Abstract
The conserved RNA helicase DDX3 is of major medical importance due to its involvement in numerous cancers, human hepatitis C virus (HCV) and HIV. Although DDX3 has been reported to have a wide variety of cellular functions, its precise role remains obscure. Here, we raised a new antibody to DDX3 and used it to show that DDX3 is evenly distributed throughout the cytoplasm at steady state. Consistent with this observation, HA-tagged DDX3 also localizes to the cytoplasm. RNAi of DDX3 in both human and Drosophila cells shows that DDX3 is required for cell viability. Moreover, using RNAi, we show that DDX3 is required for expression of protein from reporter constructs. In contrast, we did not detect a role for DDX3 in nuclear steps in gene expression. Further insight into the function of DDX3 came from the observation that its major interaction partner is the multi-component translation initiation factor eIF3. We conclude that a primary function for DDX3 is in protein translation, via an interaction with eIF3.
INTRODUCTION
Human DDX3 is a ubiquitously expressed ∼73 kD protein that belongs to the DEAD box family of ATP-dependent RNA helicases (1,2). DDX3 (also referred to as DDX3X, DBX, HLP2, DDX14, DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3, CAP-Rf, DEAD/H box-3 and helicase like protein 2) is located on the X chromosome and is highly homologous (>90%) to DDX3Y (also called DBY), which is present on the Y chromosome and expressed only in the male germ line (1,2). DDX3 has been the subject of intensive investigation because of its potential medical importance in both cancer and viral infection as well as its roles in numerous cellular processes (1–6). DDX3 is thought to be a key cellular target of Hepatitis C virus (HCV) core protein (7−9) and is required for HCV RNA replication (2,10,11). DDX3 also functions as a cellular cofactor for CRM-dependent nuclear export of HIV RNA (12). Finally, DDX3 is a component of neuronal transport granules as well as germinal granules, both of which are involved in localized mRNP translation (13–15).
Both DDX3 and its essential yeast homolog, Ded1, have ATP-dependent RNA helicase activity (12,16,17). More recently, Ded1 was also shown to be capable of displacing a protein complex from RNA in the absence of duplex unwinding (18) and to have RNA chaperone activity (19). Among the reported roles for Ded1 in yeast, the most compelling evidence exists for a direct role in translation initiation. In particular, Ded1 is present in the cytoplasm and is required for translation in vitro (20,21) and in vivo (15,20,22). Ded1 also interacts genetically with several translation initiation factors, including the well-known DEAD box RNA helicase eIF4A and the cap-binding protein eIF4E (1,20,23). Additional studies have led to the model that Ded1 is required, in addition to eIF4A, for unwinding RNA during scanning for the translation initiation codon [see refs(24,25) and references therein].
Significantly, several metazoan homologs of Ded1, including those in Drosophila (known as Belle), mouse (PL10) and human (DDX3) can rescue the lethal phenotype of a ded1 null mutant (8,14,20). Hereafter, for simplicity, we will refer to all of the metazoan homologs as DDX3. A potential function for metazoan DDX3 in translation was suggested by the observation that human DDX3 interacts directly with the HCV core protein, and this interaction inhibits translation in vitro (8). Moreover, DDX3 was detected in polysomes in Chironomus tetans (26). However, recent RNAi studies and over-expression of DDX3 in mammalian cells have led to the view that this protein does not function in translation initiation, but instead is a translation repressor (27). In a related observation, over-expression of yeast Ded1 repressed translation, and this protein is present in, and involved in, the formation of P-bodies (15). Thus, at present, it remains unclear whether DDX3 functions in translation initiation and/or translational repression.
The subcellular localization of mammalian DDX3 has also been difficult to establish. In original immunofluorescence (IF) studies in HeLa cells, DDX3 was found concentrated in distinct nuclear spots, with only low levels in the cytoplasm (7). Another study also reported that DDX3 was largely in the nucleus when subcellular fractionation of the nucleus and cytoplasm was carried out (9). However, in the same study, flag-tagged DDX3 was found in the cytoplasm, and the authors suggested that this localization might be due to the tag (9). In two other studies, DDX3 was found mostly in the cytoplasm (8,12), but entered the nucleus when cells were treated with the protein export inhibitor, leptomycin B, indicating that DDX3 shuttles (12,28,29). Thus, further clarification of the localization of DDX3 is important for understanding the function of this protein.
In this study, we raised a new antibody to DDX3. Using this antibody or HA-tagged DDX3, we find that DDX3 is predominantly cytoplasmic at steady state. To investigate the function of this protein, we carried out RNA interference of both human and Drosophila DDX3. Significantly, this analysis revealed a dramatic decrease in the levels of protein generated from reporter constructs with no apparent defects in nuclear steps in gene expression. Further insight into the function of DDX3 came from the observation that DDX3 associates with the cytoplasmic multi-subunit translation initiation factor eIF3 in an RNase insensitive manner. Together, our data indicate that DDX3 has a conserved general function in promoting translation, and this function is mediated via an eIF3-DDX3 interaction.
MATERIALS AND METHODS
Plasmids
The human DDX3X gene, described in Owsianka and Patel (7), was subcloned into pcDNA3.1 (Invitrogen, Carlsbad, CA) using Xho I and EcoR V sites. The HA tag nucleotide sequence was inserted at the 3′ end of the DDX3 gene by PCR. The N-terminal 6XHis-tagged DDX3 was constructed by cloning human DDX3X gene into the protein expression vector pQE30 (Qiagen, Chatsworth, CA) and expressed in Escherichia coli (QIAexpression, Qiagen, Chatsworth, CA). The β-globin reporter construct containing a 3′ HA tag was described (30).
Antibodies
A rabbit polyclonal antibody was raised against an N-terminal peptide [ENALGLDQQFAGLDLNSSDNQS (Genemed Synthesis, Inc., TX)] of human DDX3X. The rabbit polyclonal antibody against UAP56 was described (31). Antibodies to HA tag and eIF3b were from Santa Cruz Biotechnology, Inc., Santa Cruz, CA. Mouse monoclonal antibodies against SAF-B and β-galactosidase were purchased from Upstate USA, Inc., Charlottesville, VA and Abcam, Cambridge, MA, respectively.
RNA interference
HeLa cells were plated overnight on 35 mm dishes with glass coverslip bottoms (MatTek Corp., MA) and transfected with lipofectamine 2000 (Invitrogen) and 50 nM siRNA according to the manufacturer's protocol. The DDX3-a siRNA target sequence is 5′-GAUGCUGGCUCGUGAUUUCUU-3′ (coding region 2025–2045). The DDX3-b mixture targets 5′-GGGAAGGUUUGAUGAUCGUGG-3′ (coding region 1134−1154) and 5′-GAAGCCAGAAAAUUUUCAUAC-3′ (coding region 1708−1728). The siRNA sequences for UAP56/URH49 were described (32). For Drosophila RNA interference studies, a stable S2 cell line was first generated expressing a β-galactosidase reporter gene (pMT/V5-His-LacZ plasmid, Invitrogen). The reporter was cotransfected with a hygromycin-resistant selection vector pCoHYGRO (Invitrogen) at a ratio of 10 : 1 using calcium phosphate. The stable cell line was selected in 100 μg/ml hygromycin. For RNAi, ∼600 bp dsRNAs targeting DDX3, Elp1, Tap or LacZ were transfected into the stable line expressing the reporter using calcium phosphate (33). Two days after transfection, cells were induced by the addition of 0.5 mM CuSO4, and at 18 h postinduction, the cell pellets were lysed and used for western blots, RT–PCR and β-galactosidase activity. For RT–PCR, total RNA was extracted from S2 cells and reverse transcription performed with M-MLV reverse transcriptase (Promega, Madison, WI) using 1 μg of total RNA primed with random hexamers. One-tenth of the reaction was used as template for PCR. β-galactosidase assays were performed as described (34,35).
HeLa cell microinjection, IF and fluorescence in situ hybridization (FISH)
DNA Microinjection and FISH were performed as described (30). For microinjection, 50 ng/μl of the 3 HA-tagged -globin gene was used. For FISH, the 5 end Alexa Fluor 546 labeled β-globin probe was used (cttcatccacgttcaccttgccccacagggcagtaacggcagacttctcctcaggagtcaggtgcaccat).
IF was preformed as described (36). DDX3 (1 : 300), eIF3b (1 : 100) and HA (1 : 100) were used as primary antibodies. The secondary antibodies were rabbit Alexa-488 and mouse Alexa-647 (Invitrogen) diluted to 1 : 1000 with IF solution (10% calf serum in PBS). Images were captured with an EM-CCD camera on an inverted microscope (200M; Zeiss, Thornwood, NY) using Metamorph software (Molecular Devices, Sunnyvale, CA). For Drosophila FISH, an oligo dT probe was used at 10 ng/200 μl and the nuclear envelope visualized using FITC-conjugated wheat germ agglutinin.
LC-MS/MS analysis
LC-MS/MS data was obtained using a LTQ Orbitrap (Thermofisher, San Jose, CA) mass spectrometer. Dried peptides were resuspended in 10 μl of 5% acetonitrile/3% acetic acid and 4 μl were loaded onto a pulled fused silica microcapillary column (125 μm ID, 12 cm bed) packed with C18 reverse phase resin (Magic C18AQ, 5 μm particles; 200 Å pore size; Michrom Bioresources, Auburn, CA). Peptides were resolved using an Agilent 1100 series binary pump across a 30 min linear gradient of 8–25% acetonitrile in 0.2% formic acid at a 250 nl/min flow rate. In each data collection cycle, one full MS scan (375–1600 m/z) was acquired in the Orbitrap (6 × 104 resolution setting; automatic gain control target of 106) followed by 10 data-dependent MS/MS scans in the LTQ (AGC target 5000; threshold 3000) using the 10 most abundant ions for collision induced dissociation for fragmentation. The method dynamically excluded previously selected ions for 30 s, singly charged ions and unassigned charged states.
RESULTS
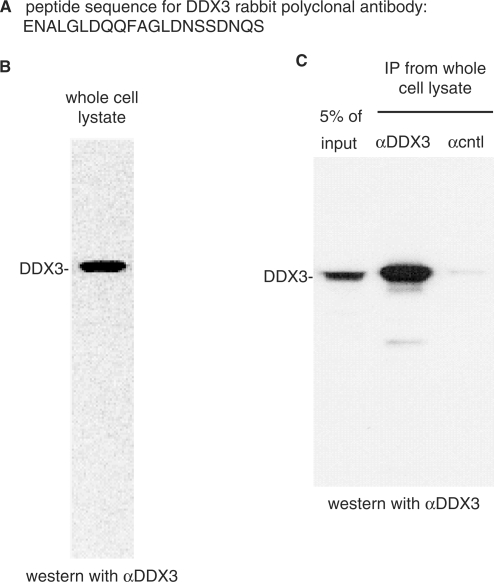
To investigate the function of human DDX3, we raised an antipeptide rabbit polyclonal antibody to a sequence near the N-terminus (Figure 1A). This antibody detected one main band by western blot in a HeLa whole cell lysate (Figure 1B). DDX3 was also specifically immunoprecipitated from whole cell lysate with the DDX3 antibody but not by a negative control antibody (α-SAP 130) (Figure 1C) or other negative control antibodies (data not shown). We conclude that our DDX3 antibody is highly specific for DDX3 and can be used for both western analysis and immunoprecipitations.
Figure 1.
Characterization of human DDX3 antipeptide antibody. (A) The N-terminal amino acid sequence of DDX3 used for raising an antipeptide rabbit polyclonal antibody. (B) Western blot of HeLa whole cell lysate using the DDX3 antibody. (C) Immunoprecipitation from whole cell lysate using α-DDX3 or α-cntl (SAP 130) antibody followed by western analysis with α-DDX3.
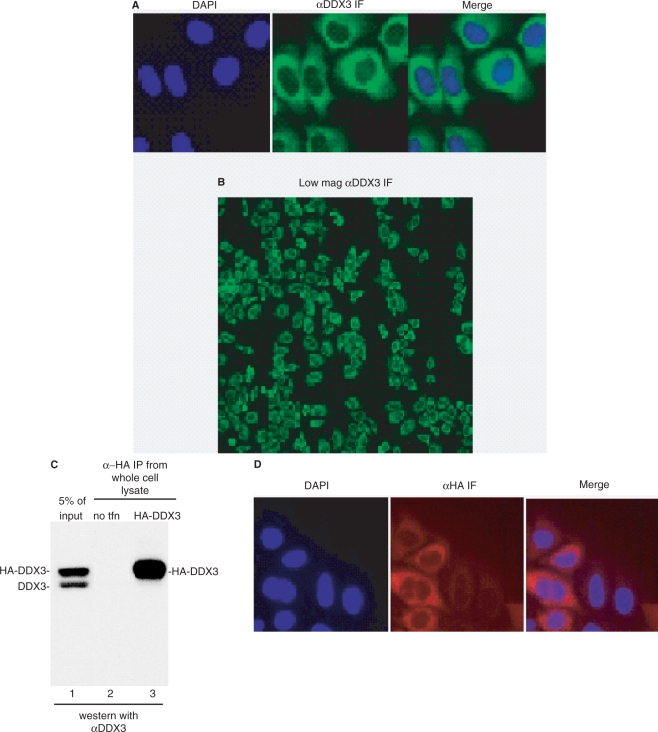
In light of variable reports on the subcellular localization of human DDX3, we next used our antibody for IF of HeLa cells. As shown in Figure 2A, this analysis revealed that DDX3 was evenly distributed throughout the cytoplasm. Moreover, when a field of cells was examined by IF using the DDX3 antibody, the cells all looked similar suggesting that DDX3 localization does not vary during the cell cycle (Figure 2B). To further verify the subcellular localization of DDX3, we expressed HA-tagged DDX3 in HeLa cells and carried out IF using an HA antibody. As shown in Figure 2C (lane 1), when a whole cell lysate of these cells was probed with our DDX3 antibody, HA-tagged DDX3 was present and expressed at a higher level than endogenous DDX3. Immunoprecipitation with an HA-antibody confirmed the identity of HA-DDX3 (Figure 2C, compare lanes 1 and 3). As shown in Figure 2D, IF using the HA-antibody revealed that HA-DDX3 is evenly distributed throughout the cytoplasm. Together, our data indicate that the steady-state localization of DDX3 is cytoplasmic.
Figure 2.
DDX3 is even distributed throughout the cytoplasm using IF. (A) DDX3 was detected in the cytoplasm by IF using our α-DDX3 antibody. (B) DDX3 localization is not cell cycle dependent. IF of HeLa cells using α-DDX3 antibody at lower (×100) magnification is shown. (C) Western analysis of whole cell lysate using α-DDX3 (lane 1). Immunoprecipitation using α-HA antibody from HeLa whole cell lysate (lane 2) or HeLa whole cell lysates expressing HA-DDX3 (lane 3) followed by western analysis with α-DDX3 antibody. (D) HA-DDX3 localized in the cytoplasm. IF of HeLa cells expressing HA-DDX3 was carried out using an HA antibody followed by an Alexa 647-conjugated mouse secondary antibody.
RNA interference of human DDX3
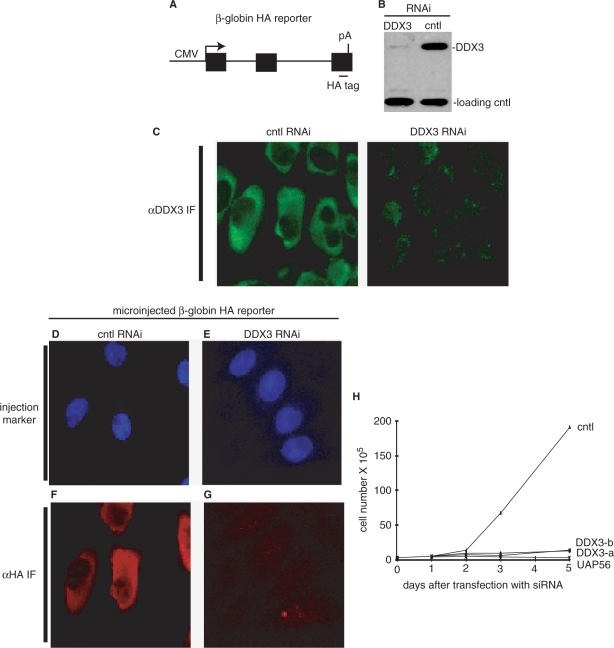
To assay for effects of DDX3 knockdown on gene expression, we employed a reporter construct, which contains the CMV promoter, an HA-tagged β-globin gene (the natural three exons and two introns) and the BGH polyA signal (Figure 3A). In previous work, we microinjected this reporter into the nucleus of HeLa cells and showed that it underwent efficient transcription, pre-mRNA processing and mRNA export (30). In addition, full-length β-globin protein was synthesized from the reporter (unpublished and see below). To prepare DDX3 knockdown cells, an siRNA targeting the coding region from 2025 to 2045 nts was transfected into HeLa cells. Western analysis showed that DDX3 protein levels were efficiently knocked down by this siRNA but not by negative control siRNAs or lipofectamine alone (Figure 3B and data not shown). We then carried out IF of the control or DDX3 knockdown cells using the DDX3 antibody, which confirmed specific knockdown of DDX3 (Figure 3C). To determine whether DDX3 knockdown affects expression of the HA-tagged β-globin reporter, we microinjected it into the nucleus of the DDX3 or control knockdown HeLa cells (Figure 3D−G). A nuclear injection marker (FITC-conjugated dextran) was coinjected to identify the cells containing the HA-tagged β-globin reporter (Figure 3D and E). After microinjection, the cells were incubated for 2.5 h to allow expression of the reporter, and then the levels of β-globin protein were determined by IF using an HA-antibody (Figure 3F and G). Strikingly, this analysis revealed that the levels of β-globin protein were severely diminished in the DDX3 knockdown cells (Figure 3G) but not in the control cells (Figure 3F). The same results were obtained using a mixture of two siRNAs targeting the coding region from 1134 to 1154 nts and from 1708 to 1728 nts (data not shown). Together, our data indicate that DDX3 functions in expression of protein-coding genes. Consistent with an important role for DDX3 in HeLa cells, our data show that the two sets of siRNAs targeting DDX3, but not the negative control, dramatically inhibited cell growth (Figure 3H). These data indicate that DDX3 has an important function in human cells.
Figure 3.
DDX3 RNA interference inhibits cell growth and expression from a β-globin reporter in HeLa cells. (A) Schematic of HA-tagged β-globin reporter. Boxes indicate exons and lines indicate introns. The CMV promoter and BGH polyA sites are shown. (B, C) Efficiency of DDX3 knockdown (using DDX3-a siRNA) in HeLa cells was examined by western analysis (B) and IF (C). Knockdown control (cntl) was lipofectamine alone. Loading control in panel b is UAP56. (D–G) Knockdown of DDX3 inhibits expression of β-globin reporter. Femtoliter aliquots of plasmid DNA (50 μg/ml) containing HA-tagged β-globin was microinjected together with a nuclear injection marker (FITC-conjugated 70 KD-dextran) into the nucleus of siRNA-treated HeLa cells. After incubation at 37°C for 2.5 h, β-globin protein expression was detected by IF using an HA antibody. (H) Knockdown of DDX3 inhibits cell growth. Two sets of siRNAs against DDX3 (DDX3-a, DDX3-b) or lipofectamine alone (cntl) were transfected into HeLa cells. Cell viability was determined using a hemocytometer after staining cells with trypan blue.
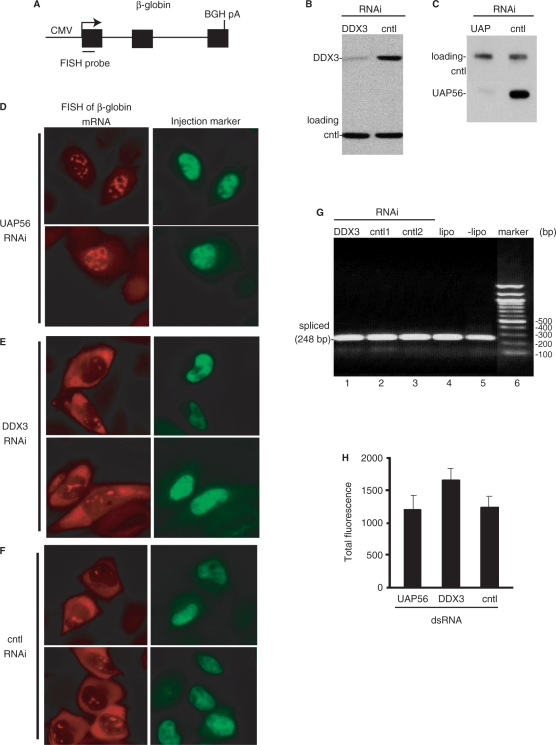
The dramatic decrease in β-globin protein levels in DDX3 knockdown cells could be due to inhibition of nuclear steps of gene expression, such as transcription, pre-mRNA processing, mRNA export or due to cytoplasmic events such as translation. To investigate whether defects in nuclear steps in gene expression could explain the dramatic loss of β-globin protein, we next microinjected the β-globin reporter (Figure 4A) into DDX3 and control knockdown HeLa cell nuclei and used FISH to determine the nucleocytoplasmic distribution of β-globin mRNA and also to quantitate total mRNA levels. The protein UAP56, which functions in mRNA export (32,37), was knocked down as a positive control. Western analysis indicated that both DDX3 (Figure 4B) and UAP56 (Figure 4C) were knocked down efficiently. As shown in Figure 4D, when UAP56 was knocked down, β-globin transcripts accumulated in the nucleus (in nuclear speckle domains, A.D. and R.R. unpublished obsvervation), indicating a block in mRNA export. In contrast, knockdown of DDX3 had no apparent effect on mRNA export, as β-globin mRNA was detected at similar levels in the cytoplasm in both DDX3 knockdown cells and the negative control (Figure 4E and F). These data indicated that the potent effect of DDX3 knockdown on β-globin protein levels (Figure 3) was not due to a defect in mRNA export. Previous work showed that proper splicing and polyadenylation are required for mRNA export (38–40). Thus, our observation that DDX3 knockdown does not affect mRNA export indicates that RNA processing defects are not likely to explain the loss of β-globin protein in the DDX3 knockdown cells. Consistent with this conclusion, knockdown of DDX3 had no effect on splicing. Specifically, RT–PCR using primers that anneal to exons 1 and 2 of the β-globin reporter revealed a band with the expected size for mature mRNA (248 bp) and no band of a size corresponding to unspliced pre-mRNA (379 bp) (Figure 4G, lane 1). The same results were obtained with the negative controls (Figure 4G, lanes 2–5). Finally, using ImageJ (41,42) for quantification of the total levels of FISH signal in the β-globin microinjection assays (Figure 4D−F), we found that the FISH signals were similar in the UAP56, DDX3 and control knockdown cells (Figure 4H), indicating that transcription and/or mRNA stability also could not account for the dramatic decrease in β-globin protein levels in DDX3 knockdown cells. Thus, our data indicate that DDX3 knockdown results in a major decrease in levels of β-globin protein without corresponding defects in nuclear processes in gene expression. Together, these data are consistent with the possibility that translation is a major function of human DDX3. However, as our data on splicing and other nuclear steps in gene expression are negative, we cannot rule out the possibility that DDX3 does play some role(s) in other steps in gene expression steps prior to translation (see Discussion section).
Figure 4.
DDX3 RNA interference has no apparent effects on nuclear steps in gene expression of β-globin reporter. (A) Schematic of β-globin reporter construct. The FISH probe is indicated (see Methods section). (B, C) The RNAi efficiencies after knockdown of DDX3 (B) or UAP56 (C) in HeLa cells were analyzed by western blotting. Loading controls were eIF4A3 (B) and CBP80 (C). Knockdown control (cntl) in panels a and b was lipofectamine alone. (D−F) The β-globin reporter construct was microinjected into the nucleus of the knockdown cells together with FITC-conjugated 70 KD-dextran as an injection marker. Thirty minutes after microinjection, α-amanitin was added to inhibit further transcription. Cells were fixed after 2 h incubation at 37°C and the distribution of β-globin mRNA was visualized by FISH using an Alexa Fluor 546 labeled probe. (G) Knockdown of DDX3 has no apparent effect on pre-mRNA splicing of β-globin reporter. After knockdown using siRNAs targeting DDX3 (lane 1), cntl1 (eIF4A3, lane 2), cntl2 (Skar, lane 3) or using lipofectamine alone (lane 4) or untreated cells (lane 5), RT–PCR was carried out on total RNA using PCR primers that flank intron 1 of the β-globin reporter. A band of the expected size for spliced mRNA (248 bp) is indicated. Marker sizes (base pair) are shown to the right of the gel. (H) Quantification of the total levels of FISH signal for β-globin mRNA was carried out using NIH ImageJ and is shown in the graph. Error bars represent standard deviations (n = 11).
RNA interference of Drosophila DDX3
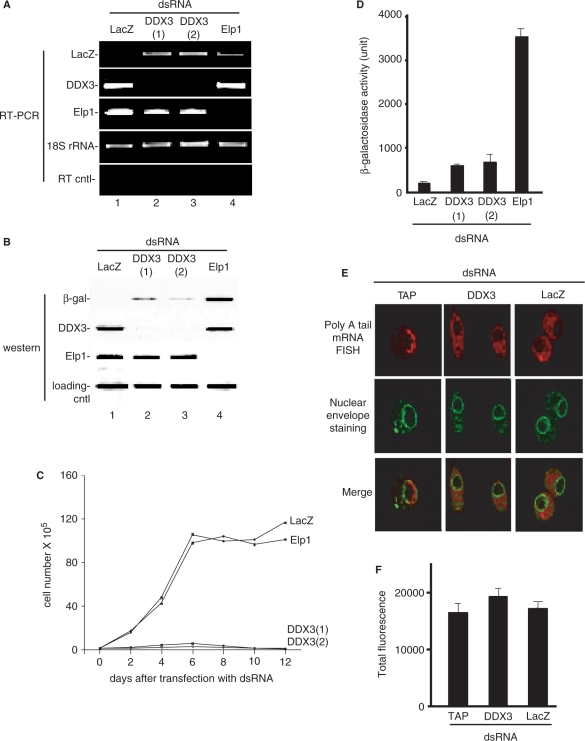
To further investigate the function of DDX3 in metazoans, we carried out RNAi of DDX3 (Belle) in Drosophila S2 cells. To do this, we first generated a stable cell line containing a copper-inducible lacZ reporter gene. RNAi of this cell line was then carried out using siRNAs against the LacZ reporter itself, DDX3 (two different siRNAs) or a negative control protein (Elp1). RT–PCR of LacZ (lane 1), DDX3 (lanes 2 and 3) and Elp1 (lane 4) revealed that the knockdowns were efficient (Figure 5A). Western analysis (Figure 5B), confirmed that β-gal (lane 1), DDX3 (lanes 2 and 3) and the negative control Elp1 (lane 4) proteins were all efficiently knocked down. We next examined cell viability over a 12-day time course after the knockdowns (Figure 5C). Significantly, the two dsRNAs targeting DDX3, but not those targeting LacZ or Elp1, dramatically inhibited cell growth. These data indicate that DDX3 is required for viability in Drosophila cells.
Figure 5.
Evidence that Drosophila DDX3 (Belle) functions in translation in S2 cells. (A, B) RT–PCR (A) and western analysis (B) of knockdown of LacZ, DDX3 and Elp1 in a stable S2 cell line containing a copper-inducible lacZ gene. 18S rRNA was used as a loading control, and RT cntl (no reverse transcription was carried out) was used as a control for RT–PCR. A loading control protein (Skar) was used for the western blot analysis. (C) Knockdown of DDX3 inhibits cell growth. Cells were transfected with the indicated dsRNAs and then cell numbers were counted each day using a hemocytometer after staining with trypan blue. (D) Protein expression levels were determined by β-galactosidase activity. Error bars represent standard deviations (n = 3). (E) FISH of mRNA export in the indicated knockdown cells using an oligo dT probe for polyA tail mRNA and FITC-conjugated wheat germ agglutinin to stain the nuclear envelope. (F) Quantification of the total levels of FISH signal for polyA tail mRNA was carried out using NIH ImageJ and is shown in the graph. Error bars represent standard deviations (n = 18).
To further investigate the cellular function of Drosophila DDX3, CuSO4 was added to the culture medium to induce LacZ reporter expression. After an 18-h induction, western analysis showed that the levels of β-gal protein were strongly reduced not only in the LacZ knockdown cells (Figure 5B, lane 1), but also in the DDX3 knockdown cells (Figure 5B, lanes 2 and 3). In contrast, the levels of β-gal were unaffected in the negative control (Elp1) knockdown cells (Figure 5B, lane 4). Consistent with this western result, β-gal enzyme activity was also decreased in the DDX3 knockdown cells but not in the negative control knockdown cells (Figure 5D). Together, these data indicate that Drosophila DDX3 functions in expression of protein-coding genes.
As observed for human DDX3 (Figure 4), we observed no effects of Drosophila DDX3 knockdown on nuclear steps in gene expression. For example, mRNA export was not affected by DDX3 knockdown, but was blocked by knockdown of the mRNA export receptor TAP (Figure 5E). Likewise, mRNA levels were not affected in DDX3 knockdown cells relative to control knockdowns, as determined by quantitating the FISH signals (Figure 5F). Finally, in DDX3 knockdown cells, only spliced Elp1 mRNA, and not unspliced pre-mRNA, was detected by RT–PCR, indicating that splicing does not explain the major loss of LacZ reporter protein observed after DDX3 knockdown (Figure 5A; compare lanes 1–3; note that the PCR primers used for Elp1 can detect both spliced and unspliced mRNA). These data are consistent with the conclusion that Drosophila DDX3 has a major general role in protein translation but not in nuclear steps in gene expression. We conclude that the function of DDX3 in promoting translation is conserved from yeast (1) to higher eukaryotes.
DDX3 interacts with eIF3
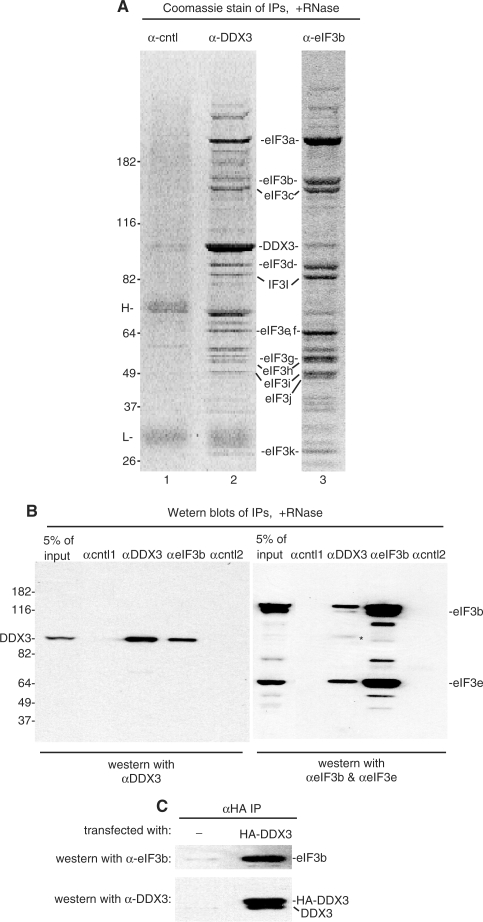
To gain additional insight into the function of DDX3, we next used our DDX3 antibody to carry out immunoprecipitations from total RNase-treated HeLa cytoplasmic extract. When the immunoprecipitate was analyzed on a Coomassie-stained gel, a specific and highly reproducible set of proteins was detected (Figure 6A, lane 2). These proteins were not detected in the negative control immunoprecipitate (α-cntl which is against the protein SAP 130) (Figure 6A, lane 1) or in any other negative control immunoprecipitates (data not shown). To identify the proteins, individual bands were analyzed by mass spectrometry. Strikingly, these data showed that DDX3 was immunoprecipitated together with at least 10 of the 13 subunits of the translation initiation factor, eIF3 (subunit j, l and m were not detected). We then carried out a reciprocal immunoprecipitation using an antibody to eIF3b. Significantly, DDX3, as well as the eIF3 subunits, were detected in this immunoprecipitate (Figure 6A, lane 3). To further verify the specificity of the DDX3-eIF3 interaction, we carried out immunoprecipitations from RNase-treated cytoplasmic extract followed by western analysis using α-DDX3, α-eIF3b and two negative control antibodies (cntl1: α-SAP 130, cntl2: α-SAF-B) (Figure 6B). These data showed that DDX3 was only present in the α-DDX3 and α-eIF3b immunoprecipitates. Likewise, eIF3 (subunits b and e) was specifically detected in the α-DDX3 and α-eIF3b immunoprecipitates but not in the negative controls (Figure 6B). Finally, when HA-DDX3 was expressed in HeLa cells followed by an immunoprecipitation using an HA antibody, western analysis revealed that eIF3b was present in the HA-DDX3 immunoprecipitate, indicating that HA-DDX3 and eIF3 also specifically interact (Figure 6C). Together, our data indicate that eIF3 is the major interaction partner of DDX3 and that these factors associate with each other via protein–protein interactions. Whether this interaction is direct or is mediated via another protein remains to be determined.
Figure 6.
DDX3 associates with the translation initiation factor, eIF3, via protein–protein interactions. (A) Immunoprecipitations were carried out using antibodies against DDX3, eIF3b or a negative control antibody (SAP 130) from HeLa cytoplasmic extract pretreated with RNase A. (B) Western blots of the indicated immunoprecipitations were first probed with α-DDX3 (left panel) followed by α-eIF3b, and finally α-eIF3e. The right panel shows the blot after probing with both α-eIF3b and α-eIF3e. The band designated by the asterisk in the right panel is remaining DDX3 signal.
DISCUSSION
In this study, we have investigated the function of human DDX3, an RNA helicase that was originally identified as a potential cellular target of HCV (7−9) and more recently as a cofactor for HIV viral RNA export (12). Our studies show that RNAi of DDX3 in HeLa cells results in a major decrease in the levels of protein expressed from a β-globin reporter construct. The same results were obtained after RNAi of DDX3 (Belle) in Drosophila cells. In contrast, we detected no apparent defects in nuclear steps in gene expression in either HeLa or Drosophila cells. Our data revealed that DDX3 is predominantly cytoplasmic at steady state. In addition, we observed a striking association between DDX3 and the multi-subunit translation initiation factor eIF3. Together, the subcellular localization of DDX3, the DDX3-eIF3 interaction data and the RNAi data all support the conclusion that DDX3 functions to promote translation, and this function is conserved from yeast (1) to humans. In a previous study, DDX3 was reported to be a translational repressor, and this effect is thought to be mediated by a direct interaction between DDX3 and eIF4E (27). We did not obtain evidence for a role of DDX3 in translation repression in our studies. Although the basis for this discrepancy is not clear, it may be due to reporter differences or cell type specific differences in DDX3 regulation, as has been observed in recent studies (4,27). In this regard, we note that our RNAi studies were carried out in HeLa cells whereas Shih et al. (27) used HuH7 cells. It is possible that DDX3 has dual roles in translation initiation and translation inhibition depending on the mRNA, the cell type or other cellular conditions. Finally, in the previous work (27), translation repression was observed after over-expression of DDX3. However, it may be difficult to interpret results from over-expressed proteins, possibly even more so with RNA helicase proteins, such as DDX3, which may have adverse effects in the cell when present in large amounts. We also did not detect an interaction between DDX3 and eIF4E in HeLa extracts using coimmunoprecipitation/western analysis (data not shown). It is possible that this interaction is much less abundant that the DDX3-eIF3 interaction, which is readily detected by Coomassie stain as well as by western.
Although our studies indicate that DDX3 functions to promote translation, previous work (1,2) indicates that DDX3 has features that are not typical of well-known translation initiation factors. In particular, DDX3 is localized in the cytoplasm as expected for a translation factor, but upon treatment of cells with leptomycin B, this protein enters the nucleus, indicating that DDX3 is a shuttling protein (12). Consistent with this observation, Daneholt and colleagues found that DDX3 associates with Balbiani Ring mRNPs cotranscriptionally in the nucleus and also with polysomes in the cytoplasm (26). DDX3 has also been detected in purified mammalian spliceosomal complexes assembled in nuclear extracts (43–45). These data have led to a model in which DDX3 is loaded onto mRNA in the nucleus and then functions in translation in the cytoplasm (26). In yeast, both biochemical and genetic evidence suggest that Ded1 plays a role, not only in translation, but also in splicing (22,44,46). In our studies, we were unable to clearly detect a specific association of DDX3 with spliceosomes by western analysis (C.-S.L. and R.R., unpublished observation). Because DDX3 is an RNA helicase, it is possible that this protein associates weakly or only transiently with spliceosomes and thus is difficult to detect by western. We were also unable to detect a role for DDX3 in splicing or other nuclear steps in gene expression in DDX3 knockdown cells. However, from these negative data, we cannot rule out the possibility that DDX3 does play some role in these steps. For example, it is possible that DDX3 functions in splicing of only certain mRNAs, such as localized mRNAs. Indeed, DDX3 is a component of germinal granules and neuronal transport granules, which function in localized mRNP translation (13,14). Thus, DDX3 may be loaded onto these localized mRNAs in the nucleus and then function in localized translation in the cytoplasm. Precedent for nuclear loading of proteins that ultimately function in the cytoplasm is exemplified by the exon junction complex (EJC), in which a specific set of proteins is loaded onto mRNA during splicing and then functions in NMD or localized translation in the cytoplasm (47–52). In addition, splicing enhances translation (53,54) and this enhancement is thought to be mediated by the EJC (55–59). Further studies are needed to determine whether DDX3 not only has a general role in translation but also has a specific role for some mRNAs in localized mRNA translation and/or in coupling splicing to translation.
ACKNOWLEDGEMENTS
We are grateful to H. Cheng and H. Lei for comments on the manuscript and to lab members for useful discussions. HeLa cells were obtained from the National Cell Culture Center (Minneapolis, MN). This work was supported by an NIH grant to R. R. Funding to pay the Open Access publication charges for this article was provided by NIH, National Institute of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Linder P. Yeast RNA helicases of the DEAD-box family involved in translation initiation. Biol. Cell. 2003;95:157–167. doi: 10.1016/s0248-4900(03)00032-7. [DOI] [PubMed] [Google Scholar]
- 2.Rosner A, Rinkevich B. The DDX3 subfamily of the DEAD box helicases: divergent roles as unveiled by studying different organisms and in vitro assays. Curr. Med. Chem. 2007;14:2517–2525. doi: 10.2174/092986707782023677. [DOI] [PubMed] [Google Scholar]
- 3.Hogbom M, Collins R, van den Berg S, Jenvert RM, Karlberg T, Kotenyova T, Flores A, Karlsson Hedestam GB, Schiavone LH. Crystal structure of conserved domains 1 and 2 of the human DEAD-box helicase DDX3X in complex with the mononucleotide AMP. J. Mol. Biol. 2007;372:150–159. doi: 10.1016/j.jmb.2007.06.050. [DOI] [PubMed] [Google Scholar]
- 4.Botlagunta M, Vesuna F, Mironchik Y, Raman A, Lisok A, Winnard P., Jr, Mukadam S, Van Diest P, Chen JH, Farabaugh P, et al. Oncogenic role of DDX3 in breast cancer biogenesis. Oncogene. 2008;27:3912–3922. doi: 10.1038/onc.2008.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang PC, Chi CW, Chau GY, Li FY, Tsai YH, Wu JC, Wu Lee YH. DDX3, a DEAD box RNA helicase, is deregulated in hepatitis virus-associated hepatocellular carcinoma and is involved in cell growth control. Oncogene. 2006;25:1991–2003. doi: 10.1038/sj.onc.1209239. [DOI] [PubMed] [Google Scholar]
- 6.Chao CH, Chen CM, Cheng PL, Shih JW, Tsou AP, Lee YH. DDX3, a DEAD box RNA helicase with tumor growth-suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer Res. 2006;66:6579–6588. doi: 10.1158/0008-5472.CAN-05-2415. [DOI] [PubMed] [Google Scholar]
- 7.Owsianka AM, Patel AH. Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology. 1999;257:330–340. doi: 10.1006/viro.1999.9659. [DOI] [PubMed] [Google Scholar]
- 8.Mamiya N, Worman HJ. Hepatitis C virus core protein binds to a DEAD box RNA helicase. J. Biol. Chem. 1999;274:15751–15756. doi: 10.1074/jbc.274.22.15751. [DOI] [PubMed] [Google Scholar]
- 9.You LR, Chen CM, Yeh TS, Tsai TY, Mai RT, Lin CH, Lee YH. Hepatitis C virus core protein interacts with cellular putative RNA helicase. J. Virol. 1999;73:2841–2853. doi: 10.1128/jvi.73.4.2841-2853.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ariumi Y, Kuroki M, Abe K, Dansako H, Ikeda M, Wakita T, Kato N. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J. Virol. 2007;81:13922–13926. doi: 10.1128/JVI.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl Acad. Sci. USA. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 13.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 14.Johnstone O, Deuring R, Bock R, Linder P, Fuller MT, Lasko P. Belle is a Drosophila DEAD-box protein required for viability and in the germ line. Dev. Biol. 2005;277:92–101. doi: 10.1016/j.ydbio.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 15.Beckham C, Hilliker A, Cziko AM, Noueiry A, Ramaswami M, Parker R. The DEAD-Box RNA Helicase Ded1p Affects and Accumulates in Saccharomyces cerevisiae P-Bodies. Mol. Biol. Cell. 2008;19:984–993. doi: 10.1091/mbc.E07-09-0954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gururajan R, Weeks DL. An3 protein encoded by a localized maternal mRNA in Xenopus laevis is an ATPase with substrate-specific RNA helicase activity. Biochim. Biophys. Acta. 1997;1350:169–182. doi: 10.1016/s0167-4781(96)00155-8. [DOI] [PubMed] [Google Scholar]
- 17.Iost I, Dreyfus M, Linder P. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J. Biol. Chem. 1999;274:17677–17683. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- 18.Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 19.Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz AM. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J. Mol. Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chuang RY, Weaver PL, Liu Z, Chang TH. Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science. 1997;275:1468–1471. doi: 10.1126/science.275.5305.1468. [DOI] [PubMed] [Google Scholar]
- 21.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burckin T, Nagel R, Mandel-Gutfreund Y, Shiue L, Clark TA, Chong JL, Chang TH, Squazzo S, Hartzog G, Ares M., Jr Exploring functional relationships between components of the gene expression machinery. Nat. Struct. Mol. Biol. 2005;12:175–182. doi: 10.1038/nsmb891. [DOI] [PubMed] [Google Scholar]
- 23.de la Cruz J, Iost I, Kressler D, Linder P. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1997;94:5201–5206. doi: 10.1073/pnas.94.10.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berthelot K, Muldoon M, Rajkowitsch L, Hughes J, McCarthy JE. Dynamics and processivity of 40S ribosome scanning on mRNA in yeast. Mol. Microbiol. 2004;51:987–1001. doi: 10.1046/j.1365-2958.2003.03898.x. [DOI] [PubMed] [Google Scholar]
- 25.Marsden S, Nardelli M, Linder P, McCarthy JE. Unwinding single RNA molecules using helicases involved in eukaryotic translation initiation. J. Mol. Biol. 2006;361:327–335. doi: 10.1016/j.jmb.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 26.Nashchekin D, Zhao J, Visa N, Daneholt B. A novel Ded1-like RNA helicase interacts with the Y-box protein ctYB-1 in nuclear mRNP particles and in polysomes. J. Biol. Chem. 2006;281:14263–14272. doi: 10.1074/jbc.M600262200. [DOI] [PubMed] [Google Scholar]
- 27.Shih JW, Tsai TY, Chao CH, Wu Lee YH. Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene. 2008;27:700–714. doi: 10.1038/sj.onc.1210687. [DOI] [PubMed] [Google Scholar]
- 28.Askjaer P, Bachi A, Wilm M, Bischoff FR, Weeks DL, Ogniewski V, Ohno M, Niehrs C, Kjems J, Mattaj IW, et al. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol. Cell Biol. 1999;19:6276–6285. doi: 10.1128/mcb.19.9.6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Askjaer P, Rosendahl R, Kjems J. Nuclear export of the DEAD box An3 protein by CRM1 is coupled to An3 helicase activity. J. Biol. Chem. 2000;275:11561–11568. doi: 10.1074/jbc.275.16.11561. [DOI] [PubMed] [Google Scholar]
- 30.Valencia P, Dias AP, Reed R. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl Acad. Sci. USA. 2008;105:3386–3391. doi: 10.1073/pnas.0800250105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapadia F, Pryor A, Chang TH, Johnson LF. Nuclear localization of poly(A)+ mRNA following siRNA reduction of expression of the mammalian RNA helicases UAP56 and URH49. Gene. 2006;384:37–44. doi: 10.1016/j.gene.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 33.Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl Acad. Sci. USA. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Errington J, Mandelstam J. Use of a lacZ gene fusion to determine the dependence pattern of sporulation operon spoIIA in spo mutants of Bacillus subtilis. J. Gen. Microbiol. 1986;132:2967–2976. doi: 10.1099/00221287-132-11-2967. [DOI] [PubMed] [Google Scholar]
- 35.Lee CS, Clarkson J, Shu JC, Campbell ID, Yudkin MD. Bacillus subtilis mutations that alter the pathway of phosphorylation of the anti-anti-sigmaF factor SpoIIAA lead to a Spo- phenotype. Mol. Microbiol. 2001;40:9–19. doi: 10.1046/j.1365-2958.2001.02353.x. [DOI] [PubMed] [Google Scholar]
- 36.Palazzo AF, Springer M, Shibata Y, Lee CS, Dias AP, Rapoport TA. The signal sequence coding region promotes nuclear export of mRNA. PLoS Biol. 2007;5:e322. doi: 10.1371/journal.pbio.0050322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gatfield D, Le Hir H, Schmitt C, Braun IC, Kocher T, Wilm M, Izaurralde E. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 2001;11:1716–1721. doi: 10.1016/s0960-9822(01)00532-2. [DOI] [PubMed] [Google Scholar]
- 38.Bird G, Fong N, Gatlin JC, Farabaugh S, Bentley DL. Ribozyme cleavage reveals connections between mRNA release from the site of transcription and pre-mRNA processing. Mol. Cell. 2005;20:747–758. doi: 10.1016/j.molcel.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 39.Cullen BR. Connections between the processing and nuclear export of mRNA: evidence for an export license? Proc. Natl Acad. Sci. USA. 2000;97:4–6. doi: 10.1073/pnas.97.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharp PA. RNA splicing and genes. Jama. 1988;260:3035–3041. [PubMed] [Google Scholar]
- 41.Girish V, Vijayalakshmi A. Affordable image analysis using NIH Image/ImageJ. Indian J. Cancer. 2004;41:47. [PubMed] [Google Scholar]
- 42.Collins TJ. ImageJ for microscopy. Biotechniques. 2007;43:25–30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
- 43.Merz C, Urlaub H, Will CL, Luhrmann R. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA. 2007;13:116–128. doi: 10.1261/rna.336807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevens SW, Ryan DE, Ge HY, Moore RE, Young MK, Lee TD, Abelson J. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- 45.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 46.Jamieson DJ, Rahe B, Pringle J, Beggs JD. A suppressor of a yeast splicing mutation (prp8-1) encodes a putative ATP-dependent RNA helicase. Nature. 1991;349:715–717. doi: 10.1038/349715a0. [DOI] [PubMed] [Google Scholar]
- 47.Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 48.Giorgi C, Moore MJ. The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin. Cell Dev. Biol. 2007;18:186–193. doi: 10.1016/j.semcdb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 49.Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- 50.Ferraiuolo MA, Lee CS, Ler LW, Hsu JL, Costa-Mattioli M, Luo MJ, Reed R, Sonenberg N. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc. Natl Acad. Sci. USA. 2004;101:4118–4123. doi: 10.1073/pnas.0400933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shibuya T, Tange TO, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat. Struct. Mol. Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- 52.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Braddock M, Muckenthaler M, White MR, Thorburn AM, Sommerville J, Kingsman AJ, Kingsman SM. Intron-less RNA injected into the nucleus of Xenopus oocytes accesses a regulated translation control pathway. Nucleic Acids Res. 1994;22:5255–5264. doi: 10.1093/nar/22.24.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsumoto K, Wassarman KM, Wolffe AP. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 1998;17:2107–2121. doi: 10.1093/emboj/17.7.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Le Hir H, Seraphin B. EJCs at the heart of translational control. Cell. 2008;133:213–216. doi: 10.1016/j.cell.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 56.Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133:303–313. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 57.Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiegand HL, Lu S, Cullen BR. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc. Natl Acad. Sci. USA. 2003;100:11327–11332. doi: 10.1073/pnas.1934877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gudikote JP, Imam JS, Garcia RF, Wilkinson MF. RNA splicing promotes translation and RNA surveillance. Nat. Struct. Mol. Biol. 2005;12:801–809. doi: 10.1038/nsmb980. [DOI] [PubMed] [Google Scholar]