Abstract
Reverse gyrase is a peculiar DNA topoisomerase, specific of thermophilic microorganisms, which induces positive supercoiling into DNA molecules in an ATP-dependent reaction. It is a modular enzyme and comprises an N-terminal helicase-like module fused to a C-terminal topoisomerase IA-like domain. The exact molecular mechanism of this unique reaction is not understood, and a fundamental mechanistic question is how its distinct steps are coordinated. We studied the cross-talk between the components of this molecular motor and probed communication between the DNA-binding sites and the different activities (DNA relaxation, ATP hydrolysis and positive supercoiling). We show that the isolated ATPase and topoisomerase domains of reverse gyrase form specific physical interactions, retain their own DNA binding and enzymatic activities, and when combined cooperate to achieve the unique ATP-dependent positive supercoiling activity. Our results indicate a mutual effect of both domains on all individual steps of the reaction. The C-terminal domain shows ATP-independent topoisomerase activity, which is repressed by the N-terminal domain in the full-length enzyme; experiments with the isolated domains showed that the C-terminal domain has stimulatory influence on the ATPase activity of the N-terminal domain. In addition, the two domains showed a striking reciprocal thermostabilization effect.
INTRODUCTION
Reverse gyrase is the only DNA topoisomerase capable of introducing positive supercoiling into DNA molecules (for recent reviews, see 1,2). Extensive genomic analysis has shown that reverse gyrase gene is the only hyperthermophilic-specific one: it is invariably present in all organisms living above 70°C, and invariably absent in organisms living at mesophilic temperatures (3,4). This unique activity reflects a distinctive arrangement of the protein, which is composed of a C-terminal topoisomerase fused to a N-terminal helicase-like domain. The C-terminal domain belongs to the universal type IA topoisomerase family, including the bacterial Topoisomerase I and Topoisomerase III from Bacteria, Eucarya and Archaea. The N-terminal domain shows conserved sequence motives typical of the SF2 helicase family, including the ATP binding site (P-loop), but no extensive sequence similarity to other known proteins. The enzyme carries out DNA-dependent ATP hydrolysis, which is essential for positive supercoiling. Although the reaction of reverse gyrase is unique, this enzyme might be viewed as a model of interaction of topoisomerases and helicases (5). For instance, RecQ helicases form, together with Topoisomerase III, complexes involved in the maintenance of genome integrity that are strikingly conserved through the evolution, from bacteria to humans (reviewed in 6,7). How the biochemical activities of these enzymes relate to their biological function is still unclear.
The actual function of reverse gyrase in live cells has not been directly determined. Because of its association with life at high temperature, it has been suggested to be involved in DNA protection from heat-induced denaturation and/or degradation. This hypothesis has been challenged by the finding that inactivation of the reverse gyrase gene in Thermococcus kodakaraensis did not result in a lethal phenotype; however, growth of the mutant strain was significantly retarded at higher temperature, thus confirming that the enzyme plays a role in the cell adaptation to thermophily (8). Recent results suggest that reverse gyrase might participate, directly or indirectly, in the cell response to DNA damage: Sulfolobus solfataricus reverse gyrase is recruited to DNA in vivo after UV irradiation (9) and is specifically degraded in cells treated with the alkylating agent MMS, in concomitance with DNA degradation (10).
Despite the fact that reverse gyrases from several organisms have been extensively studied and the crystal structure of the Archaeoglobus fulgidus enzyme has been solved (11; see 1,2, for a complete reference list), the mechanism of the positive supercoiling reaction remains elusive. As for all DNA topoisomerases, the catalytic cycle of reverse gyrase can be dissected into four steps: DNA binding, DNA cleavage, strand passage and religation of the DNA ends (reviewed in 12). Whereas the C-terminal domain of reverse gyrase is clearly responsible for strand passage, the function of the N-terminal domain and of ATP hydrolysis is not understood. At least three different biochemical mechanisms have been proposed for reverse gyrase. In the first model, ATP hydrolysis-dependent translocation of the enzyme driven by the ‘helicase’ domain produces two topological domains, one with underwound base pairs and the other with positive supercoils; selective relaxation of negative supercoiling by the topoisomerase module would result in net positive supercoiling. ATP hydrolysis would be required for enzyme translocation on DNA (13). In the second model, the two topologically distinct domains result upon enzyme binding-induced local unwinding of the double-strand; reverse gyrase would then renature unwound DNA, thus resulting in net positive supercoiling. In this case, ATP hydrolysis would trigger the enzyme affinity switch from single strand (ss) to double strand (ds) DNA (14). The third model proposes that reverse gyrase is capable of a direction-specific strand passage toward linking number increase, driven by ATP hydrolysis (11). Unfortunately, available experimental evidence does not support or even argue against one or more aspects of these models. First, no helicase activity could be observed with reverse gyrase or its N-terminal domain (15,16). Second, the proposed switch from an unwinding activity to a renaturing one has not been demonstrated; third, the mechanistic basis for a direction-specific strand passage is not clear; finally, the role of ATP hydrolysis in the process is not understood.
Previous elegant work has shown that positive supercoiling activity of the Sulfolobus acidocaldarius enzyme can be reconstituted by mixing the two separate domains (15). Starting from this observation, in the current study we have obtained the two separate domains of one of the two S. solfataricus reverse gyrase isoforms and have addressed several issues regarding the contributions of the two domains to different enzyme activities (DNA binding, ATPase, relaxation, positive supercoiling) and the function of the N-terminal domain. We show that, whereas the two domains retain their independent DNA binding and enzymatic properties, together they give rise to a completely new entity, displaying not only the reverse gyrase peculiar activity (positive supercoiling), but also its peculiar thermostability. In addition, our results suggest an intimate cross-talk between the two components in all steps of the reaction. Whereas in the whole enzyme the ATP-independent C-terminal topoisomerase activity is repressed by the N-terminal domain, the N-terminal domain has a very weak ATPase activity by its own, which is stimulated by the C-terminal domain. Plasmon resonance experiments showed physical interaction between the two domains.
MATERIALS AND METHODS
Cloning of TopR1
The TopR1 gene was amplified from S. solfataricus P2 (17) genomic DNA by using the following oligonucleotides: topR15′-His (5′-ATATGAGCTCTATGACAAGCATCAATAAAGTCCCACCCTCAAT-3′)/topR1-3′His (5′-ATATCTGCAGTCATCAACTAATTGTTTGTATTTCATTATATAATTCTT-3′). The oligonucleotides match the 5′- and the 3′-terminal ends of the coding sequence with the addition of a SacI site at the 5′-end and a PstI site at the 3′-end (underlined). The SacI/PstI fragment was cloned in pQE31 (Qiagen, Hilden, Germany) in frame with a sequence coding for a histidine tag at its N-terminus, producing pQE31-topR1, whose insert was checked by DNA sequencing. This construct, introduced in the E. coli Able C strain (Stratagene, La Jolla, CA, USA), resulted in low number of transformants, low expression level and high frequency of plasmid rearrangements, suggesting that TopR1 is highly toxic (data not shown). In order to reduce the basal expression level of TopRI from the leaky T5 promoter/lac operator in pQE31, we decided to replace this regulatory region with the more stringent T7 promoter from pET9d (Novagen, Darmstadt, Germany) as shown in Supplementary Figure 1S, obtaining pQET7-topR1.
Construction of deletion mutants
Starting from pQET7-topR1, a site-directed mutagenesis strategy was followed in order to obtain constructs expressing the separate N-terminal and C-terminal domains of TopRI. The GeneTailor™ Site-Directed Mutagenesis System (Invitrogen, Carlsbad, CA, USA) was used with the following oligonucleotides: NterPstImut (5′-TTAAGTAAAATCAAGAAAGAATAATAACTGCAGTCATCATCCC-3′)/NterPstIrev (5′-TTCTTTCTTGATTTTACTTAAACTCTCTCTTTC-3′)/CterSacImut (5′-GAAGAAAGAGAGAGTTTAAGTGAGCTCTAAGAAAGAAGG-3′)/CterSacIrev (5′-ACTTAAACTCTCTCTTTCTTCTGTTATTTTAGC-3′). The former two primers were used to introduce two stop codons (italic) and a second PstI site (underlined) immediately after the Glu607 residue of the TopRI protein. Plasmids carrying mutations were easily identified by PstI digestion. Selected mutants were digested and religated, leading to the final construct (pQET7-topR1ΔC) containing 1888 bp, coding for amminoacids 1–607 of TopR1 fused to the hexahistidine coding sequence (Nter). For the expression of the TopRI C-terminal domain (Cter), the latter two primers were used to introduce a second SacI site (underlined) immediately before the Lys605 residue. SacI digestion and religation allowed to obtain pQET7-topR1ΔN, containing 1968 bp coding for amminoacids 605–1242 of TopR1 fused to the hexahistidine coding sequence. The SacI/PstI region on both plasmids was checked by DNA sequencing.
Protein purification
TopR1 and deletion mutants were expressed in the E. coli BL21-AI strain (Stratagene). Cultures were grown overnight at 37°C in 2 l of Luria–Bertani (LB) medium supplemented with 50 μg/ml of ampicillin, 0.1% glucose. Cells were centrifuged for 20 min at 4000 r.p.m. in a Sorvall GSA rotor, resuspended in 30 ml of either phosphate (20 mM NaH2PO4, 20 mM Na2HPO4 pH 7.4) or tris buffer (10 mM Tris–HCl pH 7.4) containing 300 mM NaCI, broken by enzymatic digestion with lisozyme (1 mg/g wet cells) and benzonase (25 U/g wet cells), immediately broken using a French Press and then sonicated while kept refrigerated. The lysate was clarified by centrifugation for 30 min at 30 000 r.p.m. in a Beckman 70Ti rotor. Samples were applied to a Ni-triclotriacetic acid column (Amersham HisTrap, 1 ml), chromatographed with an AKTA FPLC system (Amersham Pharmacia Biotech, GE Healthcare, Buckingamshire, UK) and eluted with a linear gradient (0–1 M) of imidazole. Fractions containing the expected protein band and active in either topoisomerase or ATPase activity assays were pooled, concentrated with Amicon Ultra system (Millipore) and loaded onto a HR 16/60 Superdex 200 column. Active fractions were pooled, concentrated and stored at −20°C with the addition of 20% glycerol. The purity of the proteins was assessed by SDS–PAGE. Protein concentration was determined with a Bio-Rad Protein Assay Kit (Bio-Rad Pacific, USA).
Positive supercoiling assays
Standard assays were performed as reported (18,19) using plasmid pQE31 (Qiagen) as substrate, and the indicated temperatures and time spans. The activity of TopR1 and the two separate domains was found very sensitive to repeated freezing, excessive dilution and prolonged storage, due to protein degradation. As a consequence, the enzyme-specific activity of each batch could vary from time to time, thus appropriate controls were included in each experiment to correctly evaluate quantitative differences among samples and, to minimize variations within each experiment, a single mix with all common components was set up; moreover, whenever different enzyme amounts were used, total protein concentration was corrected with BSA. Samples were analysed by 2D agarose gel electrophoresis with ethidium bromide (0.01 μg/ml) in the second dimension. After electrophoresis, gels were stained with ethidium bromide (1 μg/ml), analysed and quantified under UV light with a Chemi-doc apparatus and the QuantityOne software (Bio-Rad, Hercules, CA, USA). Each assay was performed at least four times.
Measurement of the molecular interaction by SPR spectroscopy
Physical interaction between the two separate domains was analysed by the surface plasmon resonance technology (SPR) with a BIAcore® 2000 instrument (GE Healthcare). 0.6 μmol of Cter in 10 mM acetate buffer, pH 3.5, were immobilized onto a CM5 sensor chip by the Amine Coupling Kit (GE Healthcare) containing N-hydroxysuccinimide (NHS), N-ethyl-N9-[(-3dimethylamino)-propyl]-carbodiimide-hydrochloride (EDC) and ethanolamine–HCl, following the manufacturer's instructions, giving a final amount of immobilization of ca 4500 RU. Experiments were performed at either 25 or 40°C in HBS buffer (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% v/v Surfactant P20, pH 7.4). In order to avoid mass transfer effects, analyte injection time of 5 min and flow rate of 50 μl/min were used. Measurements were performed subtracting the value of the free channel on the sensor chip to eliminate any buffer bulk effect.
Electrophoretic mobility shift assays
PAGE-purified oligonucleotides (Primm s.r.l., Milano, Italy), listed in Table SI, were annealed to obtain DNA substrates shown in Table SII. Prior to annealing, downstream oligonucleotides were radiolabelled at the 5′-end using [32P]γ-ATP and T4 polynucleotide kinase. Annealing reaction contained equimolar concentration of two oligonucleotides in 20 mM Tris–acetate (pH 7.9), 10 mM magnesium acetate, 50 mM potassium acetate and 1 mM DTT. Standard binding reactions (10 μl) contained: 20 mM Tris–HCl (pH 8.0), 10% glycerol, 50 mM KCl, 0.1 mM DTT, 9 × 104 c.p.m. of labelled DNA probe (final concentration of 1.2 μg/ml) and the indicated amounts of appropriate proteins. Following incubation for 10 min at 37°C, samples were immediately loaded on non-denaturing 5% polyacrylamide gels in 0.5X TBE buffer and run at 70 V for 1.5 h at room temperature. Radioactivity was determined by autoradiography with a Storm PhosphoImager and quantified with the IQ-Mac software (Molecular Dynamics, GE Healthcare, Buckingamshire, UK).
ATPase assays
ATPase assay reaction mixture (10 μl) contained 35 mM Tris–HCl, pH 7, 0.1 mM Na2EDTA, 30 mM MgCl2, 2 mM DTT, 20 μM [32P]γ-ATP (0.5–1 μCi), and the indicated amounts of enzymes. Unless indicated, all reactions contained 0.32 nM of heat-denatured phage λ DNA. Total protein concentration was adjusted with BSA. Incubations were performed for 10 min at 80°C in a heated-top PCR machine to prevent evaporation and stopped on ice. A 1 μl aliquot of each mixture was spotted on a polyethyleneimine–cellulose thin layer plate (Macherey–Nagel, Duren, Germany), and developed in 0.5 M LiCl and 1 M formic acid. The amounts of [32P]γ-ATP hydrolyzed to [32P]-orthophosphate were quantified using a PhosphorImager (Amersham Biosciences). Steady-state kinetic constants were determined using the Malachite Green Phosphate Assay Kit (BioAssay System, Hayward, CA) adapting the manufacturer's instructions. Reactions (total volume 0.1 ml) were prepared in Activity Buffer (35 mM Tris–HCl; 0.1 mM EDTA; 30 mM MgCl2; 2 mM DDT; pH 7.0) with different concentrations of ATP (10 μM–2 mM). After addition of the appropriate amount of enzyme (10–40 ng/μl), each reaction was divided into five aliquots (20 μl) and incubated at 80°C for different times (0, 1, 5, 10, 20 min). Reactions were stopped on ice, transferred to a 96-well microtiter and mixed with 80 μl of 4-fold diluted Malachite Green Reagent in a final volume of 0.1 ml. After incubation at room temperature for 20 min, the absorbance of released inorganic phosphate was measured at 600 nm with a Multiskan RC plate reader (Thermo Labsystems, Sangrok, Korea). Each value was corrected for the spontaneous hydrolysis of ATP at the same assay temperature. A standard curve was obtained using known amounts of inorganic phosphate. For each ATP concentration, values were plotted against time to obtain the initial velocity, v0, expressed as specific activity in units/milligram of enzyme (one unit was the amount of enzyme able to hydrolyze 1 μmol of ATP to ADP and inorganic phosphate in 1 min under the conditions indicated earlier). The GraFit 3.0 program [Leatherbarrow, R. J. (1992) GraFit version 3.0, Erithacus Software Ltd, Staines, UK] was used to plot v0 values against ATP concentrations by fitting the Michaelis–Menten equation and leading to the determination of the steady-state kinetic constants.
RESULTS
Reconstitution of reverse gyrase activity from its separate domains
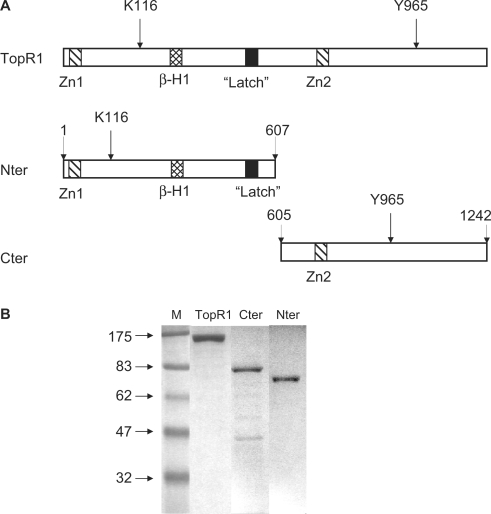
Sulfolobus solfataricus genome contains two genes coding for two reverse gyrase isoforms, named TopR1 and TopR2 (17). TopR1 shows higher sequence similarity with the reverse gyrase characterized from Sulfolobus acidocaldarius (13). The topR1 gene was amplified from the S. solfataricus P2 strain and the protein was expressed in E. coli and purified to homogeneity (Figure 1). Previous work showed that the separate N-terminal helicase-like and C-terminal topoisomerase domains of the S. acidocaldarius reverse gyrase cooperate to reconstitute the positive supercoiling reaction (15). In order to extend these results and investigate the interplay of the two domains in TopR1, we have used sequence and predicted secondary structure alignments between TopR1, the A. fulgidus and the S. acidocaldarius reverse gyrase, to design two polypeptides, one encoding the N-terminal ATPase and the other the C-terminal topoisomerase (Figure 1A). The two domains were separately expressed in E. coli and purified (Figure 1B). TopR1 and its halves were found prone to inactivation by degradation, both during and after purification. Similar behaviour was also reported for native reverse gyrase (10,16), thus suggesting that it reflects some intrinsic feature of the enzyme.
Figure 1.
(A) Schematic diagram showing the domain architecture of TopR1 and the two separate domains. The positions of the essential residues K116 in the catalytic site of the ATPase domain and Y965 in the topoisomerase domain are indicated by an arrow; the predicted Zn-finger motives and the region corresponding to the ‘latch’ in the A. fulgidus reverse gyrase are shown by internal boxes. (B) Coomassie-stained gel of purified TopR1 (1 μg), Cter and Nter (500 ng each). Fainter bands correspond to degradation products. M, molecular weight marker (kDa).
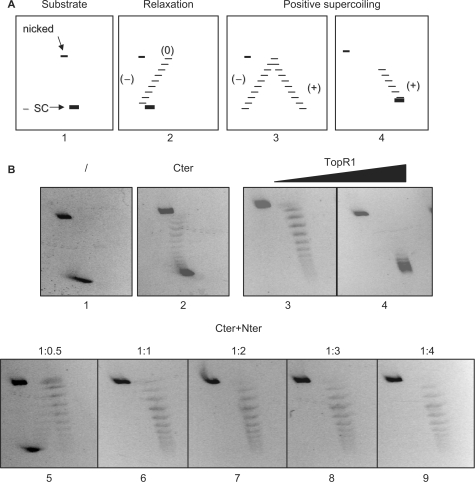
The hallmark of reverse gyrase is its ability to induce positive supercoiling in an ATP hydrolysis-dependent reaction. Topological changes in covalently closed DNA circle can be analyzed by two-dimensional agarose gel electrophoresis using an intercalant agent (e.g. ethidium bromide) in the second dimension (Figure 2). Incubation of reverse gyrase with a negatively supercoiled plasmid DNA produces topoisomers with increasing Linking number (Lk), from less negative to highly positive. As expected, Cter on its own was only able to relax negative supercoils but did not catalyze positive supercoiling reaction, regardless of the presence of ATP. Addition of Nter restored positive supercoiling activity (Figure 2B); the optimal efficiency of the reaction was obtained at stoichiometric ratio of the two domains and was not improved by further increase of Nter. The efficiency of supercoiling obtained with the reconstituted enzyme was comparable to that obtained with the same molar concentration of TopR1, and showed the same temperature dependence (Figure 3C). Whereas both enzymes were barely active below 50°C (not shown), increasingly positive topoisomers were produced raising temperature from 60°C to 90°C; however, Nter + Cter showed reduced activity at 90°C, which was due to partial inactivation of the reconstituted enzyme during the assay at this temperature (see later).
Figure 2.
Analysis of TopR1 and truncated enzymes topoisomerase activity by bi-dimensional agarose gel electrophoresis. (A) Diagram of 2D gels. Panel 1 shows the migration of the negative supercoiled (-SC) and nicked forms of the plasmid substrate. Panel 2 shows a ladder of negative to relaxed (0) topoisomers. Panel 3 shows the migration of a complete arc of negative to positive topoisomers; in panel 4 only positive topoisomers are shown. (B) Four hundred nanograms (final concentration 0.4 nM) of pQE31 plasmid DNA (ΔLk > −12) were mock incubated (/) or incubated with indicated enzymes for 10 min at 80°C in a final volume of 20 μl, and subjected to 2D agarose gel electrophoresis. All reactions contained 1 mM ATP and the following protein concentration: panel 2, Cter (30 nM); panels 3 and 4, TopR1 (30 and 60 nM, respectively); panels 5–9, Cter (30 nM) plus Nter (15, 30, 60, 90 and 120 nM, respectively). In all samples, total protein concentration was adjusted adding the required amounts of BSA.
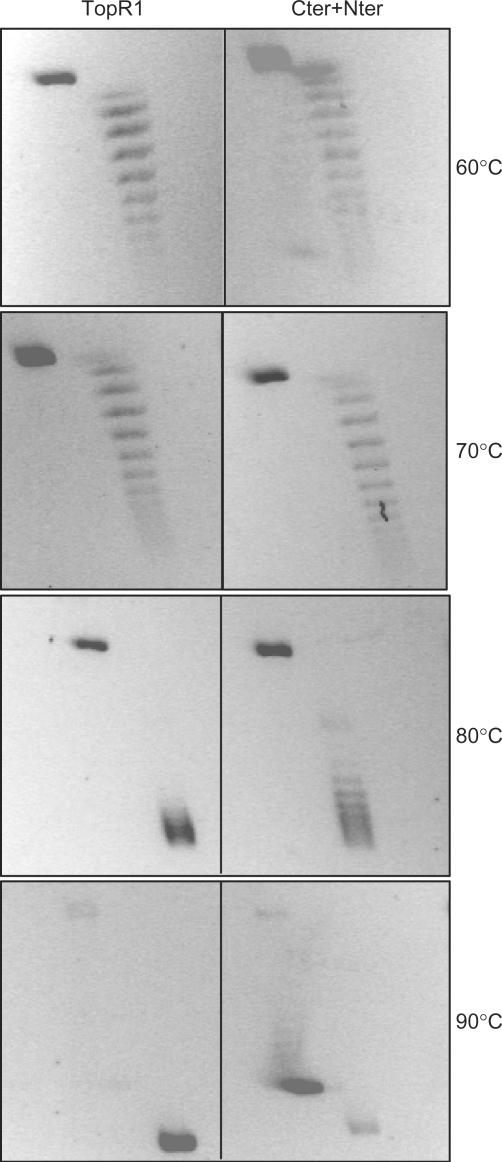
Figure 3.
Temperature dependence of positive supercoiling activity. Reactions were set up as described in the legend to Figure 2B, using TopR1 (60 nM) or Cter + Nter (60 nM each) and incubated for 10 min at the indicated temperatures.
Physical interaction between Nter and Cter
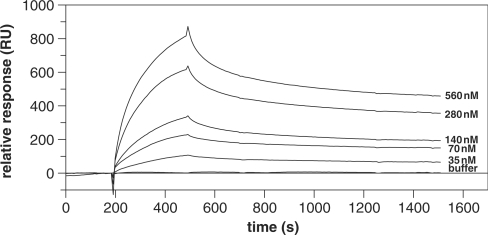
The intermolecular interaction between the two halves of TopRI was tested by surface plasmon resonance kinetic analysis. Cter was immobilized onto a sensor chip to ca 4500 response units (RU) and various concentrations of Nter (35–560 nM) were passed over. Cter and Nter specifically interacted with a relative binding affinity (KD) of about 0.1 μM (Figure 4, Table 1). The interaction was not significantly affected by the presence of 1 mM ATP, and was within the same order of magnitude when the analysis was performed at either 25 or 40°C (which is the maximal temperature allowed by the instrument). The specificity of the interaction was tested by passing over the chip similar concentrations of BSA (not shown) and of the non-specific DNA-binding protein Smj12 of S. solfataricus (20), which gave very low binding affinity value (Table 1).
Figure 4.
Binding analysis of Nter and Cter by SPR. Increasing concentrations of Nter were injected over the immobilized ligand Cter. The analysis was performed at 25°C. Each Nter concentration was injected in duplicate and average increases in RU were plotted. Each sensorgram was corrected by subtracting a sensorgram obtained from a reference flow cell.
Table 1.
Binding parameters obtained from SPR analysis
| Ligand | T (%) | Ka (M−1s−1) | Kd (s−1) | KA (M−1) | KD (M) |
|---|---|---|---|---|---|
| Nter | 25 | 1.42 × 104 | 1.87 × 10−3 | 7.59 × 106 | 1.32 × 10−7 |
| Nter + ATP | 25 | 6.90 × 103 | 1.09 × 10−3 | 6.32 × 106 | 1.58 × 10−7 |
| Nter | 40 | 4.35 × 103 | 1.52 × 10−3 | 2.87 × 106 | 3.49 × 10−7 |
| Smj12 | 25 | 8.87 × 10−1 | 3.13 × 10−3 | 2.84 × 102 | 3.53 × 10−3 |
The crystal structure of the A. fulgidus reverse gyrase revealed two obvious points of contact between the two domains, namely: (i) the latch in subdomain H3 interacts with the so-called topoisomerase insertion in subdomain T3; (ii) three strands from subdomain H1 and one strand from subdomain T1 are connected to form a β sheet (11). Results reported in this and previous paragraph suggest that proper physical and functional interactions between the two reverse gyrase domains may form even in the absence of covalent linkage between them. Thus, the separate domains and the reconstituted enzyme can be used to dissect the different activities of the wild-type protein and analyze their cooperation in the different steps of the reverse gyrase reaction.
DNA binding
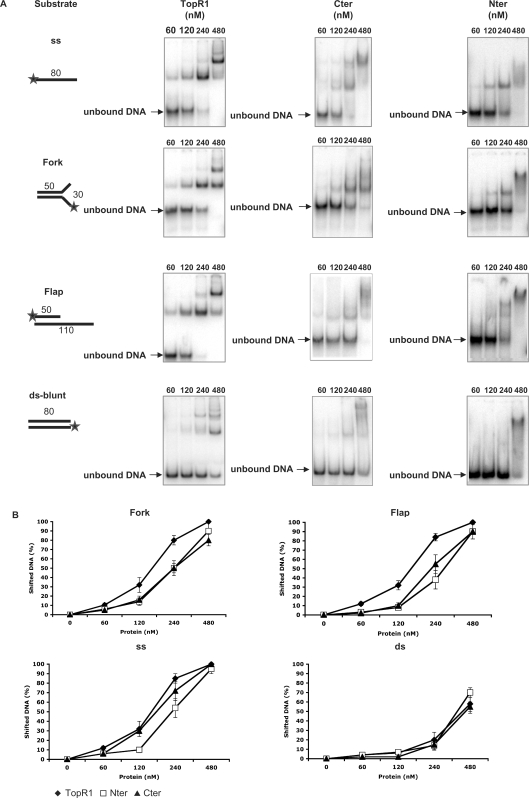
The crystal structure of the A. fulgidus reverse gyrase predicted four potential binding regions (11): a Zn finger at the N-terminus; a large region of positive potential corresponding to the latch; a β-hairpin a in the subdomain H1; a Zn-finger motif in the topoisomerase domain T1 (Figure 1). Three-dimensional electron microscopy (EM) studies of reverse gyrase from Sulfolobus tokodai showed that the enzyme contacts double strand DNA both in the helicase-like and in the topoisomerase domains (21). Positive supercoiling greatly depends upon enzyme stoichiometry, suggesting that DNA binding plays a critical role in the efficiency of the reaction (14). In order to study the contribution of each domain to DNA binding, we tested the affinity and selectivity of binding of reverse gyrase and its domains to oligonucleotide substrates with different structure (Figure 5A). TopR1, Cter and Nter formed complexes with all oligonucleotides tested, thus showing that the multiple binding regions identified by structural analysis behave as independent binding sites. Whereas all three proteins showed similar low affinity for the blunt ds probe, the extent of complex formation was different when the proteins were used with the other substrates (Figure 5A and B). The ss probe was bound with higher avidity by TopR1 and Cter, compared with Nter. Interestingly, TopR1 was more effective than the two domains in binding substrates containing ds–ss junctions (fork and flap).
Figure 5.
Analysis of TopR1 and truncated enzymes binding activity by EMSA. (A) Each panel shows the structure of the substrate used and a typical autoradiogram obtained. Stars denote the 32P-labelled end. Increasing amounts of purified TopR1 or TopR1 mutants (Cter and Nter) were incubated in the standard 10 μl reaction mixture for 10 min at 37°C. The enzyme concentration (nM) is indicated above each lane in the autoradiogram, whereas the substrate concentration was constant in all experiments (9 × 104 c.p.m./lane, 1.2 μg/ml). (B) Quantitation of binding activity. For each DNA ligand, the fraction of shifted DNA versus the amount of protein used is plotted. Binding assays were performed at least in triplicate and the results were averaged. Values are the mean ± SE of three independent experiments.
It has been previously hypothesized that the affinity of reverse gyrase for ds versus ss DNA might be modulated by nucleotide binding/hydrolysis (14); however, no supporting experimental data were presented. In addition, EM analysis could not make clear whether ATP might play a role in opening the topoisomerase gate to bind DNA (21). To test these possibilities, we repeated the EMSA experiments in the presence of either 1 mM ATP or 1 mM ADP, but found no difference in binding activity of the three proteins with any of the above-described probes (data not shown).
ATP hydrolysis
Reverse gyrase shows DNA-stimulated ATPase activity (22–24). However, the exact role of ATP hydrolysis in the reverse gyrase reaction is not understood. Despite the presence of sequence motives found in SF2 helicases, no DNA unwinding activity has ever been shown for reverse gyrase (15,19). In agreement with the previous results, neither the N-terminal domain nor the full-length protein showed helicase activity with a variety of substrates formed by untailed, 5′-tailed, 3′-tailed or doubly tailed oligonucleotides (data not shown).
It is becoming increasingly clear that not all predicted helicases actually possess true helicase activity. For instance, helicase motifs are characteristic of ATP-dependent chromatin-remodelling factors, including the SWI2/SNF2 and ISWI families that translocate along ds DNA without opening the duplex and can induce torsional stress (25,26). We tested the ability of TopR1 and Nter to translocate along ds or ss DNA by using the method of Birda and Raneny (27). No evidence that the proteins can displace streptavidin from biotinylated oligonucleotides at temperatures ranging between 60°C and 80°C could be obtained (data not shown). These results thus failed to provide evidence in support of the translocation model.
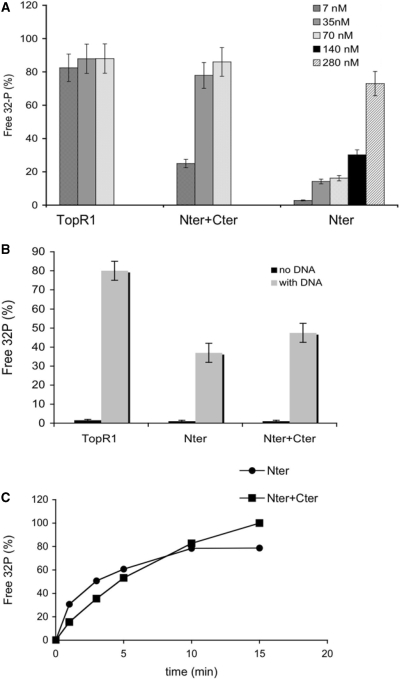
How ATP hydrolysis is coordinated with the supercoiling reaction is not clear: in the A. fulgidus enzyme, the supercoiling rate is the same in a wide range of ATP concentrations and ATPase activity continues at a linear rate even when the positive supercoiling reaction stops (28). We have analyzed the efficiency of ATP hydrolysis by TopR1, Nter and the reconstituted enzyme. Although the ATPase domain is entirely contained within Nter, this protein showed highly reduced activity as compared to the whole protein; interestingly, the activity was restored to almost normal levels by the presence of equimolar amounts of Cter (Figure 6A). Both Nter and the reconstituted enzyme showed the same dependence on DNA as the whole protein (Figure 6B). A time course of the reaction showed that Nter was active throughout the assay (Figure 6C), thus showing that stimulation of the ATPase activity by Cter is not due to a stabilization effect (see later).
Figure 6.
Cter stimulates the ATPase activity of Nter. (A) The three enzymes were assayed using 32P-γ-ATP as reported in ‘Materials and Methods’ section. Shown are the final concentrations of TopR1 and Nter, alone or in combination with equimolar amounts of Cter. In all samples total protein concentration was adjusted with BSA. Incubation was for 10 min at 80°C with the addition of ss DNA (see ‘Materials and Methods’ section for details). The fraction of free 32P versus total radioactivity, corrected for the spontaneous ATP hydrolysis, is plotted. Assays were performed at least in triplicate and the results were averaged. Values are the mean ± SE of three independent experiments. (B) Dependence on DNA of the ATPase reaction. Assays were performed and analysed as described in the legend to Figure 6A, with or without DNA. Final concentrations were: TopR1 7 nM, Nter 140 nM, Cter + Nter 14 nM each. (C) Time course of the ATPase reaction. Reaction mixtures for Nter (280 nM) and Nter+Cter (70 nM each) were set and assayed for the indicated time spans at 80°C. Assays were performed and quantified as reported in the legend to Figure 6A.
A spectrophotometric assay allowed us to determine the steady-state kinetic constants of ATP hydrolysis at 80°C for all the three enzymes (Table 2). The TopR1 ATPase activity showed a typical Michaelis–Menten behaviour and was significantly stimulated by DNA, as shown previously for Thermotoga maritima reverse gyrase (24). Nter showed a 100-fold reduction of ATP hydrolysis efficiency as compared with TopR1, with marked reduction of both KM and kcat. Addition of equimolar amounts of Cter restored the ATP hydrolysis efficiency to levels comparable to that of the wild-type enzyme. Interestingly, the KM of the reconstituted enzyme was identical to that of the full-length enzyme.
Table 2.
Steady state kinetic constants for the ATP hydrolysis at 80°C
| KM (mM) | Kcat (s−1) | Kcat/KM (mM−1s−1) | |
|---|---|---|---|
| TopRI | 77.1 ± 19.4 | 2.55 ± 0.16 | 3.30 × 10−2 |
| TopRI (no DNA) | 16.2 ± 10.2 | 0.05 ± 0.01 | 7.00 × 10−3 |
| Nter | 775.7 ± 339.3 | 0.35 ± 0.07 | 1.89 × 10−4 |
| Nter + Cter | 61.9 ± 31.2 | 0.77 ± 0.09 | 1.24 × 10−2 |
In A. fulgidus reverse gyrase, deletion of the latch or of the C-terminal ‘topoisomerase insertion’ interacting with it resulted in significant increase of the ATPase activity, suggesting that the ATP binding cleft is closed by the latch and open when the nucleotide is bound (11,28,29). Our results agree and provide further insight in the cooperation between the two domains in the modulation of the ATPase activity. Indeed, in the isolated N-terminal domain, the conformation of the ATP hydrolysis cleft might be more tightly closed than in the full-length enzyme, and might not be opened upon simultaneous binding of the nucleotide and DNA, like in reverse gyrase. The correct active site architecture allowing ATP binding/hydrolysis is restored only when Cter is present. Since deletion of the topoisomerase insertion in the A. fulgidus enzyme results in the opposite effect (increase of ATP hydrolysis), our data suggest that complex interactions between some other C-terminal region(s) and the latch (or other N-terminal regions) are required to regulate ATPase activity.
DNA relaxation
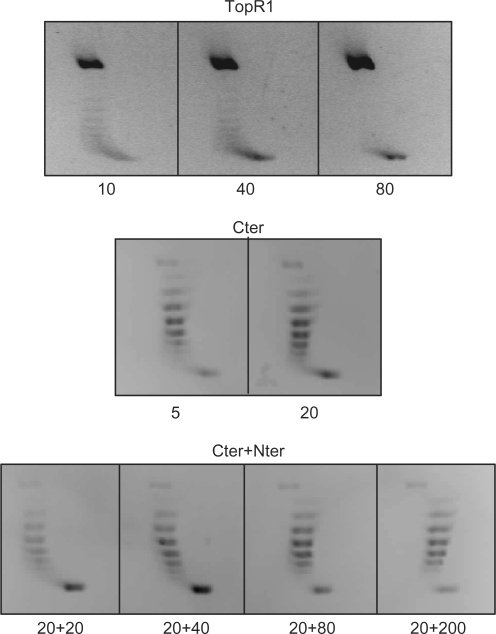
Like other reverse gyrases (15), TopR1 showed very weak ATP-independent relaxation (Figure 7); when used at very high concentration, the enzyme produced high amount of nicked plasmid, likely because simultaneous binding/cleavage by a high number of enzyme molecules impairs the ligase step and/or causes unbalance of the reaction. In contrast, Cter showed a measurable relaxation activity. These results concur with those reported for the A. fulgidus enzyme (11,28,29): crystal structure and biochemical analysis suggested that, in the absence of the nucleotide, the latch represses the relaxation activity by closing the topoisomerase gate; ATP hydrolysis would then induce a movement of the latch shifting from a close to an open position allowing strand passage. According to this model, we expected that addition of Nter, which contains the region corresponding to the latch in the S. solfataricus enzyme, would repress the relaxation activity of Cter. Surprisingly, this was not the case: Nter did not affect the Cter relaxation activity even when used at 10-fold excess (Figure 7). Our data show that, while in the presence of ATP non-covalent interactions between the two domains promote both efficient nucleotide hydrolysis (Figure 6) and communication to the C-terminal domain to obtain positive supercoiling (Figures 2 and 3), such interactions are unable to restore repression of the relaxation activity in the absence of the nucleotide. We thus assume that repression of relaxation activity and stimulation of ATPase activity/positive supercoiling are not mediated by the same interactions.
Figure 7.
DNA relaxation assay. Assays were performed and analysed as described in the legend to Figure 2, with the exception that no ATP was added; proteins were used at the indicated concentrations (nM).
Whereas in our hand the presence of Nter does not either stimulate or repress Cter relaxation activity, the S. acidocaldarius reverse gyrase N-terminal domain was reported to stimulate its C-terminal topoisomerase activity (15). This discrepancy might be due to the different stability of the two topoisomerase domains. Indeed, the C-terminal domain of the S. acidocaldarius enzyme seems highly prone to degradation, at least under the conditions used by Declais et al. (15), making it possible that the apparent stimulation of relaxation activity by the N-terminal domain is due to a stabilization effect (see ‘Thermostability’ section).
Thermostability
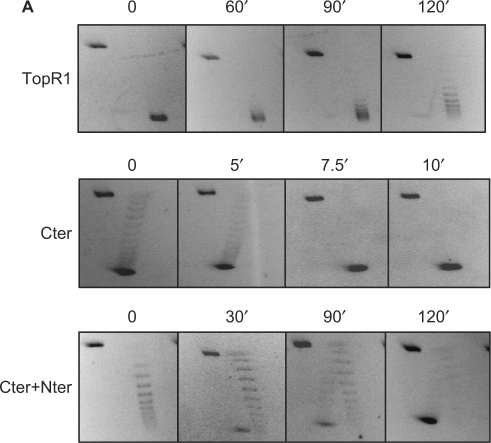
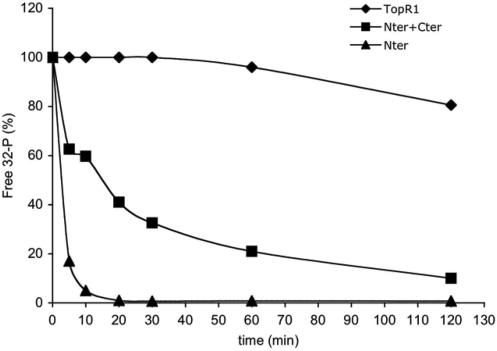
Thermal stability of the topoisomerase activity was tested after incubation of TopR1, Cter or Cter+Nter for increasing time spans at 80°C in the absence (Figure 8) or in the presence of 1 mM ATP (not shown); assays were always performed at 80°C in the presence of the nucleotide. Under these latter conditions, both TopR1 and Nter+Cter induce positive supercoiling, whereas Cter is only able to relax DNA. Although TopR1 was still active after 2 h of incubation at 80°C, the activity of the C-terminal domain was highly unstable (100% inactivation within 7.5 min at 80°C). Interestingly, the thermostability of Cter was rescued by incubation with equimolar amounts of Nter. Stabilization was seen in the absence of DNA, and addition of ATP had no effect on the stability of the three proteins (data not shown).
Figure 8.
Nter increases thermal stability of Cter topoisomerase activity. Proteins (final concentration 60 nM each) were incubated at 80°C for the indicated timespans in reaction mix without ATP. Assays (10 min at 80°C) were performed and analysed in the presence of 1 mM ATP as described in the legend to Figure 2.
In addition, whereas TopR1 ATPase activity was completely stable after 1 h of incubation at 80°C, the ATPase activity of the N-terminal domain was highly unstable (<20% residual activity after 5 min of incubation at 80°C). Addition of the C-terminal domain significantly rescued the stability of the N-terminal domain, leading to about 20% residual activity after 2 h at 80°C (Figure 9).
Figure 9.
Cter increases thermal stability of Nter ATPase activity. The indicated enzymes were incubated at 80°C for the indicated time spans in reaction mix without ATP and DNA. Final concentrations were: TopR1, 60 nM; Nter, 660 nM; Nter+Cter, 60 nM each. ATPase assays (10 min at 80°C) were performed and analysed as described in the legend to Figure 6. Values were expressed as a fraction of the activity of each protein at time = 0, which was set 100%.
DISCUSSION
The roles of the individual domains of the reverse gyrase in the catalyzed reactions were examined by comparing the properties of the full-length enzyme with those of truncated polypeptides lacking the N-terminal or the C-terminal region. Both domains were able to bind DNA independently and contribute to the highly efficient binding activity of reverse gyrase. Whereas Nter showed low binding affinity for all tested substrates, both TopR1 and Cter showed marked binding preference for ss DNA, as previously reported for other reverse gyrases (22–24) and Type IA topoisomerases (30). Both TopR1 (this work) and the A. fulgidus reverse gyrase (28,29) bind with high affinity to substrates containing both ss and ds regions. Our results show that only the full-length enzyme displays such property, thus suggesting that the combination of the independent binding sites of the two domains, and likely their simultaneous interaction with the substrate, give rise to a DNA-binding activity with different specificity. The ability to recognize substrates mimicking partially denatured regions or arrested replicative forks might have relevance for the function of the enzyme in vivo.
The C-terminal domain retained DNA relaxation properties, hallmark characteristics of type IA topoisomerases; in contrast, the isolated N-terminal domain showed only weak ATPase activity by its own. ATPase activity, as well as positive supercoiling, were restored when both fragments were mixed reconstituting the unique enzyme function. In addition, we have shown that the two domains form specific physical interactions with each other in the absence of covalent bond, DNA or ATP and these interactions have strong mutual stabilizing effect. Although Cter behaves like a classical topoisomerase IA, its ability to cooperate with Nter is unique, as E. coli Topoisomerase 3 could be neither stabilized nor induced to produce positive supercoiling by Nter (A. D'Amaro and M. Ciaramella, unpublished results).
We have shown that the physical interaction with Nter turns a universal topoisomerase IA module, with a moderate relaxing activity and unstable at high temperature, into an enzyme with unique features, unable to relax DNA in the absence of the nucleotide, capable of performing the unique positive supercoiling reaction upon ATP hydrolysis, and highly stable at 80°C. Such a profound effect is reciprocal: Cter in turn stimulates and stabilizes the ATPase module, which is almost inactive and unstable at 80°C by its own. Our data might suggest that the strong pressure imposed by the extreme environmental temperature determined the evolution of a novel activity through the fusion of two pre-existing non-thermostable modules. However, we cannot rule out the possibility that reverse gyrase originated in a thermophilic organism from already thermostable polypeptides. In this view, it is possible that the isolated domains are unstable at high temperature because they are somehow artificial constructs.
These results are consistent with a model in which the activity and stability of reverse gyrase is modulated through multiple interactions between the two domains. Interestingly, not all correct interactions can be restored when the two modules are not covalently bound: indeed, in the absence of the nucleotide the N-terminal domain is able to repress the topoisomerase activity only if covalently linked. This observation suggests that the presence of ATP might help the correct positioning of the two domains, thus resulting in positive supercoiling. While the mechanistic bases of this interaction need to be elucidated, it might provide a means to regulate the enzyme activity in vivo. We and others have found that reverse gyrase is prone to degradation both in vitro and in vivo (10,16). Degradation might provide an elegant way of controlling the enzyme activity, since the separate modules are not stable. For many multiprotein complexes, absence of one subunit renders others unstable. For example, BLAP75, an integral component of BLM complexes, is required for stability of BLM and Topo III (31) and stability of the yeast chromatin remodelling protein SWI3 is reduced in the absence of its interacting partners SWI1 and SWI2 (32).
Our results do not support a number of predictions of the different models proposed to explain the mechanics of the positive supercoiling reaction. Once more, we could not find any evidence of strand separation activity, an essential prediction of tracking models. In addition, we could not find evidence of reverse gyrase or Nter translocation along either ss or ds DNA. Moreover, in no circumstances was DNA binding influenced by nucleotides, thus suggesting that it does not require drastic ATP hydrolysis-induced conformational changes and not supporting the change of affinity model. Our data are compatible with a simpler model originally proposed by Forterre (PhD thesis, 1985; personal communication), originally based on the hypothesis that reverse gyrase binding induces local unwinding of DNA, which was experimentally confirmed later on (15). In a covalently closed circular DNA, this conformational change produces a positive compensatory superturn elsewhere. A strand passage event in the locally unwound region would increase the linking number, thus stabilizing the positive superturn. Since Declais et al. (15) have shown that local unwinding requires both domains, one should assume that only when both domains are present the enzyme gains the ability to produce a DNA bubble that is the preferred substrate for the C-terminal topoisomerase, allowing efficient strand passage toward linking number increase. However, the molecular basis of this ‘conformation-specific DNA binding’ (14) as well as the role of ATP hydrolysis remain elusive. Clearly, the mechanics of this reaction, the multiple interactions between the two domains and how the interaction with the N-terminal domain may direct strand passage unidirectionally toward linking number increase await further investigation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENT
Agenzia Spaziale Italiana (Project MoMa n. 1/014/06/0). Funding to pay the Open Access publication charges for this article was provided by Agenzia Spaziale Italiana (Project MoMa n. 1/014/06/0).
Conflict of interest statement: None declared.
REFERENCES
- 1.D’Amaro A, Rossi M, Ciaramella M. Reverse gyrase: an unusual DNA manipulator of hyperthermophilic organisms. Ital. J. Biochem. 2007;56:103–109. [PubMed] [Google Scholar]
- 2.Nadal M. Reverse gyrase: an insight into the role of DNA-topoisomerases. Biochimie. 2007;89:447–455. doi: 10.1016/j.biochi.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 3.Brochier-Armanet C, Forterre P. Widespread distribution of archaeal reverse gyrase in thermophilic bacteria suggest a complex hystory of vertical inheritance and lateral gene transfers. Archaea. 2006;2:i–xi. doi: 10.1155/2006/582916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forterre P. A hot story from comparative genomics: reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 2002;18:236–237. doi: 10.1016/s0168-9525(02)02650-1. [DOI] [PubMed] [Google Scholar]
- 5.Duguet M. When helicase and topoisomerase meet! J. Cell Sci. 1997;110:1345–1350. doi: 10.1242/jcs.110.12.1345. [DOI] [PubMed] [Google Scholar]
- 6.Khakhar RR, Cobb JA, Bjergbaek L, Hickson ID, Gasser SM. RecQ helicases: multiple roles in genome maintenance. Trends Cell Biol. 2003;13:493–450. doi: 10.1016/s0962-8924(03)00171-5. [DOI] [PubMed] [Google Scholar]
- 7.Mankouri HW, Hickson ID. The RecQ helicase-topoisomerase III-Rmi1 complex: a DNA structure-specific ‘dissolvasome’? Trends Biochem. Sci. 2007;32:538–546. doi: 10.1016/j.tibs.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Atomi H, Matsumi R, Imanaka T. Reverse gyrase is not a prerequisite for hyperthermophilic life. J. Bacteriol. 2004;186:4829–4833. doi: 10.1128/JB.186.14.4829-4833.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Napoli A, Valenti A, Salerno V, Nadal M, Garnier F, Rossi M, Ciaramella M. Reverse gyrase recruitment to DNA after UV light irradiation in Sulfolobus solfataricus. J. Biol. Chem. 2004;279:33192–33198. doi: 10.1074/jbc.M402619200. [DOI] [PubMed] [Google Scholar]
- 10.Valenti A, Napoli A, Ferrara MC, Nadal M, Rossi M, Ciaramella M. Selective degradation of reverse gyrase and DNA fragmentation induced by alkylating agent in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 2006;34:2098–2108. doi: 10.1093/nar/gkl115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez AC, Stock D. Crystal structure of reverse gyrase: insights into the positive supercoiling of DNA. EMBO J. 2002;21:418–426. doi: 10.1093/emboj/21.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 13.Confalonieri F, Elie C, Nadal M, de La Tour C, Forterre P, Duguet M. Reverse gyrase: a helicase-like domain and a type I topoisomerase in the same polypeptide. Proc. Natl Acad. Sci. USA. 1994;90:4753–4757. doi: 10.1073/pnas.90.10.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh TS, Capp C. Nucleotide- and stoichiometry-dependent DNA supercoiling by reverse gyrase. J. Biol. Chem. 2005;280:20467–20475. doi: 10.1074/jbc.M502739200. [DOI] [PubMed] [Google Scholar]
- 15.Declais AC, Marsault J, Confalonieri F, de La Tour CB, Duguet M. Reverse gyrase, the two domains intimately cooperate to promote positive supercoiling. J. Biol. Chem. 2000;275:19498–19504. doi: 10.1074/jbc.M910091199. [DOI] [PubMed] [Google Scholar]
- 16.Nadal M, Couderc E, Duguet M, Jaxel C. Purification and characterization of reverse gyrase from Sulfolobus shibatae. Its proteolytic product appears as an ATP-independent topoisomerase. J. Biol. Chem. 1994;269:5255–5263. [PubMed] [Google Scholar]
- 17.She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC, Clausen IG, Curtis BA, De Moors A, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl Acad. Sci. USA. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Napoli A, Zivanovic Y, Bocs C, Buhler C, Rossi M, Forterre P, Ciaramella M. DNA bending, compaction and negative supercoiling by the architectural protein Sso7d of Sulfolobus solfataricus. Nucleic Acids Res. 2002;30:2656–2662. doi: 10.1093/nar/gkf377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Napoli A, Valenti A, Salerno V, Nadal M, Garnier F, Rossi M, Ciaramella M. Functional interaction of reverse gyrase with single-strand binding protein of the archaeon Sulfolobus. Nucleic Acids Res. 2005;33:564–576. doi: 10.1093/nar/gki202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Napoli A, Kvaratskelia M, White MF, Rossi M, Ciaramella M. A novel member of the bacterial-archaeal regulator family is a non-specific DNA-binding protein and induces positive supercoiling. J. Biol. Chem. 2001;276:10745–10752. doi: 10.1074/jbc.M010611200. [DOI] [PubMed] [Google Scholar]
- 21.Matoba K, Mayanagi K, Nakasu S, Kikuchi A, Morikawa K. Three-dimensional electron microscopy of the reverse gyrase from Sulfolobus tokodaii. Biochem. Biophys. Res. Commun. 2002;297:749–755. doi: 10.1016/s0006-291x(02)02255-6. [DOI] [PubMed] [Google Scholar]
- 22.Shibata T, Nakasu S, Yasui K, Kikuchi A. Intrinsic DNA-dependent ATPase activity of reverse gyrase. J. Biol. Chem. 1987;262:10419–10421. [PubMed] [Google Scholar]
- 23.Nakasu S, Kikuchi A. Reverse gyrase; ATP-dependent type I topoisomerase from Sulfolobus. EMBO J. 1985;4:2705–2710. doi: 10.1002/j.1460-2075.1985.tb03990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jungblut SP, Klostermeier D. Adenosine 5′-O-(3-thio)triphosphate (ATPgammaS) promotes positive supercoiling of DNA by T. maritima reverse gyrase. J. Mol. Biol. 2007;371:197–209. doi: 10.1016/j.jmb.2007.05.031. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, Cairns BR, Peterson CL, Bustamante C. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol. Cell. 2006;24:559–568. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat. Struct. Mol. Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- 27.Byrd AK, Raney KD. Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat. Struct. Mol. Biol. 2004;11:531–538. doi: 10.1038/nsmb774. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez AC. Investigating the role of the latch in the positive supercoiling mechanism of reverse gyrase. Biochemistry. 2003;42:5993–6004. doi: 10.1021/bi034188l. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez AC. Studies of a positive supercoiling machine. Nucleotide hydrolysis and a multifunctional “latch” in the mechanism of reverse gyrase. J. Biol. Chem. 2002;277:29865–29873. doi: 10.1074/jbc.M202853200. [DOI] [PubMed] [Google Scholar]
- 30.Changela A, DiGate RJ, Mondragon A. Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature. 2001;411:1077–1081. doi: 10.1038/35082615. [DOI] [PubMed] [Google Scholar]
- 31.Yin J, Sobeck A, Xu C, Meetei AR, Hoatlin M, Li L, Wang W. BLAP75, an essential component of Bloom's syndrome protein complexes that maintain genome integrity. EMBO J. 2005;24:1465–1476. doi: 10.1038/sj.emboj.7600622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterson CL, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.