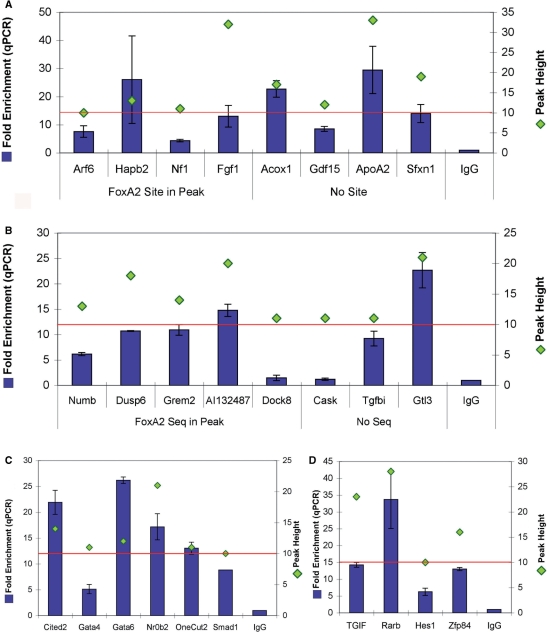
Figure 4.
QPCR fold enrichment of annotated genes containing Foxa2-binding site peaks. All genes shown contained SAGE tag(s) indicating that they were expressed in the adult liver. QPCR fold enrichment and peak height are shown for each target; the red line indicates the peak height threshold of 10 XSETs. As previously, IgG enrichment served as the control. (A) Annotated genes that have characterized roles in liver function. Peaks are divided into those that contained a Foxa2-binding sequence and those that did not. All eight targets showed fold enrichments above 4. (B) Annotated genes that have not previously been shown to be expressed in the liver, with peaks divided into those that contained a Foxa2-binding sequence and those that did not. Dock8 and Cask, which did not show enrichment, were the only targets out of 41 peaks tested that were not enriched. The peak height of these two targets was close to the peak height threshold. The remaining six targets all showed fold enrichments above 6. (C) Transcription factors that have a characterized role in liver function in literature. All six targets showed fold enrichments above 5. (D) Transcription factors with uncharacterized roles in the liver. Expression of these transcription factors in the liver is a novel finding. All four targets showed fold enrichments above 5.
As shown earlier, >15% of genes associated with Foxa2-binding sites are involved in transcription as classified by GO. This observation supports Foxa2 having a role in regulating downstream transcription factors, consistent with results that show regulatory relationships between a core group of six liver transcription factors (9,12,13). To identify novel regulatory cascades involving Foxa2, we measured qPCR fold enrichments for 10 Foxa2 peaks that were associated with genes encoding transcription factor proteins. Those genes with characterized roles in the liver included GATA binding protein 4 (Gata4), GATA binding protein 6 (Gata6), nuclear receptor subfamily 0, group B, member 2 (Nr0b2), one cut domain family member 2 (Onecut2), MAD homolog 1 (Smad1) and Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 (Cited2) (78–82). Those without known roles in the liver included TG-interacting factor (TGIF), retinoic acid receptor, beta (Rarb), hairy and enhancer of split 1 (Hes1) and zinc-finger protein 84 (Zfp84). Peak heights ranged from the minimum threshold of 10 up to 28; qPCR enrichment was observed for all 10 peaks, ranging from 5.1 for Gata4 to 33.7 for Rarb (Figure 4C and D). These results further supported a role for Foxa2 in transcriptional cascades in the liver and suggest that peaks associated with transcription factor genes in our dataset had a high probability of representing in vivo Foxa2 cross-transcription factor promoter binding sites.
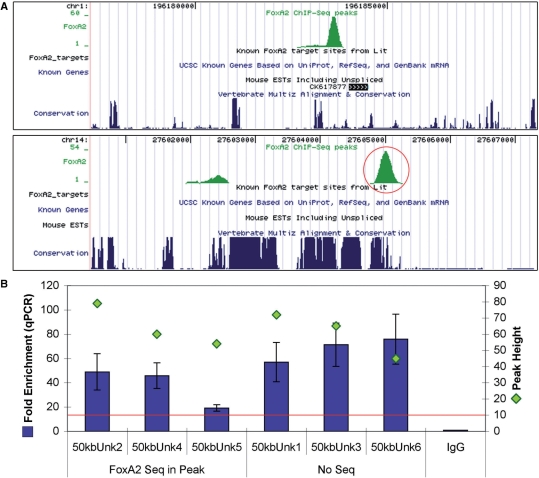
One of the strengths of our genome-wide analysis of Foxa2-binding sites was that it had the capability of identifying novel gene regions that were potentially regulated by Foxa2. As mentioned earlier, 20.5% of Foxa2-binding sites were located >50 kb from any annotated gene (Table 1). We used qPCR to determine the fold enrichment of six of these peaks, whose heights ranged from 45 to 79. As for the groups described earlier, the peaks included both those with and without Foxa2-binding sequences. All six peaks overlapped, or were near, regions of high inter-species sequence conservation and none of the six were located over microsatellite repeats. Figure 5A shows two example peaks; one close to a mouse EST (unknown #4) and the other adjacent to several highly conserved regions (unknown #5). Fold enrichments by qPCR ranged from 19.1 for unknown #5 to 75.9 for unknown #6 (Figure 5B).
Figure 5.
Examples of peaks located >50 kb away from annotated genes. (A) UCSC genome browser mm8 screenshots of two remote peaks in regions containing no known genes. The first peak (unknown #4) lies close to a mouse EST, while the second peak (unknown #5) overlaps and is close to highly conserved sequence regions. (B) Graph of qPCR results of all six peaks lying in unknown gene regions. Four peaks contained Foxa2-binding sequences and two peaks did not. As previously, both fold enrichment by qPCR and peak height are shown. Fold enrichments are relative to IgG control. All six peaks showed fold enrichments above 19.
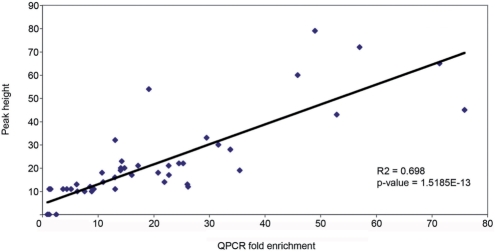
In summary, we tested 41 identified peaks by ChIP coupled with qPCR, corresponding to heights ranging from 10 to 79 from our high-confidence dataset. We also tested six negative sites selected from gaps between peaks of height 2. None of the six negative sites showed qPCR enrichment, while 39 of 41 peaks (95%) showed enrichments greater than 3-fold. Both of the peaks that showed no enrichment by qPCR (Cask and Dock8) had a height of 11, close to our minimal height cutoff. However, eight of the 10 sites chosen for qPCR investigation that had peak heights of 10 or 11 did show enrichment, indicating that, even at and near the peak height threshold of 10, at least 80% of peaks represented in vivo Foxa2-binding sites. We plotted the mean fold enrichment of three biological replicates for each qPCR-validated site against the height of the corresponding peak (Figure 6). Significantly, we observed a strong positive correlation (R2 = 0.698, P = 1.5E-13), demonstrating that peak height was a good indicator of the likely fold enrichment in biological replicates as measured by qPCR. We also tested an additional 33 sites by ChIP–qPCR consisting of both sites from peaks below the height 10 threshold cutoff, as well as sites predicted to show Foxa2 binding from previous publications where no peak was seen (e.g. Hhex) (data not shown). From the 81 total sites tested, using peaks of height 10 and above as true positives, we calculated a false negative rate of 8.9% and a positive predictive value (PPV) of 94.4%. Together, these results indicate that our dataset, which included peaks that varied widely with respect to location within a gene, height and the presence or absence of a Foxa2-binding sequence, represents a high confidence set of Foxa2-binding sites in the adult mouse liver.
Figure 6.
Positive correlation between fold enrichment by qPCR and peak height. Each data point represents the average fold enrichment of three separate Foxa2 ChIP experiments. As indicated, taller peaks showed greater fold enrichments. For this dataset, peak height can be considered to be a predictor of expected fold enrichment.
Foxa2 knockdown affects target gene expression
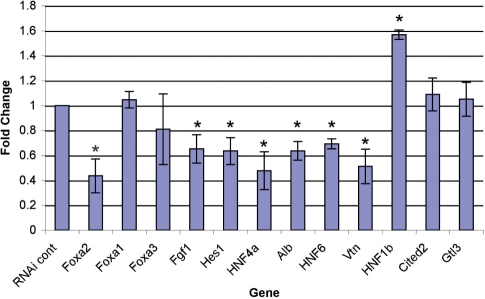
To determine whether Foxa2 binding affects gene expression levels of targets identified by ChIP–Seq, we knocked down Foxa2 in the mouse hepatocellular carcinoma cell line Hepa1-6 by using siRNA pools. This cell line has properties of the developing liver and thus does not express all targets investigated above. However, we were able to identify a subset of targets which were expressed. We investigated the expression of nine of these, including both known (Alb, HNF4α, HNF6) and novel (HNF1β, Hes1, Fgf1, Vtn, Cited2, Gtl3) targets, following Foxa2 knockdown. As shown in Figure 7, we were able to decrease Foxa2 expression by ∼50%, while not significantly affecting levels of Foxa1 or Foxa3 gene expression. Foxa2 protein knockdown was also confirmed by western blotting (data not shown). The knockdown of Foxa2 resulted in significant decrease in expression of six of the nine targets (Alb, HNF4α, HNF6, Hes1, Fgf1, Vtn) and an increase in expression of one—HNF1β. Expression of Cited2 and Gtl3 remained unchanged upon Foxa2 knockdown. These data suggest that many of the Foxa2-binding sites identified can function to regulate gene expression.
Figure 7.
Foxa2 knockdown in Hepa1-6 cells and its effect on expression of targets identified by ChIP–Seq. Foxa2 siRNA treatment results in ∼50% knockdown of Foxa2 expression compared to control siRNA. Upon Foxa2 knockdown, expression of both known (Alb, HNF4α) and novel (HNF6, Hes1, Fgf1, Vtn) decreased. Expression of HNF1β increased, while expression of Cited2 and Gtls3 remained unchanged. The expression of Foxa1 was unaffected by Foxa2 knockdown, indicating the specificity of the Foxa2 siRNA. *P < 0.05.
Foxa2 peaks contained binding motifs for potential coregulatory transcription factors
Foxa2 can cooperate with other transcription factors to regulate gene expression. To identify potential cofactors, we used 363 vertebrate TFBS models from TRANSFAC Professional v9.3 to scan sequences under the 3016 peaks that contained an exact match to our 10-mer Foxa2-binding sequence (see Methods section). The sequences scanned extended 250 bp on either side of a peak's Foxa2-binding sequence(s). The 20 TFBS models with the highest enrichment rankings are shown in Table 2; each of these had an estimated enrichment P < 0.05. While the list included Fox TFBS models, as expected, it also included the transcription factors HNF4, HNF1, CoupTF, GATA4 and Pax6, which are all known to cooperate with Foxa2 (12,83,84). In addition, the list identified nine novel potential cofactors that included a wide variety of transcription factor types. We determined whether individual transcription factors were expressed in the mouse adult liver using a combination of SAGE data or reverse transcription coupled with PCR. Twelve out of the top 20 transcription factors were expressed in the liver, and a further four transcription factor families were represented by TFBS models.
Table 2.
The top 20 enrichment-ranked TRANSFAC v9.3 TFBS models that co-occurred within ±250 bp of Foxa2-binding sequences within peaks or occurred in within ±250 bp of a peak maximum in peaks without Foxa2 sites
| TFBS model | Model # | No. of peaks | Z-score | TF in liver? | No. of genes | Level 5 GO Biological Process | Percentage (%) |
|---|---|---|---|---|---|---|---|
| Peaks with exact FoxA2 sequence | |||||||
| FOX_Q2 | M00809 | 995 | 231.3 | Family | 621 | Cellular metabolic process | 25.0 |
| PAX6_Q2 | M00979 | 138 | 161.3 | No | 102 | Biopolymer modification/Cellular protein metabolic process | 29.0 |
| HFH3_01 | M00289 | 448 | 140.9 | No | 302 | RNA metabolic process/Cellular protein metabolic process | 21.4 |
| MAZR_01 | M00491 | 139 | 106.3 | Yes (PCR) | 83 | RNA metabolic process | 30.8 |
| HNF3B_01 | M00131 | 569 | 82.4 | Yes (tag) | 340 | Cellular protein metabolic process | 26.2 |
| HNF4_Q6_01 | M01031 | 791 | 58.8 | Yes (tag) | 462 | Cellular protein metabolic process | 24.0 |
| MAZ_Q6 | M00649 | 245 | 56.8 | Yes (tag) | 162 | Regulation of cellular metabolic process | 29.4 |
| HNF4_DR1_Q3 | M00764 | 754 | 53.3 | Yes (tag) | 462 | RNA metabolic process | 22.9 |
| SMAD3_Q6 | M00701 | 97 | 44.0 | Yes (PCR) | 64 | Cellular protein metabolic process | 33.3 |
| COUPTF_Q6 | M01036 | 479 | 43.8 | Family | 270 | Cellular protein metabolic process | 21.3 |
| PAX_Q6 | M00808 | 706 | 41.5 | Family | 433 | Cellular protein metabolic process | 26.5 |
| HNF1_Q6 | M00790 | 844 | 41.2 | Yes (tag) | 465 | Regulation of cellular metabolic process | 21.5 |
| GATA4_Q3 | M00632 | 1285 | 37.6 | Yes (tag) | 700 | Cellular protein metabolic process | 24.9 |
| HNF1_C | M00206 | 1029 | 34.8 | Yes (tag) | 549 | Cellular protein metabolic process | 29.7 |
| EVI1_04 | M00081 | 1028 | 33.5 | Yes (PCR) | 565 | Cellular protein metabolic process | 25.9 |
| PPARG_03 | M00528 | 522 | 32.2 | Yes (tag) | 310 | RNA metabolic process | 19.3 |
| LDSPOLYA_B | M00317 | 240 | 30.5 | No | 129 | RNA metabolic process | 28.8 |
| FREAC2_01 | M00290 | 34 | 30.5 | No | 23 | Regulation of cellular metabolic process/RNA metabolic process/Transcription | 43.8 |
| KROX_Q6 | M00982 | 13 | 29.0 | Family | 11 | Cell surface receptor linked signal transduction | 50 |
| FOXJ2_01 | M00422 | 124 | 27.6 | Yes (tag) | 75 | RNA metabolic process/Regulation of cellular metabolic process | 23.2 |
| Peaks with no FoxA2 sequence | |||||||
| MAZR_01 | M00491 | 138 | 82.9 | Yes (PCR) | 77 | Cellular protein metabolic process | 29.2 |
| PAX6_Q2 | M00979 | 69 | 57.0 | No | 44 | Intracellular signaling cascade | 31.8 |
| MAZ_Q6 | M00649 | 269 | 47.0 | Yes (tag) | 164 | RNA metabolic process/Transcription/Regulation of cellular metabolic process | 31.2 |
| KROX_Q6 | M00982 | 25 | 39.0 | Family | 20 | Regulation of cellular metabolic process/RNA metabolic process/Transcription/Cellular protein metabolic process | 33.3 |
| HNF4_Q6_01 | M01031 | 576 | 21.7 | Yes (tag) | 361 | Cellular protein metabolic process | 25.7 |
| UF1H3BETA_Q6 | M01068 | 2 | 19.9 | No | 1 | 15 different termsa | 100 |
| HNF4_DR1_Q3 | M00764 | 537 | 17.4 | Yes (tag) | 321 | Cellular protein metabolic process | 21.9 |
| SMAD3_Q6 | M00701 | 51 | 15.5 | Yes (PCR) | 31 | Intracellular signaling cascade/RNA metabolic process | 19.2 |
| NRF1_Q6 | M00652 | 5 | 14.9 | Yes (tag) | 6 | Response to oxidative stress/Carboxylic acid metabolic process/Cellular protein metabolic process/Mitochondrion organization and biogenesis/Macromolecule biosynthetic process/DNA metabolic process | 33.3 |
| COUPTF_Q6 | M01036 | 333 | 14.7 | Family | 225 | Cellular protein metabolic process | 26.6 |
| MIF_01 | M00279 | 6 | 12.3 | Yes (PCR) | 4 | Cytoskeleton organization and biogenesis/Establishment of cellular localization | 100 |
| SMAD4_Q6 | M00733 | 46 | 11.8 | Yes (tag) | 29 | Phosphate metabolic process/Cellular protein metabolic process | 28.6 |
| ATF_B | M00338 | 4 | 11.7 | Family | 3 | RNA metabolic process/Regulation of cellular metabolic process/Transcription | 100 |
| PPARG_03 | M00528 | 423 | 11.1 | Yes (tag) | 246 | Cellular protein metabolic process | 24.8 |
| PAX_Q6 | M00808 | 483 | 10.9 | Family | 287 | Cellular protein metabolic process | 30.9 |
| SF1_Q6 | M00727 | 118 | 10.9 | Yes (tag) | 83 | Regulation of cellular metabolic process | 32.6 |
| BRN2_01 | M00145 | 75 | 10.8 | No | 21 | Regulation of cellular metabolic process | 50 |
| RREB1_01 | M00257 | 9 | 10.4 | Yes (tag) | 7 | Regulation of cellular metabolic process/RNA metabolic process/Transcription | 50 |
| SP3_Q3 | M00665 | 59 | 10.0 | Yes (tag) | 36 | Cellular protein metabolic process | 47.6 |
| LHX3_01 | M00510 | 11 | 8.3 | No | 5 | Regulation of cellular metabolic process/RNA metabolic process/Transcription | 66.7 |
Included in the table are: (i) the number of peaks that contained the TFBS model; (ii) the Z-score, which represents the difference between the MATCH v2.6 PWM scan enrichment of real peak sequences and the mean for randomized sequences, divided by the standard deviation for randomized sequences (see Methods section); (iii) an indication of whether the transcription factor is expressed in the adult liver, based either on SAGE tag(s) from our adult liver library or RT–PCR amplification in the adult liver; (iv) the number of annotated genes that contained at least one peak within its extended gene region (10 kb upstream of the TSS to 1 kb downstream of the TTS); (v) the ‘level 5’ Gene Ontology Biological Process category assigned to the largest subgroup within these gene groups and (vi) the corresponding percentage of genes in the subgroup. TFBS models that co-occurred with a Foxa2-binding sequence included both transcription factors that have previously been demonstrated to interact with Foxa2 (HNF4 and members of the Coup-TF family) and also novel potential interacting transcription factors (Smad3 and Maz). Ten of the TFBS models that co-occurred with Foxa2-like binding sequences in peaks containing such sequences also occurred in peaks that did not contain Foxa2-like binding sequences.
aRegulation of biological process; cellular metabolic process; macromolecule metabolic process; primary metabolic process; cell organization and biogenesis; regulation of metabolic process; regulation of cellular process; organelle organization and biogenesis; biopolymer metabolic process; nucleobase, nucleoside, nucleotide and nucleic acid metabolic process; chromosome organization and biogenesis; regulation of cellular metabolic process; RNA metabolic process; transcription; DNA metabolic process.
We generated distribution plots of the distance between TFBS model sites and the Foxa2-binding sequence and found that 14 out of the top 20 had a significantly nonuniform distribution (P = 1.60E-181–0.03). GATA4 motifs had a relatively uniform distribution across the 500 bp surrounding the Foxa2-binding sequences (Figure 8A), while Pax6 motifs were primarily excluded from the region of the Foxa2-binding sequence out to a distance of ∼40 bp (Figure 8B). In contrast, HNF1 motifs were clustered around the Foxa2-binding sequence (Figure 8C). The distribution of cotranscription factor motifs relative to Foxa2 sequences may reflect the type of interaction that occurs between the two transcription factors.
Figure 8.
The distribution of TFBS sequence models relative to the corresponding Foxa2-binding sequence for three of the top 20 cotranscription factors in peaks containing a Foxa2-binding sequence. (A) The distribution of GATA4 TFBS models (Transfac model M00632). Motifs for this transcription factor show a uniform distribution across the 500 bp sequence. (B) The distribution of Pax6 TFBS models (Transfac model M00979). Pax6 motifs clustered at a distance of ∼40 bp away from the Foxa2-binding sequence (P = 1.1E-3). (C) The spatial distribution of HNF1 TFBS models (TRANSFAC model M00790). HNF1 motifs are spatially concentrated near the Foxa2-binding sequence (P = 5.7E-9).
We used the same approach to identify motifs that were enriched in the 2494 peaks that contained no Foxa2-binding sequence match, to either the 10-mer model ±1 bp mismatch or the full 14-mer model ±2 bp mismatch (24). We examined 500 bp sequences that were centered on each peak's maximum for the presence of each of the 363 TRANSFAC TFBS motifs. The HNF3beta (i.e. Foxa2) TRANSFAC model was assigned a very low rank (251 out of 363 models) indicating that our site-based scanning approach for identifying Foxa2-binding sequences was consistent with a TRANSFAC PWM scanning approach. Ten of the 20 highest ranked TFBS models in peaks without Foxa2-binding sites were also among the top 20 models in the peaks that contained Foxa2-binding sequences, and the genes for six of these transcription factors were expressed in the mouse adult liver (Table 2). Of the ten transcription factors corresponding to models that were exclusive to peaks without a Foxa2-binding sequence, five were expressed in the liver. These data provide strong evidence for transcription factor interactions and identify several transcription factors as future targets for investigating Foxa2 interaction.
DISCUSSION
ChIP–Seq identified Foxa2-binding sites throughout the genome
Using ChIP–Seq, we have identified over 11 000 highly confident in vivo Foxa2-binding sites in the adult liver. We were able to show a strong positive correlation between peak height in the ChIP–Seq data and the fold enrichment of binding sites as measured using ChIP–qPCR. This demonstrates that peak height can be an indicator of expected fold enrichment and supports our definition of a high-confidence dataset. The stringency used in analysis of Foxa2-binding sequences identified high affinity, high quality Foxa2-binding regions. However, it is likely that a more relaxed binding motif would have identified a greater number of lower confidence Foxa2-binding sequences in peaks.
A previous study using ChIP-chip to investigate TFBS in the liver identified 574 Foxa2-bound promoters using a microarray that tiled 10 kb around the TSS of 4000 mouse genes (11). Of the 579 ChIP–Seq peaks present in the regions defined in the ChIP-chip study, >50% of them overlapped with Foxa2-bound regions in the thresholded arrayed dataset (Supplementary Figure 5) (11). This level of overlap is consistent with previous comparisons between ChIP–Seq and ChIP-chip (18). However, since ChIP–Seq can cost effectively sample the entire genome, it has the ability to identify binding sites that occur in regions not often included in promoter microarrays. For example, our study identified over 2300 peaks occurring greater than 50 kb away from any annotated gene. We further observed that 952 of these peaks overlapped regions of high conservation, suggesting that the functions of many of these Foxa2-binding sites have been evolutionarily conserved. These peaks may regulate nearby genes that have not yet been annotated or they may mark enhancer regions that are located far from the genes that they regulate. Alternatively, these sites could represent regions where Foxa2 binds to regulate the chromatin state (8). A further possibility is that these sites are nonfunctional, representing either fortuitous binding or regulatory regions that have been lost through evolution. Further analysis of these regions is necessary to fully define the roles of these Foxa2-binding sites. These remote peaks demonstrate that genome-wide analysis of TFBS is critical for a full understanding of in vivo gene regulatory mechanisms.
Identification of in vivo Foxa2-binding sites suggests mechanisms of gene regulation
We observed that close to 45% of genes shown to be expressed in the liver by SAGE are associated with a Foxa2 peak, suggesting that a large number of these genes are potentially regulated by Foxa2. Although Foxa2 is known to be a major regulator of the hepatocyte phenotype (14), the extent of coverage of these liver-expressed genes by in vivo Foxa2-binding sites is surprising. Our results may actually underestimate the number of genes regulated by Foxa2, since genes that are expressed at levels not detected by SAGE, and therefore not represented by a tag, are not included. Of interest, Foxa2 has been shown to function by controlling DNA accessibility for transcriptional machinery in the embryo. Through its ability to bind to core histone proteins present in closed chromatin, it can render DNA accessible to additional transcription factor binding and therefore initiate transcriptional activity (8). The large number of binding sites for Foxa2 in the liver may reflect a role for Foxa2 in maintaining the competence for gene expression critical for hepatocyte function rather than a direct regulatory relationship. Of interest, we observed Foxa2 binding in the second intron of Gata4 and the cooccurrence of the GATA4 TFBS model with Foxa2-binding sites, suggesting a coregulatory interaction between these two factors. Given that GATA4 is also able to bind closed chromatin with Foxa2, this supports the role for both in the regulation of chromatin structure (8). Another possibility for the large number of Foxa2-binding sites observed close to liver expressed genes may also represent fortuitous binding of Foxa2 in regions of open chromatin rather than functional binding.
While our analysis suggested a major role for Foxa2 in liver function, deletion of Foxa2 specifically in the liver has shown this transcription factor is not essential for adult liver functions (85). However, combination knockout and conditional knockout studies have shown that, at least in the embryo, either Foxa2 or the closely related family member, Foxa1, is necessary for embryonic liver development (86). In addition, the three FoxA family members recognize similar binding sequences (5), suggesting that at least Foxa1 and Foxa2 can function redundantly. The level of redundancy between these three family members reflects the large roles that they play in the formation and functioning of the liver. Our analysis of Foxa2 knockdown in the Hepa1-6 cell line indicates that Foxa2 directly regulates the level of gene expression for at least some of novel gene targets identified that are associated with the Foxa2-binding sites. Additionally, given that Foxa1 expression levels were unaffected by Foxa2 knockdown in these experiments, this suggests that for a subset of genes Foxa1 is unable to completely replace reduction of Foxa2 with respect to gene expression regulation. These genes may be sensitive to reduction of FoxA, since Foxa1 is expressed at lower levels than Foxa2 in these cells. Alternatively, these genes may be preferentially regulated by Foxa2. The expression of some genes associated with Foxa2 binding was not affected by Foxa2 knockdown, indicating one of several possibilities suggested above including replacement of Foxa2 function by Foxa1, a role for Foxa2 in maintaining open chromatin rather than direct regulation of gene expression, or the presence of other transcription factors which are more critical to the regulation of these genes.
In addition to the global analysis of Foxa2-binding sites, our data suggest novel mechanisms of regulation at the level of individual genes. For example, we observed that the HNF4α enhancer site that binds Foxa2 is located in the first intron of a Riken gene (0610008F07Rik) on the opposite chromosomal strand, encoding a hypothetical, unclassified protein. This Riken gene is expressed in our adult liver SAGE library. Thus, this Foxa2-binding site may be a dual regulatory site involved in the promotion of transcription of two genes, a phenomenon which has been previously documented in other systems (87). Another example is seen with Afp, where there is no peak over the characterized Foxa2 site in the enhancer region that regulates embryonic expression of Afp. However, a peak was observed lying between the characterized site and the Afp TSS, ∼5 kb upstream of the Afp TSS. Since Afp is not expressed in adult liver, this indicates that the binding of Foxa2 is not sufficient for Afp transcription and suggests that Foxa2 binding at this alternative site may act in a repressive manner. Interestingly, previous studies have demonstrated that the Afp gene is repressed in the adult liver via a competitive mechanism between FoxA and p53 (88). The occurrence of alternate sites for the same transcription factor acting as an inducer or a repressor depending on the site it binds to, has been well documented in literature. For example, the transcription factor Sox9 occupies different inductive or repressive binding sites on the collagen 2a1 gene depending on the activation state of chondrocytes (89).
Foxa2 functions in a transcriptional network
A number of ChIP-chip studies that characterize cross-regulation between the promoters of several well characterized liver transcription factors have shown a high level of interaction between a core group of thirteen transcription factors important to liver development and function—HNF1α, HNF1β, HNF4α, Foxa1, Foxa2, HNF6, C/EBPγ, COUP-TFII, LRH1, USF1, PXR, FXRα and GATA6 (9,12,14). Our data showed in vivo Foxa2-binding sites within the extended gene region of nine of these 13 core transcription factors, including three transcription factors that had not previously been shown to be bound by Foxa2, indicating a high level of potential cross regulation by Foxa2 in the liver. We also documented the presence of Foxa2-binding sites associated with many additional transcription factors expressed in the adult liver. These results suggest that in the adult liver, Foxa2 is involved in additional transcriptional cascades that contribute to liver function.
In addition to Foxa2-binding sites in the promoters of other transcription factors, Foxa2 has been shown to interact with transcription factor proteins to coordinately regulate gene expression. Studies have indicated simultaneous occupation of gene promoters by Foxa2 in combination with HNF4α, HNF6, CREB1, USF1, HNF1α or GATA4 suggesting cooperative interaction between Foxa2 and these transcription factors (12,31). To extend our knowledge of transcription factors that could interact with Foxa2 in the adult liver, we determined the 20 most highly enriched cooccurring TFBS models that occurred within 250 bp of Foxa2 sites. This group included many transcription factors known to interact with Foxa2, including HNF4, HNF1 and GATA4, as well as novel transcription factors such as Maz, Mazr and Smad3, all of which are expressed in the adult liver as shown by SAGE or RT–PCR. Of interest, several TFBS models were observed in both peaks with and without Foxa2-binding sequences. These included Pax6, Maz, Mazr, Smad3, HNF4 and Pparg, of which all but Pax6 are expressed in the adult liver. One possibility for the absence of a Foxa2-binding site in some peaks is a physical regulatory interaction, whereby Foxa2 itself is bound not directly to the DNA, but to a cointeracting transcription factor at a close enough distance to enable protein–protein chemical cross-linking. Smad3 is thought to be primarily involved in acute liver damage via TGFβ signaling, while Pparg is involved in lipid metabolism (90,91). Additionally, Pparg may inhibit TGFβ/Smad3 signaling in liver fibrosis (92). Evi1 has been previously shown to be expressed in the adult liver; however, neither Maz nor Mazr have been reported to be expressed in the liver. The functional role of these three transcription factors in liver gene regulation is currently unknown (93). The fact that many of these top cooccurring transcription factors are expressed and have roles in the adult liver further supports the likelihood that they can interact with Foxa2.
Although Pax6 is not expressed in the liver, it is expressed in the closely related pancreatic tissue. Notably, the Pax6 TFBS model was found amongst the top 20 enriched cooccurring transcription factors in both peaks with and without an exact Foxa2-binding site. In the pancreas, Foxa2 and Pax6 interact to regulate the activation of genes such as glucagon and Pdx1 (83,94). Of interest, the location of the Pax6 TFBS model in relation to Foxa2-binding sequences showed that the Pax6-binding site was consistently greater than 40 bp away from Foxa2 in either direction (Figure 7B). Thus, although Foxa2- and Pax6-binding sites are found in close proximity, at least 40 bp must be present between the two sites, suggesting that Pax6 and Foxa2 can bind together to regulate gene expression in the pancreas or that noncompetitive inhibitory mechanisms limit the binding of both factors simultaneously.
This study has presented novel Foxa2 regulatory mechanisms in the mouse adult liver by determining genome wide binding sites. We have also shown a high level of coverage of known functional binding sites as well as identifying several potential novel interactions with annotated genes and unannotated genomic regions. Finally, we have shown a potentially wide-ranging role for Foxa2 in regulating gene expression in the function of the adult liver. Overall, this study demonstrates the advantages of using ChIP–Seq technology to characterize transcription factor binding in tissue, and paves the way for further investigations of in vivo transcription factor networks.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the support of the MORGEN project team managers, Amanda Kotzer and Joanne Johnson. This study received funding support from the Genome British Columbia and Genome Canada. P.A.H., M.M. and S.J. are Scholars of the Michael Smith Foundation for Health Research. M.M. is a Terry Fox Young Investigator. P.A.H. is a Canadian Institutes for Health Research New Investigator. Funding to pay the Open Access publication charges for this article was provided by Genome Canada and Genome BC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ang SL, Wierda A, Wong D, Stevens KA, Cascio S, Rossant J, Zaret KS. The formation and maintenance of the definitive endoderm lineage in the mouse: involvement of HNF3/forkhead proteins. Development. 1993;119:1301–1315. doi: 10.1242/dev.119.4.1301. [DOI] [PubMed] [Google Scholar]
- 2.Rausa FM, Tan Y, Zhou H, Yoo KW, Stolz DB, Watkins SC, Franks RR, Unterman TG, Costa RH. Elevated levels of hepatocyte nuclear factor 3beta in mouse hepatocytes influence expression of genes involved in bile acid and glucose homeostasis. Mol. Cell. Biol. 2000;20:8264–8282. doi: 10.1128/mcb.20.21.8264-8282.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolfrum C, Asilmaz E, Luca E, Friedman JM, Stoffel M. Foxa2 regulates lipid metabolism and ketogenesis in the liver during fasting and in diabetes. Nature. 2004;432:1027–1032. doi: 10.1038/nature03047. [DOI] [PubMed] [Google Scholar]
- 4.Costa RH, Grayson DR, Darnell J.E., Jr Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and alpha 1-antitrypsin genes. Mol. Cell. Biol. 1989;9:1415–1425. doi: 10.1128/mcb.9.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai E, Prezioso VR, Smith E, Litvin O, Costa RH, Darnell J.E., Jr HNF-3A, a hepatocyte-enriched transcription factor of novel structure is regulated transcriptionally. Genes Dev. 1990;4:1427–1436. doi: 10.1101/gad.4.8.1427. [DOI] [PubMed] [Google Scholar]
- 6.Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- 7.Friedman JR, Kaestner KH. The Foxa family of transcription factors in development and metabolism. Cell Mol. Life Sci. 2006;63:2317–2328. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 9.Kyrmizi I, Hatzis P, Katrakili N, Tronche F, Gonzalez FJ, Talianidis I. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev. 2006;20:2293–2305. doi: 10.1101/gad.390906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao H, Falt S, Sandelin A, Gustafsson JA, Dahlman-Wright K. Genome-wide identification of estrogen receptor {alpha} binding sites in mouse liver. Mol. Endocrinol. 2008;22:10–22. doi: 10.1210/me.2007-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat. Genet. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Odom DT, Dowell RD, Jacobsen ES, Nekludova L, Rolfe PA, Danford TW, Gifford DK, Fraenkel E, Bell GI, Young RA. Core transcriptional regulatory circuitry in human hepatocytes. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100059. 2006.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rada-Iglesias A, Wallerman O, Koch C, Ameur A, Enroth S, Clelland G, Wester K, Wilcox S, Dovey OM, Ellis PD, et al. Binding sites for metabolic disease related transcription factors inferred at base pair resolution by chromatin immunoprecipitation and genomic microarrays. Hum. Mol. Genet. 2005;14:3435–3447. doi: 10.1093/hmg/ddi378. [DOI] [PubMed] [Google Scholar]
- 15.Rubins NE, Friedman JR, Le PP, Zhang L, Brestelli J, Kaestner KH. Transcriptional networks in the liver: Hepatocyte nuclear factor 6 function is largely independent of Foxa2. Mol. Cell. Biol. 2005;25:7069–7077. doi: 10.1128/MCB.25.16.7069-7077.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith AD, Sumazin P, Zhang MQ. Tissue-specific regulatory elements in mammalian promoters. Mol. Syst. Biol. 2007;3:73. doi: 10.1038/msb4100114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teufel A, Weinmann A, Krupp M, Budinger M, Galle PR. Genome-wide analysis of factors regulating gene expression in liver. Gene. 2007;389:114–121. doi: 10.1016/j.gene.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 18.Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, et al. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 2007;3:e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 20.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 21.Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, et al. Global mapping of c-myc binding sites and target gene networks in human B cells. Proc. Natl Acad Sci USA. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barski A, Cuddapah S, Cui K, Roh T.-Y, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 24.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 25.Montgomery SB, Griffith OL, Sleumer MC, Bergman CM, Bilenky M, Pleasance ED, Prychyna Y, Zhang X, Jones SJ. ORegAnno: an open access database and curation system for literature-derived promoters, transcription factor binding sites and regulatory variation. Bioinformatics. 2006;22:637–640. doi: 10.1093/bioinformatics/btk027. [DOI] [PubMed] [Google Scholar]
- 26.Vlieghe D, Sandelin A, De Bleser PJ, Vleminckx K, Wasserman WW, van Roy F, Lenhard B. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–D97. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siddiqui AS, Khattra J, Delaney AD, Zhao Y, Astell C, Asano J, Babakaiff R, Barber S, Beland J, Bohacec S, et al. A mouse atlas of gene expression: large-scale digital gene-expression profiles from precisely defined developing C57BL/6J mouse tissues and cells. Proc. Natl Acad. Sci. USA. 2005;102:18485–18490. doi: 10.1073/pnas.0509455102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Helden J, Andre B, Collado-Vides J. A web site for the computational analysis of yeast regulatory sequences. Yeast. 2000;16:177–187. doi: 10.1002/(SICI)1097-0061(20000130)16:2<177::AID-YEA516>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 30.Bailly A, Torres-Padilla ME, Tinel AP, Weiss MC. An enhancer element 6 kb upstream of the mouse HNF4alpha1 promoter is activated by glucocorticoids and liver-enriched transcription factors. Nucleic Acids Res. 2001;29:3495–505. doi: 10.1093/nar/29.17.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bossard P, Zaret KS. GATA transcription factors as potentiators of gut endoderm differentiation. Development. 1998;125:4909–4917. doi: 10.1242/dev.125.24.4909. [DOI] [PubMed] [Google Scholar]
- 32.Cheng A, Zhang M, Crosson SM, Bao ZQ, Saltiel AR. Regulation of the mouse protein targeting to glycogen (PTG) promoter by the FoxA2 forkhead protein and by 3′,5′-cyclic adenosine 5′-monophosphate in H4IIE hepatoma cells. Endocrinology. 2006;147:3606–3612. doi: 10.1210/en.2005-1513. [DOI] [PubMed] [Google Scholar]
- 33.Costa RH, Grayson DR. Site-directed mutagenesis of hepatocyte nuclear factor (HNF) binding sites in the mouse transthyretin (TTR) promoter reveal synergistic interactions with its enhancer region. Nucleic Acids Res. 1991;19:4139–4145. doi: 10.1093/nar/19.15.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denson LA, McClure MH, Bogue CW, Karpen SJ, Jacobs HC. HNF3beta and GATA-4 transactivate the liver-enriched homeobox gene, hex. Gene. 2000;246:311–320. doi: 10.1016/s0378-1119(00)00082-2. [DOI] [PubMed] [Google Scholar]
- 35.Groupp ER, Crawford N, Locker J. Characterization of the distal alpha-fetoprotein enhancer, a strong, long distance, liver-specific activator. J. Biol. Chem. 1994;269:22178–22187. [PubMed] [Google Scholar]
- 36.Hatzis P, Talianidis I. Regulatory mechanisms controlling human hepatocyte nuclear factor 4alpha gene expression. Mol. Cell. Biol. 2001;21:7320–7330. doi: 10.1128/MCB.21.21.7320-7330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackson DA, Rowader KE, Stevens K, Jiang C, Milos P, Zaret KS. Modulation of liver-specific transcription by interactions between hepatocyte nuclear factor 3 and nuclear factor 1 binding DNA in close apposition. Mol. Cell. Biol. 1993;13:2401–2410. doi: 10.1128/mcb.13.4.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nitsch D, Boshart M, Schutz G. Activation of the tyrosine aminotransferase gene is dependent on synergy between liver-specific and hormone-responsive elements. Proc. Natl Acad. Sci. USA. 1993;90:5479–5483. doi: 10.1073/pnas.90.12.5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Brien RM, Noisin EL, Suwanichkul A, Yamasaki T, Lucas PC, Wang JC, Powell DR, Granner DK. Hepatic nuclear factor 3- and hormone-regulated expression of the phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein 1 genes. Mol. Cell. Biol. 1995;15:1747–1758. doi: 10.1128/mcb.15.3.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qian X, Samadani U, Porcella A, Costa RH. Decreased expression of hepatocyte nuclear factor 3 alpha during the acute-phase response influences transthyretin gene transcription. Mol. Cell. Biol. 1995;15:1364–1376. doi: 10.1128/mcb.15.3.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samadani U, Costa RH. The transcriptional activator hepatocyte nuclear factor 6 regulates liver gene expression. Mol. Cell. Biol. 1996;16:6273–6284. doi: 10.1128/mcb.16.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimizu S, Miyamoto Y, Hayashi M. Cell-type dependency of two Foxa/HNF3 sites in the regulation of vitronectin promoter activity. Biochim. Biophys. Acta. 2002;1574:337–344. doi: 10.1016/s0167-4781(02)00232-4. [DOI] [PubMed] [Google Scholar]
- 43.Antes TJ, Levy-Wilson B. HNF-3 beta, C/EBP beta, and HNF-4 act in synergy to enhance transcription of the human apolipoprotein B gene in intestinal cells. DNA Cell Biol. 2001;20:67–74. doi: 10.1089/104454901750070265. [DOI] [PubMed] [Google Scholar]
- 44.Arenzana N, Rodriguez de Cordoba S. Promoter region of the human gene coding for beta-chain of C4b binding protein. hepatocyte nuclear factor-3 and nuclear factor-I/CTF transcription factors are required for efficient expression of C4BPB in HepG2 cells. J. Immunol. 1996;156:168–175. [PubMed] [Google Scholar]
- 45.Beck S, Sommer P, dos Santos Silva E, Blin N, Gott P. Hepatocyte nuclear factor 3 (winged helix domain) activates trefoil factor gene TFF1 through a binding motif adjacent to the TATAA box. DNA Cell Biol. 1999;18:157–164. doi: 10.1089/104454999315547. [DOI] [PubMed] [Google Scholar]
- 46.Berg DT, Gerlitz B, Sharma GR, Richardson MA, Stephens EJ, Grubbs RL, Holmes KC, Fynboe K, Montani D, Cramer MS, et al. FoxA2 involvement in suppression of protein C, an outcome predictor in experimental sepsis. Clin. Vaccine Immunol. 2006;13:426–432. doi: 10.1128/CVI.13.3.426-432.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ceelie H, Spaargaren-Van Riel CC, De Jong M, Bertina RM, Vos HL. Functional characterization of transcription factor binding sites for HNF1-alpha, HNF3-beta (FOXA2), HNF4-alpha, Sp1 and Sp3 in the human prothrombin gene enhancer. J. Thromb. Haemost. 2003;1:1688–1698. doi: 10.1046/j.1538-7836.2003.00393.x. [DOI] [PubMed] [Google Scholar]
- 48.Cha JY, Kim H, Kim KS, Hur MW, Ahn Y. Identification of transacting factors responsible for the tissue-specific expression of human glucose transporter type 2 isoform gene. Cooperative role of hepatocyte nuclear factors 1alpha and 3beta. J. Biol. Chem. 2000;275:18358–18365. doi: 10.1074/jbc.M909536199. [DOI] [PubMed] [Google Scholar]
- 49.Christoffels VM, Grange T, Kaestner KH, Cole TJ, Darlington GJ, Croniger CM, Lamers WH. Glucocorticoid receptor, C/EBP, HNF3, and protein kinase A coordinately activate the glucocorticoid response unit of the carbamoylphosphate synthetase I gene. Mol. Cell. Biol. 1998;18:6305–6315. doi: 10.1128/mcb.18.11.6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crowe AJ, Sang L, Li KK, Lee KC, Spear BT, Barton MC. Hepatocyte nuclear factor 3 relieves chromatin-mediated repression of the alpha-fetoprotein gene. J. Biol. Chem. 1999;274:25113–25120. doi: 10.1074/jbc.274.35.25113. [DOI] [PubMed] [Google Scholar]
- 51.Delesque-Touchard N, Park SH, Waxman DJ. Synergistic action of hepatocyte nuclear factors 3 and 6 on CYP2C12 gene expression and suppression by growth hormone-activated STAT5b. Proposed model for female specific expression of CYP2C12 in adult rat liver. J. Biol. Chem. 2000;275:34173–34182. doi: 10.1074/jbc.M004027200. [DOI] [PubMed] [Google Scholar]
- 52.Harnish DC, Malik S, Kilbourne E, Costa R, Karathanasis SK. Control of apolipoprotein AI gene expression through synergistic interactions between hepatocyte nuclear factors 3 and 4. J. Biol. Chem. 1996;271:13621–13628. doi: 10.1074/jbc.271.23.13621. [DOI] [PubMed] [Google Scholar]
- 53.Jung D, Hagenbuch B, Fried M, Meier PJ, Kullak-Ublick GA. Role of liver-enriched transcription factors and nuclear receptors in regulating the human, mouse, and rat NTCP gene. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;286:G752–G761. doi: 10.1152/ajpgi.00456.2003. [DOI] [PubMed] [Google Scholar]
- 54.Lantz KA, Vatamaniuk MZ, Brestelli JE, Friedman JR, Matschinsky FM, Kaestner KH. Foxa2 regulates multiple pathways of insulin secretion. J. Clin. Invest. 2004;114:512–520. doi: 10.1172/JCI21149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lemaigre FP, Durviaux SM, Rousseau GG. Liver-specific factor binding to the liver promoter of a 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase gene. J. Biol. Chem. 1993;268:19896–19905. [PubMed] [Google Scholar]
- 56.Li C, Zhu NL, Tan RC, Ballard PL, Derynck R, Minoo P. Transforming growth factor-beta inhibits pulmonary surfactant protein B gene transcription through SMAD3 interactions with NKX2.1 and HNF-3 transcription factors. J. Biol. Chem. 2002;277:38399–38408. doi: 10.1074/jbc.M203188200. [DOI] [PubMed] [Google Scholar]
- 57.Raynal JF, Dugast C, Le Van Thai A, Weber MJ. Winged helix hepatocyte nuclear factor 3 and POU-domain protein brn-2/N-oct-3 bind overlapping sites on the neuronal promoter of human aromatic L-amino acid decarboxylase gene. Brain Res. Mol. Brain Res. 1998;56:227–237. doi: 10.1016/s0169-328x(98)00048-5. [DOI] [PubMed] [Google Scholar]
- 58.Roux J, Pictet R, Grange T. Hepatocyte nuclear factor 3 determines the amplitude of the glucocorticoid response of the rat tyrosine aminotransferase gene. DNA Cell Biol. 1995;14:385–396. doi: 10.1089/dna.1995.14.385. [DOI] [PubMed] [Google Scholar]
- 59.Sawaya PL, Luse DS. Two members of the HNF-3 family have opposite effects on a lung transcriptional element; HNF-3 alpha stimulates and HNF-3 beta inhibits activity of region I from the clara cell secretory protein (CCSP) promoter. J. Biol. Chem. 1994;269:22211–22216. [PubMed] [Google Scholar]
- 60.Scassa ME, Guberman AS, Ceruti JM, Canepa ET. Hepatic nuclear factor 3 and nuclear factor 1 regulate 5-aminolevulinate synthase gene expression and are involved in insulin repression. J. Biol. Chem. 2004;279:28082–28092. doi: 10.1074/jbc.M401792200. [DOI] [PubMed] [Google Scholar]
- 61.Unterman TG, Fareeduddin A, Harris MA, Goswami RG, Porcella A, Costa RH, Lacson RG. Hepatocyte nuclear factor-3 (HNF-3) binds to the insulin response sequence in the IGF binding protein-1 (IGFBP-1) promoter and enhances promoter function. Biochem. Biophys. Res. Commun. 1994;203:1835–1841. doi: 10.1006/bbrc.1994.2401. [DOI] [PubMed] [Google Scholar]
- 62.Vallet V, Antoine B, Chafey P, Vandewalle A, Kahn A. Overproduction of a truncated hepatocyte nuclear factor 3 protein inhibits expression of liver-specific genes in hepatoma cells. Mol. Cell. Biol. 1995;15:5453–5460. doi: 10.1128/mcb.15.10.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verschuur M, de Jong M, Felida L, de Maat MP, Vos HL. A hepatocyte nuclear factor-3 site in the fibrinogen beta promoter is important for interleukin 6-induced expression, and its activity is influenced by the adjacent -148C/T polymorphism. J. Biol. Chem. 2005;280:16763–16771. doi: 10.1074/jbc.M501973200. [DOI] [PubMed] [Google Scholar]
- 64.Wang JC, Stromstedt PE, O'Brien RM, Granner DK. Hepatic nuclear factor 3 is an accessory factor required for the stimulation of phosphoenolpyruvate carboxykinase gene transcription by glucocorticoids. Mol. Endocrinol. 1996;10:794–800. doi: 10.1210/mend.10.7.8813720. [DOI] [PubMed] [Google Scholar]
- 65.Williamson EA, Wolf I, O’Kelly J, Bose S, Tanosaki S, Koeffler HP. BRCA1 and FOXA1 proteins coregulate the expression of the cell cycle-dependent kinase inhibitor p27(Kip1) Oncogene. 2006;25:1391–1399. doi: 10.1038/sj.onc.1209170. [DOI] [PubMed] [Google Scholar]
- 66.Wiwi CA, Waxman DJ. Role of hepatocyte nuclear factors in transcriptional regulation of male-specific CYP2A2. J. Biol. Chem. 2005;280:3259–3268. doi: 10.1074/jbc.M409294200. [DOI] [PubMed] [Google Scholar]
- 67.Zhu QS, Xing W, Qian B, von Dippe P, Shneider BL, Fox VL, Levy D. Inhibition of human m-epoxide hydrolase gene expression in a case of hypercholanemia. Biochim. Biophys. Acta. 2003;1638:208–216. doi: 10.1016/s0925-4439(03)00085-1. [DOI] [PubMed] [Google Scholar]
- 68.Khattra J, Delaney AD, Zhao Y, Siddiqui A, Asano J, McDonald H, Pandoh P, Dhalla N, Prabhu AL, Ma K, et al. Large-scale production of SAGE libraries from microdissected tissues, flow-sorted cells, and cell lines. Genome Res. 2007;17:108–116. doi: 10.1101/gr.5488207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Al-Shahrour F, Díaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of gene ontology terms to groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 70.Brannan CI, Perkins AS, Vogel KS, Ratner N, Nordlund ML, Reid SW, Buchberg AM, Jenkins NA, Parada LF, Copeland NG. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- 71.Choi-Miura NH, Otsuyama K, Sano Y, Saito K, Takahashi K, Tomita M. Hepatic injury-specific conversion of mouse plasma hyaluronan binding protein to the active hetero-dimer form. Biol. Pharm. Bull. 2001;24:892–896. doi: 10.1248/bpb.24.892. [DOI] [PubMed] [Google Scholar]
- 72.Duncan SA. Transcriptional regulation of liver development. Dev. Dyn. 2000;219:131–142. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1051>3.3.co;2-e. [DOI] [PubMed] [Google Scholar]
- 73.Fleming MD, Campagna DR, Haslett JN, Trenor C.C., III, Andrews NC. A mutation in a mitochondrial transmembrane protein is responsible for the pleiotropic hematological and skeletal phenotype of flexed-tail (f/f) mice. Genes Dev. 2001;15:652–657. doi: 10.1101/gad.873001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fan CY, Pan J, Chu R, Lee D, Kluckman KD, Usuda N, Singh I, Yeldandi AV, Rao MS, Maeda N, et al. Hepatocellular and hepatic peroxisomal alterations in mice with a disrupted peroxisomal fatty acyl-coenzyme A oxidase gene. J. Biol. Chem. 1996;271:24698–24710. doi: 10.1074/jbc.271.40.24698. [DOI] [PubMed] [Google Scholar]
- 75.Hsiao EC, Koniaris LG, Zimmers-Koniaris T, Sebald SM, Huynh TV, Lee SJ. Characterization of growth-differentiation factor 15, a transforming growth factor beta superfamily member induced following liver injury. Mol. Cell. Biol. 2000;20:3742–3751. doi: 10.1128/mcb.20.10.3742-3751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jung J, Zheng M, Goldfarb M, Zaret KS. Initiation of mammalian liver development from endoderm by fibroblast growth factors. Science. 1999;284:1998–2003. doi: 10.1126/science.284.5422.1998. [DOI] [PubMed] [Google Scholar]
- 77.Suzuki T, Kanai Y, Hara T, Sasaki J, Sasaki T, Kohara M, Maehama T, Taya C, Shitara H, Yonekawa H, et al. Crucial role of the small GTPase ARF6 in hepatic cord formation during liver development. Mol. Cell. Biol. 2006;26:6149–6156. doi: 10.1128/MCB.00298-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clotman F, Jacquemin P, Plumb-Rudewiez N, Pierreux CE, Van der Smissen P, Dietz HC, Courtoy PJ, Rousseau GG, Lemaigre FP. Control of liver cell fate decision by a gradient of TGF beta signaling modulated by onecut transcription factors. Genes Dev. 2005;19:1849–1854. doi: 10.1101/gad.340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim JY, Kim HJ, Kim KT, Park YY, Seong HA, Park KC, Lee IK, Ha H, Shong M, Park SC, et al. Orphan nuclear receptor small heterodimer partner represses hepatocyte nuclear factor 3/Foxa transactivation via inhibition of its DNA binding. Mol. Endocrinol. 2004;18:2880–2894. doi: 10.1210/me.2004-0211. [DOI] [PubMed] [Google Scholar]
- 80.Qu X, Lam E, Doughman YQ, Chen Y, Chou YT, Lam M, Turakhia M, Dunwoodie SL, Watanabe M, Xu B, et al. Cited2, a coactivator of HNF4alpha, is essential for liver development. EMBO J. 2007;26:4445–4456. doi: 10.1038/sj.emboj.7601883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shen H, Huang G, Hadi M, Choy P, Zhang M, Minuk GY, Chen Y, Gong Y. Transforming growth factor-beta1 downregulation of Smad1 gene expression in rat hepatic stellate cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2003;285:G539–G546. doi: 10.1152/ajpgi.00436.2002. [DOI] [PubMed] [Google Scholar]
- 82.Zhao R, Watt AJ, Li J, Luebke-Wheeler J, Morrisey EE, Duncan SA. GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Mol. Cell. Biol. 2005;25:2622–2631. doi: 10.1128/MCB.25.7.2622-2631.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gauthier BR, Schwitzgebel VM, Zaiko M, Mamin A, Ritz-Laser B, Philippe J. Hepatic nuclear factor-3 (HNF-3 or Foxa2) regulates glucagon gene transcription by binding to the G1 and G2 promoter elements. Mol. Endocrinol. 2002;16:170–183. doi: 10.1210/mend.16.1.0752. [DOI] [PubMed] [Google Scholar]
- 84.Legraverend C, Eguchi H, Strom A, Lahuna O, Mode A, Tollet P, Westin S, Gustafsson JA. Transactivation of the rat CYP2C13 gene promoter involves HNF-1, HNF-3, and members of the orphan receptor subfamily. Biochemistry. 1994;33:9889–9897. doi: 10.1021/bi00199a010. [DOI] [PubMed] [Google Scholar]
- 85.Sund NJ, Ang SL, Sackett SD, Shen W, Daigle N, Magnuson MA, Kaestner KH. Hepatocyte nuclear factor 3beta (Foxa2) is dispensable for maintaining the differentiated state of the adult hepatocyte. Mol. Cell. Biol. 2000;20:5175–5183. doi: 10.1128/mcb.20.14.5175-5183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on foxa transcription factors. Nature. 2005;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- 87.Zanotto E, Shah ZH, Jacobs HT. The bidirectional promoter of two genes for the mitochondrial translational apparatus in mouse is regulated by an array of CCAAT boxes interacting with the transcription factor NF-Y. Nucleic Acids Res. 2007;35:664–677. doi: 10.1093/nar/gkl1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nguyen TT, Cho K, Stratton SA, Barton MC. Transcription factor interactions and chromatin modifications associated with p53-mediated, developmental repression of the alpha-fetoprotein gene. Mol. Cell. Biol. 2005;25:2147–2157. doi: 10.1128/MCB.25.6.2147-2157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kypriotou M, Fossard-Demoor M, Chadjichristos C, Ghayor C, de Crombrugghe B, Pujol JP, Galera P. SOX9 exerts a bifunctional effect on type II collagen gene (COL2A1) expression in chondrocytes depending on the differentiation state. DNA Cell Biol. 2003;22:119–129. doi: 10.1089/104454903321515922. [DOI] [PubMed] [Google Scholar]
- 90.Gauldie J, Bonniaud P, Sime P, Ask K, Kolb M. TGF-beta, Smad3 and the process of progressive fibrosis. Biochem. Soc. Trans. 2007;35:661–664. doi: 10.1042/BST0350661. [DOI] [PubMed] [Google Scholar]
- 91.Schadinger SE, Bucher NL, Schreiber BM, Farmer SR. PPARgamma2 regulates lipogenesis and lipid accumulation in steatotic hepatocytes. Am. J. Physiol. Endocrinol. Metab. 2005;288:E1195–E1205. doi: 10.1152/ajpendo.00513.2004. [DOI] [PubMed] [Google Scholar]
- 92.Zhao C, Chen W, Yang L, Chen L, Stimpson SA, Diehl AM. PPARgamma agonists prevent TGFbeta1/Smad3-signaling in human hepatic stellate cells. Biochem. Biophys. Res. Commun. 2006;350:385–391. doi: 10.1016/j.bbrc.2006.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wimmer K, Vinatzer U, Zwirn P, Fonatsch C, Wieser R. Comparative expression analysis of the antagonistic transcription factors EVI1 and MDS1-EVI1 in murine tissues and during in vitro hematopoietic differentiation. Biochem. Biophys. Res. Commun. 1998;252:691–696. doi: 10.1006/bbrc.1998.9588. [DOI] [PubMed] [Google Scholar]
- 94.Samaras SE, Cissell MA, Gerrish K, Wright CV, Gannon M, Stein R. Conserved sequences in a tissue-specific regulatory region of the pdx-1 gene mediate transcription in pancreatic beta cells: Role for hepatocyte nuclear factor 3 beta and Pax6. Mol. Cell. Biol. 2002;22:4702–4713. doi: 10.1128/MCB.22.13.4702-4713.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.