Abstract
We describe the use of ATP caged with [7-(diethylamino)coumarin-4-yl]methyl (DEACM) for light-controlled in vitro transcription reactions. Polymerization is blocked when DEACM is bonded to the gamma phosphate group of the ATP molecule. Controlled light irradiation releases ATP and transcription is initiated. In order to provide full control over the process, conditions involved in substrate release, nucleotide availability after release and the effect of the released coumarin in RNA polymerization were assessed in further detail. Together, our data provide the first direct evidence of control over enzymatic polymerization of nucleic acids through light. This approach may provide researchers with a unique tool for the study of biological processes at a molecular level.
INTRODUCTION
Controlled temporal and spatial release of biomolecules from photolabile precursors, commonly known as caged molecules, is increasingly becoming an important tool in biology studies. The rationale of using caged compounds is straightforward: the molecule of interest is rendered biologically inactive (caged) by chemical modification with a protecting group that can be removed with light by irradiation of a suitable wavelength (1,2). This promotes the release of the biologically active molecule, generating a time-controlled burst in concentration with tight spatial control (3).
The caged molecule must fulfill several critical conditions: (i) the resulting byproducts (after photocleavage), when used at the chosen concentration, should not facilitate or hamper the reaction; (ii) the rate of uncaging must be faster than the process under study, i.e. the rate at which the biomolecule is released should not be the rate limiting step and (iii) the efficiency of uncaging should be high in order to avoid long irradiation times and deleterious effects to the biological samples. Till now, the choice of caging agents released by light is still restricted to a small number of molecules. Most are UV absorbing molecules with low photocleavage efficiencies and small extinction coefficients, e.g. 2-nitrobenzyl (4,5) and nitrophenylalkyl derivatives (6). The hydroxyphenancyl derivatives show enhanced photochemical properties but also absorb in the UV region (7). Quinoline (8,9) and coumarin (10–12) derivatives present higher extinction coefficients, high photochemical quantum yields and absorb in the visible region (<420 nm). Coumarin derivatives also have the advantage of fast photocleavage kinetics and wavelength tunability through changes in the position and/or nature of the chemical residues attached to the coumarin moiety (3,13). Despite the advantages over the UV absorbing groups, these quinoline or coumarin-based compounds have generally low water solubility. Strategies to cage virtually any constituent or biomolecule have been proposed [for a review see refs (14,15) and references therein]. In particular, caging of nucleotides—ATP and cAMP—has been applied in living cells for physiologic response studies, where the unaltered nucleotide is released by UV or visible irradiation (11,16).
Nucleotides, the natural substrates of DNA and RNA polymerases, appear like a straightforward application of the caging technique to DNA and/or RNA enzymatic synthesis. Utilization of caged nucleotides in nucleic acid polymerization reactions may provide new tools in molecular biology studies. However, thus far, the use of caged nucleotides for controlled polymerization of nucleic acids has not been reported.
Here, we describe the use of ATP caged with [7-(diethylamino)coumarin-4-yl]methyl (DEACM) for light-controlled in vitro transcription reactions (Figure 1). Polymerization is blocked when DEACM is bonded to the gamma phosphate group of the ATP molecule. Controlled light irradiation releases ATP and transcription can resume. DEACM release from the active molecule through visible light irradiation prevents UV-associated nucleic acid and protein damage. In order to provide full control over the process, conditions involved in substrate release, nucleotide availability after release and the effect of the released coumarin in RNA polymerization were assessed in further detail.
Figure 1.
Photolysis of DEACM-ATP and ATP release. (A) Structure of DEACM-ATP and respective photoproducts. After irradiation with 390 nm light, the ester bond between DEACM and ATP is cleaved, generating DEACM-OH and free ATP. (B) Representation of the light-controlled transcription process. ATP (gray circle) is not available as substrate for transcription due to efficient caging by DEACM (black ellipse). Irradiation with visible light releases ATP (a) allowing transcription to be resumed (b), yielding full-length RNA products. RNA polymerase is represented by a gray pacman.
MATERIALS AND METHODS
Materials
All chemicals were purchased from Sigma-Aldrich, St. Louis, MO, USA in the highest purity available and used without further purification. T7 RNA Polymerase and Revert-Aid™ M-MuLV Reverse Transcriptase were purchased from Fermentas, Vilnius, Lithuania. DNase I and SYBR® GreenER Real-Time PCR Kit purchased from Invitrogen, Karlsbad, CA, USA. All oligonucleotides were purchased from STAB Vida, Portugal.
DEACM-ATP synthesis and purification
P3-[7-(dimethylamino)coumarin-4-yl]methyl adenosine 5′-triphosphate trisodium salt (DEACM-ATP) was synthesized as described by Geiβler et al., method B (11).
A Hitachi-Merck HPLC L6200A Pump with an L-4500 Diode Array Detector using a Polystirene-Divinylbenzene (PLRP-S, Polymer Labs, Darmstadt, Germany) column—analytical column: 4.6 mm × 15 mm, 8 µm, 300 Å; semi-preparative column: 7.4 mm × 15 mm, 8 µm, 300 Å, was employed for separation and purification of DCEAM-ATP. Eluent A was tributylammonium acetate buffer in water, 5 mM, pH 6.9; eluent B was methanol. Semi-preparative gradient started with 20 min at 30% of B in A; with an increase to 100% B after 21 min, and finished after 26 min at 100% of B. Separations were run at a flow rate of 3 ml/min and the column temperature was 35°C. After peak separation and collection, samples were lyophilized, resuspended in water and stored in the dark at −20°C. A purity of >95% was determined by HPLC. All solutions were protected from light and DEACM-ATP manipulations were made in a dark chamber under red-light illumination.
Photochemical and photophysical studies
All spectroscopic measurements and irradiations were performed in a 60 µl quartz fluorescence cuvette (0.3 cm optical path) at 21°C. Absorption spectra were recorded on a Shimadzu UV-2501PC spectrophotometer. Fluorescence measurements of aerated solutions were performed on a Horiba-Jobin-Yvon SPEX Fluorolog 3.22 spectrofluorimeter. All spectra collected with 1.5 nm slit bandwidth for excitation and emission, with correction files.
Fluorescence quantum yields (Φf) were determined by the relative method versus coumarin 1 (7-diethylamino-4-methylcoumarin) degassed solution in ethanol (Φf = 0.730) (17). The optical densities of DEACM-ATP, 7-diethylamino-4-hydroxymethylcoumarin (DEACM-OH) solutions in water at pH 7.0 and that of the standard were adjusted to identical values (0.08–0.12) at excitation wavelength (386 nm). Correction for the refractive index was included in the calculation.
DEACM-ATP photochemical quantum yields (ΦChem), defined as the number of DEACM-OH molecules formed by each photon absorbed by DEACM-ATP, were carried out in water at pH 7.0. Sequential irradiation times of DEACM-ATP solution (between 49 and 450 µM) were carried out and the resulting DEACM-OH determined by HPLC using a Polystirene-Divinylbenzene (PLRP-S, Polymer Labs, Darmstadt, Germany) column (4.6 mm × 15 mm, 8 µm, 300 Å). Eluent A was tributylammonium acetate buffer in water, 5 mM, pH 6.9; eluent B was methanol. Eluent gradient started with 2 min at 30% of B in A, with an increase to 90% B after 2.5 min, and finished after 10 min at 90% of B. Separations were run at a flow rate of 2 ml/min, at 35°C column temperature. Product separation was monitored at 380 nm and DEACM-OH (retention time = 6.32 min) concentration determined by peak area quantification. DEACM-OH irradiations were carried out in a Horiba-Jobin-Yvon Spex Fluorolog 1681 Spectrometer with a 450 W xenon arc lamp, monochromated for the excitation wavelength (390 nm, 18 nm slit bandwidth). The actinometry of the irradiation setup was performed with the concentrated potassium ferrioxalate actinometer (18) and an intensity of 1.36 × 10−8 Einstein/min was measured. Photochemical quantum yields were calculated as the slope of the linear regression obtained by plotting DEACM-OH moles formed (ΔnDEACM-OH) as a function of irradiation time (Δt), according to Equation (1).
The fraction of light absorbed by the solution (1 − 10−AT) was calculated from the absorption spectra. The fraction of that light which was absorbed by DEACM-ATP (ADEACM-ATP/AT) was derived from DEACM-OH and DEACM-ATP concentrations as determined by HPLC and their respective extinction coefficients.
Transcription template cloning and purification
A 110-bp fragment of Exon 7 of the human p53 gene (TP53 tumor protein p53 [Homo sapiens], GenBank accession no. X54156) was PCR amplified using primers E7p53Fw: 5′-gttggctctgactgtaccac-3′ and E7p53Rev: 5′-ctggagtcttccagtgtgatg-3′. PCR amplification was carried out in 20 µl using 0.5 µM of primers, 0.2 mM dNTPs with 0.5 U Taq DNA polymerase (Amersham Biosciences, GE Healthcare, Europe, GmbH) on a Tpersonal Thermocycler (Whatman Biometra, Germany). Following denaturation at 95°C for 5 min, amplification was performed for 25 cycles, each cycle consisting of 95°C for 30 s, annealing at 54°C for 30 s, extension at 72°C for 30 s, with a final extension step at 72°C for 5 min. The resulting product was re-amplified using a T7 promoter-E7p53Fw primer (5′-taatacgactcactatagggagagttggctctgactgtaccac-3′) and subsequently cloned in pJET1.2 (CloneJET™ PCR Cloning Kit, Fermentas, Vilnius, Lithuania) according to manufacturer's protocol. The resulting 133-bp fragment was PCR amplified using primers E7p53Rev and T7 primer (5′-taatacgactcactatagggaga-3′) as described above, purified through SureClean® purification kit (Bioline, London, UK) and used as template for in vitro transcription reactions. The cloned fragment was confirmed by direct sequencing using ABI Prism 3100 and ABI Prism Big Dye technology (Applied Biosystems, Foster, CA, USA).
In vitro transcription
Standard in vitro transcription using 100 ng of template was performed with 20 U of T7 RNA Polymerase (Fermentas, Vilnius, Lithuania) according to the manufacturer's protocol. Reactions were incubated 1 h at 37°C, followed by heat inactivation of enzyme for 15 min at 75°C. DNA template digestion, ensuring absence of DNA after transcription, was performed with 27 U of DNase I (Invitrogen, Karlsbad, CA, USA) for 1 h at 37°C, which was subsequently heat inactivated for 15 min at 75°C. In assays involving caged-ATP, ATP was substituted by the equivalent amount of DEACM-ATP. Products were analyzed on 3% agarose gel electrophoresis in 1 × TBE buffer. The in vitro transcription reactions with radiolabeled α-32P-UTP (Perkin Elmer, 800 Ci/ml, 10 mCi/ml), followed the same procedure as described earlier, to which 1.5 µl of α-32P-UTP were added and reactions incubated 1 h at 37°C. The resulting products were denaturated for 2 min at 75°C and analyzed on a 6% polyacrylamide/8M urea gel electrophoresis in 1 × TBE. Denaturing PAGE was run at 70 W for 2 h. Results were visualized in an Optical Scanner Storm™ 860 Instrument (Molecular Dynamics, GE Healthcare, Europe, GmbH).
Reverse transcription (RT) and real-time PCR reaction
RT was performed with Revert-Aid™ M-MuLV Reverse Transcriptase (Fermentas, Vilnius, Lithuania) according to manufacturer's specifications, using 0.25 µM of E7p53Rev primer, annealing at 42°C for 1 h.
Real-time PCR assays were performed in a Corbett Research Rotor-Gene RG3000 using SYBR® GreenER Real-Time PCR kit (Invitrogen, Karlsbad, CA, USA) according to manufacturer's specifications. Reactions were performed in a total volume of 25 µl with 0.5 µM of primers E7p53Fw and E7p53Rev. Following a preincubation at 50°C for 2 min and denaturation at 95°C for 10 min, Real-time PCR was performed for 40 cycles, each cycle consisting of 95°C for 30 s, annealing at 54°C for 30 s, extension at 72°C for 30 s, with a final extension step at 72°C for 5 min.
RESULTS AND DISCUSSION
Photochemical properties
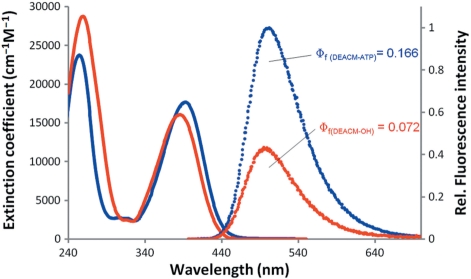
The photocleavage of DEACM-ATP upon irradiation at 390 nm presents almost exclusively DEACM-OH and ATP as products. The absorbed light promotes cleavage of the C–O bond linking the 4-methyl group of 7-diethylamino-4-methylcoumarin and the ATP phosphate ester group, yielding free ATP and DEACM-OH after solvolysis of the generated carbocation (3) (Figure 1). The absorption and emission spectra of DEACM-ATP and DEACM-OH are shown in Figure 2.
Figure 2.
Absorption and emission spectra of DEACM-ATP and DEACM-OH. Measurements of DEACM-ATP (18.5 µM, blue lines) and DEACM-OH (20.8 µM, red lines) were taken in water pH 7.0. Absorption intensities are represented as extinction coefficient as function of wavelength (full lines). Relative fluorescence intensities with excitation at 385 nm (dashed lines) were corrected for respective absorptions at excitation wavelength. Fluorescence quantum yields (Φf) for both molecules are also indicated.
DEACM-ATP long-wavelength absorption band (λmax = 392 nm, εmax = 17700/M/cm) presents a 7 nm red shift and a slightly higher extinction coefficient with respect to the alcohol (DEACM-OH, λmax = 385 nm, εmax = 16000/M/cm), in line with what is observed for other (coumarin-4-yl)methyl caged esters and corresponding alcohols (19). The differences in absorption between DEACM-ATP and DEACM-OH provide the means for ATP release monitorization. Emission bands present very close maxima, but DEACM-ATP fluorescence intensity (fluorescence quantum yield—Φf = 0.166) is larger than that found for DEACM-OH in water (Φf = 0.072). Absorption and emission bands of 7-aminocoumarines are susceptible to medium polarity (20,21) and while for in vitro applications, the solution polarity is usually known, inside living cells significant polarity differences may occur and fluorescence monitoring could be compromised.
The profile of ATP release with irradiation time (and the efficiency of ATP release upon irradiation—ΦChem) was measured in water pH 7.0 for different concentrations of caged ATP (Figure 3). The increase in caged nucleotide concentration leads to a significant decrease of the photochemical yield. An important practical implication is that for higher DEACM-ATP concentrations, longer irradiation times are needed to attain the same fraction of free ATP. For low concentrations of DEACM-ATP, quantitative release of ATP can be attained in a few minutes (for 90% conversion, 10 min irradiation of a 50 µM solution or 15 min for a 100 µM solution). For higher concentrations of DEACM-ATP, however, quantitative release of ATP is time-consuming and the large irradiation times seriously compromise biological applications.
Figure 3.
Irradiation profiles of four different concentrations of DEACM-ATP in water, pH 7.0. DEACM-OH molar fraction obtained after discrete irradiation times with 390 nm light were determined by HPLC (see Materials and methods section). For each initial DEACM-ATP concentration, the respective photochemical quantum yield is also indicated.
This concentration quenching effect is an important drawback, for the reason that for quantitative release with short irradiation times, very diluted solutions of DEACM-ATP are necessary. This may originate some limitations for in vitro application as higher concentrations of nucleotides are usually needed to perform reactions.
In vitro transcription
For in vitro transcription, 50 µM of a ribonucleotide mix (CTP, UTP and GTP) was employed. In control samples, absence of ATP impeded transcription (negative control) and incubation with ATP (50 µM) yielded a full-length transcription product readily detected (positive control). ATP was then substituted by DEACM-ATP and the mixture submitted to several irradiation times (Figure 4A). In the sample not subjected to irradiation no product formation was observed, indicating DEACM efficient caging of ATP molecules blocking RNA polymerization. After irradiation, full transcription product could be detected, indicating that ATP had been released. The quantity of transcript reaches its maximum with 5 min of irradiation and is significantly lower than in the positive control, even though DEACM-ATP and ATP concentration are identical.
Figure 4.
In vitro transcription using DEACM-ATP. (A) Agarose gel electrophoresis of in vitro transcription reaction with 50 µM of CTP, GTP and UTP. Positive control—transcription with 50 µM ATP. Negative control—transcription in absence of ATP. Irradiated samples—transcription containing 50 µM of DEACM-ATP before irradiation; duration of irradiation with 390 nm visible light is indicated for each sample: 0 min (nonirradiated), 2 min, 5 min and 35 min. (B) Denaturing PAGE analysis of in vitro transcription reaction with 50 µM of CTP, GTP and α-32P-UTP. Increasing ATP concentrations were employed for each sample, ranging from 0 µM (right lane) to 50 µM (left lane). (C) Denaturing PAGE analysis of in vitro transcription reaction with 50 µM of CTP, GTP, α-32P-UTP and DEACM-ATP. Each sample was subjected to increasing irradiation times, from 0 min (right lane) to 35 min (left lane). Released ATP concentrations estimated by HPLC (see Materials and methods section).
A gradual increase in product formation is observed with increasing irradiation time (and consequently ATP release) for concentrations up to 33 µM of released ATP. For higher concentrations, transcription is somehow hampered. In order to assess whether the observed direct relation between released ATP and transcription was associated with formation of full-length transcript, sequential concentrations of added ATP (Figure 4B) and released ATP (resulting from increasing irradiation times of DEACM-ATP, Figure 4C) were used in transcription reactions, and analyzed by denaturing PAGE. These results indicate that DEACM-ATP irradiation and photo-released ATP yield full-length transcripts. Residual product formation could be detected for the nonirradiated sample, which is probably due to ambient light during manipulation in transcription reactions. Even though most sample handling was made in the dark chamber, extra care must be taken during all procedure so as to minimize unwanted release from precursor.
Further confirmation that full-length transcripts were attained was shown by RT–real-time PCR, which also confirmed product specificity and allowed for quantification. This step takes advantage of a sequence-specific amplification primer (E7p53Rev primer), which hybridizes to the RNA template produced, allowing for a clear distinction between residual nonspecific transcription and full-length-specific products. The relative amount of initial template in each sample was determined with relation to the sample with the highest product quantity—25 µM of released ATP (values obtained directly relate to quantities generated in transcription), and the attained results can be seen in Figure 5. Amplification products were additionally submitted to melting curve analysis and a dispersion of <0.5°C was obtained for the DEACM-ATP samples (data not shown). Taken together, these results clearly denote that specific full-length RNA is quantitatively generated after ATP photo release. Once again, a direct relation between transcript production and released ATP occurs up to a certain concentration of ATP (ca. 25 µM), after which a decrease in product formation is observed.
Figure 5.
Relative quantification of full-length transcription products as function of ATP released after DEACM-ATP irradiation. Transcript quantification by RT–real-time PCR as function of estimated released-ATP. Relative product quantity was normalized in relation to the sample showing the highest product quantity (25 µM ATP) (see Materials and methods section).
The transcription assays show the presence of an inhibitory effect within the reaction, indicating that a photolysis byproduct of DEACM-ATP irradiation interferes with transcription. The cause of such hindrance seems to be the DEACM-OH generated after DEACM-ATP photocleavage. Clearly, DEACM-ATP does not impede transcription as at maximal DEACM-ATP concentration (before irradiation), residual transcription can still be detected (Figure 4C). ATP released from caged-precursors suffers no chemical alteration [see refs (11,19 and references therein], which in the present case is evidenced by successful initiation of transcription. Conversely, increasing concentrations of DEACM-OH after a certain point lead to decreasing transcription yields. For concentrations above 100 µM DEACM-OH, no transcript formation was observed. This was confirmed by performing control transcription reactions with 50 µM ATP to which increasing amounts of DEACM-OH were added (Figure 6A). In addition, fluorescence spectra of DEACM-OH in the presence of T7 RNA polymerase show a 2 nm red shift and a 20% decrease of intensity (Figure 6B), indicating that DEACM-OH is in a different environment (20,21). This observation suggests a partition of DEACM-OH into the protein, which might be responsible for enzyme inhibition (20–24). No spectral interaction was observed between template DNA and DEACM-OH. Also, DEACM-OH presents a poor solubility in water (between 10−4 M and 10−5 M), which could be the reason for the observed partition and resulting inhibitory effect over T7 RNA polymerase. This inhibitory effect of DEACM-OH limits the concentration range of DEACM-ATP that can be used for light-controlled in vitro transcription reactions.
Figure 6.
Effect of DEACM-OH in transcription reactions. (A) Transcription levels in reactions containing unmodified ATP, CTP, GTP and UTP in presence of increasing DEACM-OH concentrations. ‘Blank’ refers to reaction in absence of DEACM-OH. (B) Emission spectra of DEACM-OH (2.3 µM, black line) and DEACM-OH in presence of T7 RNA Polymerase (gray line). Both spectra performed in phosphate buffer 10 mM, 0.1 M NaCl, pH 7.0.
CONCLUSIONS
We have successfully demonstrated the use of ATP caged with DEACM for light-controlled in vitro transcription reactions. When bound to ATP, DEACM was shown to efficiently impede transcription. Upon irradiation, ATP is released and the transcription reaction resumes, yielding full-length RNA products. The release of ATP is concentration dependent, with the photochemical quantum yield quenched for high concentrations of DEACM-ATP. Higher concentrations of caged-ATP significantly increase the irradiation times needed for complete photolysis. The released byproduct, DEACM-OH, seems to inhibit in vitro transcription for concentrations higher than 25 µM, probably due to the partition of DEACM-OH into the protein, evidenced by the red shift and concomitant decrease in intensity of the fluorescence spectra of DEACM-OH in presence of T7 RNA polymerase. The nature of interaction between DEACM-OH and T7 RNA polymerase, and possible inhibition mechanisms still require further clarification.
To our knowledge, this is the first time that photo manipulation of nucleic acids polymerization is described. Furthermore, polymerization control was accomplished through visible light irradiation, thus preventing UV-associated nucleic acid and protein damage incompatible with potential biotechnological applications.
ACKNOWLEDGEMENTS
The authors would like to acknowledge Dr A. Jorge Parola for fruitful suggestions related to photochemistry procedures, Dr João Noronha for Mass Spectra analysis, Dr Isabel Sá Nogueira and Dr José Inácio for assistance with α-32P-UTP experiments. This work was supported by Fundação para a Ciência e Tecnologia (REQUIMTE, CIGMH, PTDC/BIO/66514/2006, PTDC/SAU-BEB/66511/2006, FRH/BD/24276/2005 to A.V.P.); Fundação Calouste Gulbenkian (Ref. 76436). Funding to pay the Open Access publication charges for this article was provided by Faculdade de Ciências e Tecnologia, Universidade Nova de Lisboa.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kapland JH, Forbush B, Hoffman JF. Rapid photolytic release of adenosine 5′-triphosphate from a protected analog – utilization by Na-K pump of human red blood-cell ghosts. Biochemistry. 1978;17:1929–1935. doi: 10.1021/bi00603a020. [DOI] [PubMed] [Google Scholar]
- 2.Adams SR, Tsien RY. Controlling cell chemistry with caged compounds. Annu. Rev. Physiol. 1993;55:755–784. doi: 10.1146/annurev.ph.55.030193.003543. [DOI] [PubMed] [Google Scholar]
- 3.Schade B, Hagen V, Schmidt R, Herbrich R, Krause E, Eckardt T, Bendig J. Deactivation behavior and excited-state properties of (coumarin-4-yl)methyl derivatives. 1. Photocleavage of (7-methoxycoumarin-4-yl)methyl-caged acids with fluorescence enhancement. J. Org. Chem. 1999;64:9109–9117. doi: 10.1021/jo010692p. [DOI] [PubMed] [Google Scholar]
- 4.Wang L, Corrie JET, Wootton JF. Photolabile precursors of cyclic nucleotides with high aqueous solubility and stability. J. Org. Chem. 2002;67:3474–3478. doi: 10.1021/jo020040g. [DOI] [PubMed] [Google Scholar]
- 5.Corrie JET. Synthesis, photochemistry and enzymology of 2-O-(2-nitrobenzyl)-D-glucose, a photolabile derivative of D-glucose. J. Chem. Soc. Perkin Trans. 1. 1993;18:2161–2166. [Google Scholar]
- 6.Walbert S, Pfleiderer,W., Steiner UE. Photolabile protecting groups for nucleosides: mechanistic studies of the 2-(2-nitrophenyl)ethyl group. Helv. Chim. Acta. 2001;84:1601–1611. [Google Scholar]
- 7.Givens RS, Weber JFW, Conrad PG, Orosz G, Donahue SL, Thayer SA. New phototriggers 9: p-hydroxyphenacyl as a C-terminal photoremovable protecting group for oligopeptides. J. Am. Chem. Soc. 2000;122:2687–2697. [Google Scholar]
- 8.Fedoryak OD, Dore TM. Brominated hydroxyquinoline as a photolabile protecting group with sensitivity to multiphoton excitation. Org. Lett. 2002;4:3419–3422. doi: 10.1021/ol026524g. [DOI] [PubMed] [Google Scholar]
- 9.Zhu Y, Pavlos CM, Toscano JP, Dore TM. 8-bromo-7-quinoline as a photoremovable protecting group for physiological use: mechanism and scope. J. Am. Chem. Soc. 2006;128:4267–4276. doi: 10.1021/ja0555320. [DOI] [PubMed] [Google Scholar]
- 10.Ando H, Furuta T, Tsien RY, Okamoto H. Photo-mediated gene activation using caged RNA/DNA in zebrafishembryos. Nat. Genet. 2001;28:317–325. doi: 10.1038/ng583. [DOI] [PubMed] [Google Scholar]
- 11.Geiβler D, Kresse W, Wiesner B, Bendig J, Kettenmann H, Hagen V. DMACM-Caged adenosine nucleotides: ultrafast phototriggers for ATP, ADP and AMP activated by long-wavelength irradiation. ChemBioChem. 2003;4:162–170. doi: 10.1002/cbic.200390027. [DOI] [PubMed] [Google Scholar]
- 12.Schonleber RO, Bendig J, Hagen V, Giese B. Rapid photolytic release of cytidine 5′-diphosphate from a coumarin derivative: a new tool for the investigation of ribonucleotide reductases. Bioorg. Med. Chem. 2002;10:97–101. doi: 10.1016/s0968-0896(01)00254-1. [DOI] [PubMed] [Google Scholar]
- 13.Eckardt T, Hagen V, Schade B, Schmidt R, Schweitzer C, Bendig J. Deactivation behavior and excited-state properties of (coumarin-4-yl)methyl derivatives. 2. Photocleavage of selected (coumarin-4-yl)methyl-caged adenosine cyclic 3′,5′-monophosphates with fluorescence enhancement. J. Org. Chem. 2002;67:703–710. doi: 10.1021/jo010692p. [DOI] [PubMed] [Google Scholar]
- 14.Ellis-Davies GCR. Caged compounds: photorelease technology for control of cellular chemistry and physiology. Nat. Methods. 2007;4:619–628. doi: 10.1038/nmeth1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayer G, Heckel A. Biologically active molecules with a ‘light switch’. Angew. Chem. Int. Ed. 2006;45:4900–4921. doi: 10.1002/anie.200600387. [DOI] [PubMed] [Google Scholar]
- 16.Hagen V, Frings S, Wiesner B, Helm S, Kaupp UB, Bendig J. [7-(dialkylamino)coumarin-4-yl]methyl-caged compounds as ultrafast and effective long-wavelength phototriggers of 8-bromo-substituted cyclic nucleotides. ChemBioChem. 2003;4:434–442. doi: 10.1002/cbic.200300561. [DOI] [PubMed] [Google Scholar]
- 17.Du H, Fuh RA, Li J, Corkan A, Lindsey JS. PhotochemCAD: a computer-aided design and research tool in photochemistry. Photochem. Photobiol. 1998;68:141–142. [Google Scholar]
- 18.Montalti M, Credi A, Prodi L, Gandolfi MT. (2006) Handbook of Photochemistry, 3rd edn. CRC Press, Taylor and Francis Group, Boca Raton, FL, USA. [Google Scholar]
- 19.Schmidt R, Geiβler D, Hagen V, Bendig J. Mechanism of photocleavage of (coumarin-4-yl)methyl esters. J. Phys. Chem. 2007;111:5768–5774. doi: 10.1021/jp071521c. [DOI] [PubMed] [Google Scholar]
- 20.Barik A, Kumbhakar M, Nath S, Pal H. Evidence for the TICT mediated nonradiative deexitation process for the excited coumarin-1 dye in high polarity protic solvents. Chem. Phys. 2005;315:277–285. [Google Scholar]
- 21.Seixas de Melo JS, Becker RS, Maçanita AL. Photophysical behavior of coumarins as a function of substitution and solvent: experimental evidence for the existence of a lowest lying 1(n,π*) state. J. Phys. Chem. 1994;98:6054–6058. [Google Scholar]
- 22.Barik A, Nath S, Pal H. Effect of solvent polarity on the photophysical properties of coumarin-1 dye. J. Chem. Phys. 2003;119:10202–10208. [Google Scholar]
- 23.Gustavsson T, Cassara L, Gulbinas V, Gurzadyan G, Mialocq JC, Pommeret S, Sorgius M, van der Meulen P. Femtosecond spectroscopic study of relaxation processes of three amino-substituted coumarin dyes in methanol and dimethyl sulfoxide. J. Phys. Chem. A. 1998;102:4229–4245. [Google Scholar]
- 24.Arbeloa TL, Arbeloa FL, Tapia MJ, Arbeloa IL. Hydrogen-bonding effect on the photophysical properties of 7- aminocoumarin derivatives. J. Phys. Chem. 1993;97:4704–4707. [Google Scholar]