Abstract
Multiple segmental and selective isotope labeling of RNA with three segments has been demonstrated by introducing an RNA segment, selectively labeled with 13C9/15N2/2H(1′, 3′, 4′, 5′, 5′′)-labeled uridine residues, into the central position of the 20 kDa ε-RNA of Duck Hepatitis B Virus. The RNA molecules were produced via two efficient protocols: a two-step protocol, which uses T4 DNA ligase and T4 RNA ligase 1, and a one-pot protocol, which uses T4 RNA ligase 1 alone. With T4 RNA ligase 1 all not-to-be-ligated termini are usually protected to prevent formation of side products. We show that such labor-intensive protection of termini is not required, provided segmentation sites can be chosen such that the segments fold into the target structure or target-like structures and thus are not trapped into stable alternate structures. These sites can be reliably predicted via DINAMelt. The simplified NMR spectrum provided evidence for the presence of a U28 H3-imino resonance, previously obscured in the fully labeled sample, and thus of the non-canonical base pair U28:C37. The demonstrated multiple segmental labeling protocols are generally applicable to large RNA molecules and can be extended to more than three segments.
INTRODUCTION
Uniform and residue-type selective 13C/15N isotope labeling was introduced in the early 90s (1) and has proven to be a major advance in NMR studies of structure and dynamics of RNA molecules (2–12). This type of isotope labeling can be used to reduce resonance overlap in the NMR spectra and simplify resonance assignment by recording multi-dimensional hetero-nuclear and isotope-filtered NMR spectra. Residue-type selective 13C/15N labeling combined with (selective) deuteration (13–17) has subsequently been introduced to further reduce the complexity of the NMR spectra. Nevertheless, with a few exceptions (18–20), the application of NMR to RNA has remained restricted to molecules <16 kDa (48 nucleotides) mainly due to persisting resonance overlap for larger RNAs. To be able to study RNAs of this size or larger, the divide and conquer approach has been applied (7,21), i.e. NMR investigations have focused on smaller domains of the larger RNA (3,7). Although the high stability of many local RNA elements makes this a valid strategy, the isolated domain may adopt a structure that differs from the full RNA and potential 3D features remain unknown. Segmental labeling of RNA (20–26) further reduces the number of resonances in NMR spectra, so that specific domains within the structural and dynamical context of the full RNA molecule can be studied. The main challenge is the synthesis of sufficient amounts of segmental labeled RNA. The simplest approach is to transcribe segments of the full RNA with T7 RNA polymerase (27) separately and subsequently ligate the segments. However, several problems, e.g. multimerization and formation of alternate structures during ligation, have to be overcome and so far, only five examples of the synthesis of segmental labeled RNA for NMR studies have been described (20–26).
Segmental labeled RNA can be obtained by ligation of two RNA segments hybridized to a complementary ssDNA using T4 DNA ligase (26,28). T4 DNA ligase selectively ligates the nicked RNA on the RNA–DNA hybrid so that protection of the 3′- and 5′-termini to prevent formation of side products is not required. Segmental labeled RNA can also be prepared by ligation of RNA segments with T4 RNA ligase 1 (10,29). Single-stranded RNA is efficiently ligated by T4 RNA ligase 1 and recently it has been shown that also nicked dsRNA and nicked RNA on a RNA–DNA hybrid can be ligated (30). However, the promiscuous nature of T4 RNA ligase 1 requires the protection of those 5′-PO4 and 3′-OH termini that should not be ligated to prevent formation of side products, as circular RNA and multimers. The 3′-terminus can be protected by oxidation of its cis-diol group with periodate (24,31) and the 5′-terminus by dephosphorylation with alkaline phosphatase (22,32). Another protection method is flanking the termini with either a 3′-hammerhead or a 5′-hammerhead ribozyme (33). After ribozyme cleavage, the RNA molecules are directly protected against ligation by the presence of either a 2′3′-cyclic-phosphodiester on the 3′-end or a hydroxyl on the 5′-end. This approach was applied by Kim et al. (21) and Tzakos et al. (25) for the preparation of RNA labeled on two segments. Finally, single (or di-) nucleotide labeled RNA can be obtained by ligation of a 3′5′-diphosphorylated single (or di-) nucleotide to an RNA segment with T4 RNA ligase 1, followed by ligation to another RNA strand (22–24). To avoid the labor-intensive purification of RNA intermediates in the original method, a purification-free method was developed (24), although deprotection of intermediates is still required.
In all described examples, the RNA molecule of interest is prepared by ligation of one labeled RNA segment or one 3′5′-diphosphorylated residue to another RNA segment with T4 DNA ligase or T4 RNA ligase 1. However, it is highly desirable to label one or more segments within a larger RNA. The problem encountered with further extension after a first ligation is the required removal of the protection group before the RNA ligation product can be used in a second ligation. It seemed therefore difficult to produce RNA molecules labeled on more than two segments in amounts sufficient for NMR structural analysis.
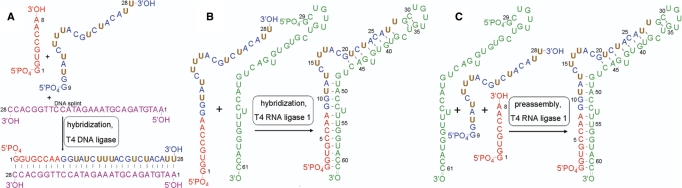
In this contribution, an efficient and general applicable two-step ligation method is described, in which a RNA segment selective labeled with 13C9/15N2/2H(1′, 3′, 4′, 5′, 5′′)-labeled uridine residues is introduced into the central position of a larger RNA molecule (Figure 1A and B) by ligation with T4 DNA ligase and T4 RNA ligase 1. The method does not require extensive protection and deprotection of the involved termini. We further demonstrate that under well-chosen conditions this multiple segmental labeling can be achieved in a one-pot ligation reaction (Figure 1C). Both the two-step and one-pot ligation methods are demonstrated on the 61 nt (20 kDa) RNA encapsidation signal of Duck Hepatitis B RNA Virus (ε-DHBV) (34–38). The segmentation pattern is chosen so that one segment encompasses the structurally and functionally important internal loop of the ε-DHBV RNA (37,38).
Figure 1.
Schematic representation of the multiple segmental labeling approaches. (A) First step of the two-step ligation: segments G1-A8 RNA (red) and G9-U28 RNA (blue/brown) are hybridized to a complementary DNA splint (purple) and ligated by T4 DNA ligase. (B) Second step of the two-step ligation: segment G1-U28 RNA is hybridized to segment G29-C61 (green) and ligated by T4 RNA ligase 1 into the full ε-DHBV RNA. (C) One-pot ligation: segments G1-A8, G9-U28 and G29-C61 are putatively preassembled to resemble the full ε-DHBV RNA and ligated by T4 RNA ligase 1. Brown uridine residues of the G9-U28 RNA segment are 13C9/15N2/2H(1′, 3′, 4′, 5′, 5′′)-stable isotope labeled.
MATERIALS AND METHODS
Design of segmentation pattern for ε-DHBV RNA
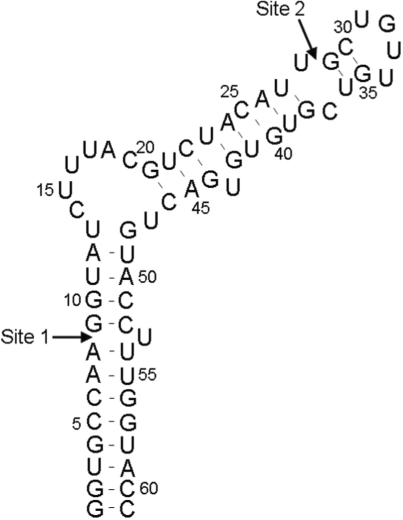
The predicted thermodynamically most stable secondary structure of the full-length ε-DHBV RNA sequence was used as basis for the choice of the ligation sites, which are shown in Figure 2. The sites were chosen such that one segment encompasses the structurally and functionally important internal loop (37,38). Simultaneously, the individual segments should be produced efficiently using T7 RNA polymerase and were therefore taken to start with a G-residue. Finally, it is crucial that the segments hybridize correctly. The thermodynamic stability towards intramolecular folds, duplex formation and intermolecular hybridization of each segment or of mixtures of two segments was therefore calculated, for which the melting profile module of the DINAMelt web server was used (39). Presentations of the folds were obtained from the two-state hybridization module (39). Folding simulations were carried out under ionic conditions as set by DINAMelt, i.e. 1 M NaCl and no MgCl2. In each calculation, the RNA strand concentration was set to 10 μM, corresponding to the strand concentrations used in the ligation experiments.
Figure 2.
Predicted most stable structure of full ε-DHBV RNA by DINAMelt. Arrows indicate the chosen segmentation sites (see text).
Preparation of ε-DHBV RNA segments
Prior to RNA transcription, 13C9/15N2/2H(1′, 3′, 4′, 5′, 5′′)-labeled UTP residues were in vitro synthesized from 450 μmol 13C6/2H7-D-glucose and 440 μmol 13C4/15N2-uracil (Cambridge Isotope Laboratories, Andover, MA, USA) using the enzymes of the glycolysis and pentose phosphate pathway (3,13,16,17) and desalted by affinity chromatography using Affi boronate gel (Bio-Rad Laboratories B.V., Veenendaal, The Netherlands). Purified nucleotides were lyophilized and dissolved to 100 mM in sterilized Milli-Q water (Millipore, Amsterdam, The Netherlands).
The full-length ε-DHBV (G1-C61) RNA sample labeled with 23 13C9/15N2/2H(1′, 3′, 4′, 5′, 5′′)-labeled uridine residues was prepared by in vitro transcription from a partially single-stranded DNA template using T7 RNA polymerase as described by Milligan et al. (27) and Cromsigt et al. (13). Segments G1-A8, G29-C61 and selectively U9-13C9/15N2/2H(1′, 3′, 4′, 5′, 5′′)-labeled G9-U28, were prepared similarly, as well as an unlabeled full-length ε-DHBV (G1-C61) RNA sample. Segments G1-C61, G9-U28 and G29-C61 were electroeluted from denaturing 20% polyacrylamide gel (PAGE) using an Elutrap device (Schleicher & Schuell GmbH, Dassel, Germany) and segment G1-A8 was purified by freeze/thaw extraction of crushed gel. Each RNA segment was recovered by ethanol precipitation and washed in a Centricon-YM3 filter (Millipore) with 20 mM sodium phosphate buffer (pH 7.5), containing 1 M NaCl and 50 mM EDTA and subsequently with Milli-Q water. The concentration of the RNA segments was determined by UV, lyophilized and dissolved to 100 μM in Milli-Q water, except the U23-labeled and unlabeled full-length ε-DHBV (G1-C61) RNA, which were directly dissolved to 0.3 mM in NMR buffer (H2O : D2O 93 : 7, no buffer salts were added, pH 6).
Characterization of interactions of RNA segments by non-denaturing (native) PAGE
The samples for a native PAGE contained 2 µl of each 100 µM RNA segment in T4 RNA ligase 1 buffer [50 mM HEPES (pH 8.0), 10 mM MgCl2, 10 mM DTT and 0.5 mM ATP] in a total volume of 10 µl. The combinations of RNA segments as described in Table 1 were prepared, except those with the DNA splint and G9-C61. The samples were heated to 90°C, slowly cooled down to room temperature, mixed with 2 µl of 30% glycerol and loaded onto a native 8% PAGE containing 10 mM MgCl2. The gel was electrophorized in 1 × TBE at 300 V and 10 mA.
Table 1.
Predicted fold and stability of hybridization interactions of RNA segments and experimental verification
| Segments in the two-step ligation | Predicted stability at 37°C |
Native PAGE | |
|---|---|---|---|
| Hybrid/Duplex (ΔG) | Intramolecular (ΔG) | ||
| G1-A8 + DNA splint | −4.8 (++, +) | NA | ND |
| G9-U28 + DNA splint | −22.5 (++, +) | NA | ND |
| G1-A8 | −1.8 | −0.5 | Duplexed |
| G9-U28 | −1.5 | −2.1 | >90% monomer, <10% duplexed |
| G1-U28 | −5.1 | −3.7 | Duplexed |
| G29-C61 | −6.3 | −9.6 | >90% monomer, <10% duplexed |
| G1-U28 + G29-C61 | −14.7 (+, +)a | NA | Hybridized |
| One-pot ligation | |||
| G1-A8 + G29-C61 | −4.3 (+, +)a | NA | No interaction |
| G9-U28 + G29-C61 | −7.4 (+, −)a | NA | No interaction |
| G9-C61 | Not formed | −14.9 (+, −) | ND |
| G9-C61 | Not formed | −12.9 (+, +) | ND |
| G1-A8 + G9-U28 + G29-C61 | Not predictable | NA | Smeared |
ΔG values are in kcal/mol at 37°C, 1 M NaCl, no Mg2+ and assuming a 10 μM strand concentration; (++, +), 100% base pairing, correct hybrid; (+, +), 50–100% base pairing, correct hybrid; (+, −), 50–100% base pairing, wrong hybrid; ND, not determined, NA, not applicable.
aPredicted structure displayed in Figure 3. Lowest values are given in bold for identification of the expected interaction.
Two-step ligation of ε-DHBV RNA segments
In the first step of the two-step ligation (Figure 1A), the unlabeled G1-A8 RNA segment was ligated to the U9-13C9/15N2/2H(1′, 3′, 4′, 5′, 5′′)-labeled G9-U28 RNA segment by T4 DNA ligase (Fermentas GmbH, St. Leon-Rot, Germany) using a complementary 28-nt ssDNA splint. A series of test reactions, in which temperature, RNA concentration, enzyme concentration and reaction time were varied, was carried out to optimize the ligation yield prior to preparative ligation. The effect of heating the reaction mixture 2 min at 95°C and cooling down to 25°C (heat-annealing) prior to ligation was also investigated. All ligation reactions were carried out in T4 DNA ligase buffer [40 mM Tris–HCl (pH 8.0), 10 mM MgCl2, 10 mM DTT and 0.5 mM ATP]. Subsequently, a 50 ml preparative ligation reaction was carried out containing 15 µM G1-A8 RNA, 10 µM G9-U28 RNA and 10 µM 28-nt DNA splint. The reaction mixture was heat-annealed and subsequently incubated at 37°C for 3 h after the addition of 12.5 kU of T4 DNA ligase. The G1-U28 RNA ligation product was purified from denaturing PAGE as described before.
In the second step of the two-step ligation (Figure 1B), the purified G1-U28 RNA was ligated to the unlabeled G29-C61 segment by T4 RNA ligase 1 (Fermentas). All ligation reactions were conducted in T4 RNA ligase 1 buffer (see native PAGE) and prior to ligation experiments, the 3′-end of the G29-C61 segment was oxidized with NaIO4. A 200-fold molar excess of freshly prepared 3.0 mM NaIO4 was added and incubated for 20 min in the dark. The G29-C61 RNA was ethanol precipitated and dissolved to 100 µM in Milli-Q water. A small series of test reactions was carried out with the G1-U28 RNA and the G29-C61 RNA to optimize input RNA concentration, T4 RNA ligase 1 concentration and reaction time. In addition, the effect of heat-annealing prior to ligation was investigated. After optimizations, a 20 ml preparative ligation reaction containing 10 µM G1-U28 RNA, 20 µM G29-C61 RNA and 8 kU T4 RNA ligase was carried out for 3 h at 37°C after heat-annealing. The segmental labeled full-length ε-DHBV RNA was purified from denaturing PAGE as described before.
One-pot ligation reaction of ε-DHBV RNA segments
Similar optimization experiments for the one-pot ligation (Figure 1C) reaction were carried out as for the two-step ligation described earlier. After optimizations, a 10 ml preparative ligation reaction in T4 RNA ligase buffer containing 10 µM G9-U28 RNA, 10 µM of 3′-end oxidized G29-C61 RNA and 15 µM G1-A8 RNA was carried out. The reaction mixture was heat-annealed and incubated at 37°C for 3 h after the addition of 4 kU of T4 RNA ligase 1. The ligation product corresponding to the full-length ε-DHBV RNA was extracted from denaturing PAGE and purified as described before.
NMR experiments
Lyophilized segmental labeled U9-ε-DHBV RNA obtained from the two-step ligation was dissolved in H2O : D2O 93 : 7 to a final concentration of 0.3 mM, pH 6 (no buffer salts were added), equal to the concentration of the uniformly labeled sample which turned out to be optimal. NMR samples were heated to 90°C and snap cooled on ice to ensure the monomeric state of the RNA molecules. NMR data of U23-ε-DHBV and U9-ε-DHBV were acquired at 800 MHz and 500 MHz, 1H-frequency, respectively at 5°C on Varian Unity Inova NMR spectrometers, using cryo-cooled probes. The 1H-15N-HSQC-spectrum of the U23-ε-DHBV was recorded at 800 MHz with 3200 (t2) × 200 (t1) points for a spectral width of 20 kHz (1H) and 1.5 KHz (15N) while the 1H-15N-HSQC-spectrum of the U9-ε-DHBV was recorded at 500 MHz with 1680 (t2) × 128 (t1) points for a spectral width of 12 kHz (1H) and 650 Hz (15N). Each t1-point was recorded with 16 scans at 800 MHz and 64 scans at 500 MHz. A 800 MHz (1H, 1H) NOESY spectrum on the unlabeled sample was recorded with 500 (t1) × 4000 (t2) points for spectral width of 17 kHz (t1) by 20 kHz (t2) and a mixing time of 200 ms at 5°C. Spectra were processed with NMRpipe (40) and resonances were assigned using the program SPARKY (T.D. Goddard and D.G. Kneller, University of San Francisco).
RESULTS
Design of segmentation pattern for ε-DHBV RNA and native PAGE of RNA segments
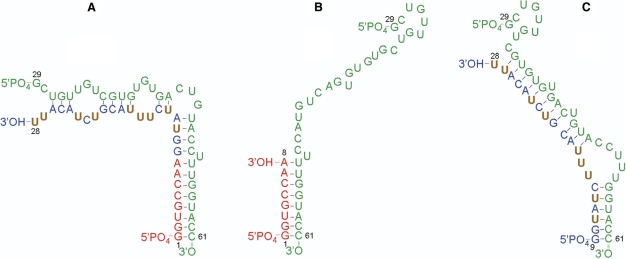
To design the optimal segmentation pattern for the two-step (Figure 1A and B) and one-pot ligations (Figure 1C), DINAMelt was used to predict the fold and hybridization state of the RNA segments. Table 1 provides an overview of the predicted folds and stabilities of the RNA segments and hybridization products. For the first step in the two-step ligation (Figure 1A), DINAMelt predicted segments G1-A8 and G9-U28 to be completely and correctly base paired with the DNA splint (Table 1), and thus accessible for ligation by T4 DNA ligase. In the second step, the ligation product of the first step, G1-U28, is to be ligated to G29-C61 by T4 RNA ligase 1. We aim to avoid labor-intensive protection of termini as much as possible and therefore, only the 3′-end of G29-C61 was protected by periodate treatment and the 5′-end of the G1-U28 was not protected. To prevent or reduce the formation of side products, it is required that in the mixture of G1-U28 and G29-C61, the hybridization product that allows for native ligation should form as much as possible. The most stable hybridization product of G1-U28 and G29-C61 predicted by DINAMelt is shown in Figure 3A. It resembles but is not identical to that of full-length G1-C61 ε-DHBV RNA (Figure 1C), differing only somewhat in the base pairing in the upper stem loop. Nevertheless, the 3′-OH of G1-U28 and the 5′-PO4 of G29-C61 are in close proximity, so that they should be easily accessible and can be efficiently and correctly ligated by T4 RNA ligase 1. The ΔG data in Table 1 indicate that duplex formation or internal folding of G1-U28 and of G29-C61 do not compete with the formation of the G1-U28/G29-C61 hybrid. DINAMelt indeed predicted that the G1-U28/G29-C61 hybrid is predominantly formed (ca. 70%, with 10 μM of each G1-U28 and G29-C61, at 37°C). Some folded G29-C61 monomer is also predicted to be present (ca. 16%). More importantly, some free folded G1-U28 is also predicted to form (ca. 11%) as well as some duplex G1-U28 (ca. 3%). The latter folds of G1-U28 may give rise to side products, e.g. circular G1-U28, because of its unprotected 5′-terminus. However, at the chosen concentration and buffer conditions DINAMelt predicted the absence of free or duplexed G1-U28 RNA in the ligation mixture in the presence of a 2-fold molar excess of G29-C61, so that only the desired ligation could take place. Therefore, a 2-fold molar excess of G29-C61 RNA was employed in the second step of the two-step ligation (see Materials and methods section).
Figure 3.
Predicted hybridizations of RNA segments in the ligation reactions: (A) Segment G1-U28 hybridized with segment G29-C61. (B) Segment G1-A8 hybridized with segment G29-C61. (C) Segment G9-U28 hybridized with segment G29-C61.
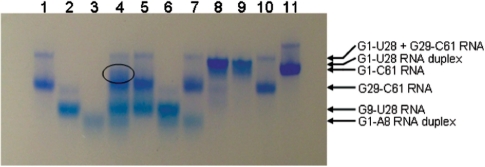
The predicted hybrids in the second step of the two-step ligation were experimentally verified on native PAGE (Figure 4). On the native PAGE, formation of the G1-U28/G29-C61 hybrid in the mixture of G1-U28 RNA + G29-C61 RNA is evidenced by the presence of a strong band (Figure 4, lane 8). This band migrates slightly slower than the band of G1-C61 full-length RNA (Figure 4, lane 11). Whether the G1-U28 duplex is present in the mixture is difficult to assess, because the G1-U28 duplex (Figure 4, lane 9) migrates at a speed comparable with that of the G1-U28/G29-C61 hybrid (Figure 4, lane 8). Some free-folded G29-C61 was also predicted to be present in the mixture. Indeed, a weak band (Figure 4, lane 8) migrates at the speed of the free folded monomer (Figure 4, lanes 1 and 10). In conclusion, the predictions appear to be born out by the native PAGE and the chosen ligation sites were therefore deemed appropriate for obtaining full-length ε-DHBV RNA via the two-step ligation approach.
Figure 4.
Native gel electrophoresis on 8% polyacrylamide gel of putative preassembled RNA complexes. RNA segments were applied in T4 RNA ligase buffer: lane 1, G29-C61; 2, G9-U28; 3, G1-A8; 4, G1-A8 + G9-U28 + G29-C61; 5, G29-C61 + G9-U28; 6, G9-U28 + G1-A8; 7, G29-C61 + G1-A8; 8, G1-U28 + G29-C61; 9, G1-U28; 10, G29-C61; 11, G1-C61. Bands are visualized with Stains-All (Acros Organics, Geel, Belgium).
For the one-pot ligation reaction using three RNA segments, we also aimed to employ as little protection of termini as possible. In this case, protection of the termini of the middle segment is not possible, as this would prevent synthesis of the desired product. As in the two-step synthesis, only the 3′-end of G29-C61 was protected by periodate treatment. A similar DINAMelt-native PAGE analysis was performed to investigate possible formation of side products. Segment G1-A8 is predicted not to form intramolecular structures, but it is able to form a duplex (Table 1). DINAMelt showed that this duplex melts at a relatively low temperature (ca. 40°C), so that at 37°C and a strand concentration of 10 μM, ca. 25% is unfolded and the rest is duplex. Segment G9-U28 is predicted to be dominantly folded as a monomer (Table 1; at 37°C and 10 μM strand concentration DINAMelt showed fractions of duplex : folded monomer : unfolded monomer ≈ 0 : 0.9 : 0.1 respectively). These predictions were confirmed by native PAGE, i.e. G1-A8 migrates as a single band somewhat smeared out, which suggests the single band to be the duplex (Figure 4, lane 3, Table 1), while G9-U28 showed little duplex formation (Figure 4, lanes 1 and 2, Table 1). G29-C61 is predicted to mostly form a folded monomer, while the duplex is only slightly less stable (Table 1). Both have imperfect base pairings. At 37°C and 10 μM strand concentration, DINAMelt predicted the fractions of duplex, folded monomer and unfolded molecules to be 0, 1.0 and 0, respectively. Indeed, G29-C61 is found to be mostly as monomer on native PAGE (Figure 4, lane 1, Table 1).
For the G1-A8 + G29-C61 mixture, a hybrid was predicted that has the native full base-pairing (Figure 3B and Table 1). This hybridization product has a stability that is lower than that of the G29-C61 duplex and folded G29-C61 (Table 1). As expected, the DINAMelt concentration and melting simulation at 37°C and 10 μM strand concentration predicted that the G1-A8 + G29-C61 mixture contained mostly folded G29-C61 monomer and duplex G1-A8, with some hybridization product present. The mixture of G1-A8 + G29-C61 on the native PAGE (Figure 4, lane 7) indeed showed the free folded G29-C61 band (lane 1) as well as the G1-A8 duplex band (lane 3). In addition, a weak band just above that of free folded G29-C61 is seen, which may be of the G1-A8/G29-C61 hybridization product. Because only the hybridization product of G1-A8 and G29-C61 has the correct base pairing, correct ligation probably still occurs in the presence of G9-U28 despite the low expected concentration of the bimolecular complex.
For the mixture of G9-U28 + G29-C61, a stable hybridization product (Figure 3C, Table 1) of G9-U28 with G29-C61 is predicted to be present. The DINAMelt concentration plots predict this product to be present in the mixture at significant but rather low concentration, while the free folded G29-C61 is predicted to dominate. On the native PAGE of the mixture (Figure 4, lane 5) indeed, bands corresponding to the bimolecular complex are observed together with the individual segments (Figure 4, lanes 1 and 2). The base pairing of the predicted hybridization product of G9-U28 + G29-C61 is not the desired native one (Figure 3C). Nevertheless, the 3′-terminus of G9-U28 and the 5′-terminus G29-C61 are in close proximity and appear accessible for ligation, so that the intermediate G9-C61 product can be formed. The efficiency of subsequent ligation of G9-C61 to G1-A8 via this fold (Figure 3C) is expected to be reduced, because it does not allow for hybridization with G1-A8. However, the native fold of G9-C61 (as discernable from Figures 1 or 2) is only slightly less stable (Table 1) and conformational exchange between the two folds may occur.
Finally, mixtures of all three segments have been analyzed on native PAGE (Figure 4) prior to ligation. A smeared band (Figure 4, lane 4, circled) appears, which migrates slightly slower than segment G29-C61. That this band indeed results from hybridization interactions between all three segments is suggested by the observation of its absence when omitting either one of the segments from the mixture (Figure 4, lanes 5–7). This observation together with the analysis of the mixtures with two segments as well as of the individual segments suggests that the correct hybridization interactions could be present, i.e. interactions that could result in full-length ε-DHBV RNA product in a one-pot ligation of the three segments. We therefore proceeded with the one-pot ligation reactions using the segmentation sites as described. Note that the products seen in the one-pot ligation reaction will give further insight into the presence of hybridizations between the three segments.
Synthesis of ε-DHBV RNA segments
The in vitro synthesis of 13C9/15N2/2H(1′, 3′, 4′, 5′, 5′′)-UTP from 450 μmol 13C6/2H7-D-glucose and 440 μmol 13C4/15N2-uracil yielded 390 μmol UTP, which corresponded to a yield of 89% as calculated from the input amount of uracil.
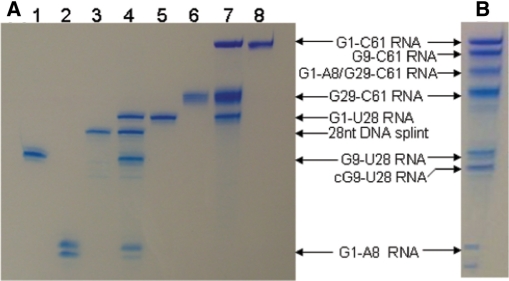
The synthesis of the RNA segments from synthetic partially duplexed DNA templates using T7 RNA polymerase, yielded 17 mg of G1-A8 RNA, 4.2 mg of U9-13C9/15N2/2H(1′,3′,4′,5′,5′′) labeled G9-U28 RNA, 8.4 mg of G29-C61 RNA and 2.2 mg of U23-13C9/15N2/2H(1′,3′,4′,5′,5′′) labeled G1-C61 full-length ε-DHBV RNA. As evidenced by analytical denaturing PAGE (Figure 5A, lanes 1, 2 and 6), the purified RNA segments showed no degenerative products and comprised a single band for each RNA segment, except for segment G1-A8 RNA. This segment is a 1:1 mixture of the G1-A8 segment and a 9-nt segment G1-A8 (n+ 1).
Figure 5.
Course of the multiple segmental labeling. (A) The two-step multiple segmental labeling. Lanes 1–8 contain the following samples: (1) 1 µg G9-U28 RNA, (2) 1 µg G1-A8 RNA, (3) 1 µg 28 nt ssDNA splint, (4) 28 nt ssDNA splinted ligation of G1-A8 RNA to G9-U28 RNA (5) purified G1-U28 RNA, (6) 1 µg G29-C61 RNA, (7) G1-U28 RNA to G29-C61 RNA ligation (8) 1 µg of purified G1-C61 ε-DHBV RNA. (B) The one-pot ligation of G1-A8 RNA, G9-U28 RNA and G29-C61 RNA into G1-C61 ε-DHBV RNA. Bands are visualized with Stains-All (Acros).
Two-step ligation of ε-DHBV RNA segments
Optimization experiments for the DNA splinted ligation of G1-A8 RNA to G9-U28 RNA revealed that the optimal strand concentrations were 15 µM of G1-A8 RNA, 10 µM of G9-U28 RNA and 10 µM of 28-nt DNA splint. The optimal ligation temperature was 37°C (very little ligation product was observed at 25°C and no product was detectable at 16 and 4°C). Time course optimization experiments showed that the minimal reaction time was at least 3 h and prolongation up to 6 h did not increase the ligation yield. The optimal T4 DNA ligase concentration was 0.25 U/μl of reaction mixture. Heat-annealing the reaction mixture prior to ligation had no effect on the ligation yield.
The preparative ligation reaction (Figure 5A, lane 4) was carried out under optimal conditions with 0.8 µmol (2.11 mg) of G1-A8 RNA and 0.53 µmol (3.5 mg) of G9-U28 RNA. This yielded 0.26 µmol of purified G1-U28 RNA (Figure 5A, lane 5), which corresponds to a yield of 49% as calculated from the input amount of G9-U28 RNA. Degenerative or side products were not produced (Figure 5A, lane 4). Not all of each of the segments was consumed in the ligation reaction, as evident from the presence of unligated G9-U28 and G1-A8 and G1-A8 (n + 1) segments (Figure 5A, lane 4). This is probably due to the presence of G1-A8 (n + 1) in the mixture (see Discussion section).
Optimization reactions for the second ligation step of G1-U28 RNA to G29-C61 RNA showed optimal ligation with 10 µM of G1-U28 RNA and a 2-fold molar excess of G29-C61 RNA (20 μM), as predicted by DINAMelt. Heat-annealing the reaction mixture prior to ligation slightly increased the yield of the ligation. Again, the optimal ligation yield was obtained at 37°C after 3 h. The optimal input of T4 RNA ligase 1 was 0.4 U/µl of reaction mixture.
The preparative ligation reaction (Figure 5A, lane 7) was carried out with 0.2 µmol (1.85 mg) of G1-U28 RNA and 0.4 µmol (4.36 mg) of G29-C61 RNA. This yielded 0.105 µmol (2.11 mg) of pure G1-C61 RNA and no side products (Figure 5A, lane 8). The yield of 0.105 µmol (2.11 mg) corresponds to a ligation yield of 52.5%, as calculated from the input amount of G1-U28 RNA. The overall ligation yield for the two-step ligation was 26%.
One-pot ligation of ε-DHBV RNA segments
Optimization experiments for the one-pot ligation reaction of ε-DHBV RNA revealed an optimal yield with 15 µM of G1-A8 RNA, 10 µM of G9-U28 RNA and 10 µM of G29-C61 RNA. The best ligation yield was obtained at 37°C after heat-annealing, but comparable amounts of full-length ε-DHBV RNA were also obtained at 25 and 16°C. Incubation at 4°C yielded no full-length product. Time course optimizations showed that the optimal reaction time was 3 h. The optimal T4 RNA ligase 1 concentration was 0.4 U/μl of reaction mixture. Side products consisting of circular G9-U28 RNA, incorrectly ligated 41-nt G1-A8 + G29-C61 RNA and a substantial amount of G9-C61 RNA lacking the G1-A8 segment were formed, but multimerization of RNA segments was not observed (Figure 5B).
The preparative one-pot ligation reaction under optimal conditions, i.e. with 0.15 µmol G1-A8, 0.1 µmol G9-U28 and 0.1 µmol G29-C61, yielded 0.015 µmol (0.3 mg) of full-length G1-C61 RNA. This corresponds to a yield of 15% as calculated from the input amount of G9-U28 RNA. Similar patterns of side products were observed as for the optimization reactions.
NMR experiments
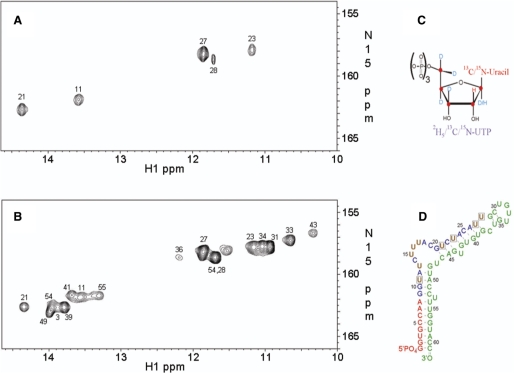
Figure 6A and B show the (1H-imino,15N) HSQC spectra of the multiple segmental labeled construct (U9-ε-DHBV, prepared with the two-step ligation method) and of the U23-13C9/15N2/2H(1′, 3′, 4′, 5′, 5′′)-labeled full-length ε-DHBV (U23-ε-DHBV), respectively. The HSQC spectrum of U9-ε-DHBV is considerably simplified showing only the imino resonances of the two U:A and two G:U base pairs in the G9-U28 segment. The simplification confirms the correct assignment of the U:A base pairs (U11 and U21), as well as the assignment of the G:U base pairs (U23 and U27) in the U23-ε-DHBV RNA. The U23-ε-DHBV RNA resonance assignments are based on an imino proton sequential walk in the (1H,1H) NOESY spectrum of full-length ε-DHBV RNA combined with the (1H-imino,15N) HSQC spectrum of the (full-length) U23-ε-DHBV RNA and confirmed by comparison with the resonance assignments in shorter RNA constructs, one encompassing only the primer loop and one encompassing the apical stem-loop (38). These latter spectra demonstrate that U13:G48 do not display imino resonances (38). Most importantly, the HSQC spectrum of U9-ε-DHBV unveils the presence of an imino resonance (Figure 6A), obscured under the intense U58 imino resonance of the G4:U58 base pair in the HSQC spectrum of U23-ε-DHBV (Figure 6B). Comparison of the HSQC spectra of U9-ε-DHBV and U23-ε-DHBV and given the knowledge that U13:G48 does not show a U-imino resonance, it follows that this resonance must be from U28 of the U28:C37 non-canonical base pair.
Figure 6.
(A) The (1H-imino,15N) HSQC NMR spectrum of the segmental labeled U9-ε-DHBV: the nine uridine residues in the G9-U28 segment are 13C9/15N2/2H(1′, 3′, 4′, 5′, 5′′)-labeled (brown residues in Figure 6D) (B) The (1H-imino,15N) HSQC NMR spectrum of U23-ε-DHBV. Sequence-specific assignment for the ε-DHBV G1-C61 imino protons was achieved from 2D (1H, 1H) NOESY and (1H-imino,15N) HSQC experiments on a nonlabeled sample and the U23-ε-DHBV sample, respectively, using standard assignment procedures (3,9), confirmed and completed by comparison with assignments from the primer loop and apical stem-loop. The imino resonances are labeled with their corresponding uridine residue number when assigned; the residue assignments on the loop residues U33 and U34 and on the bulged U43 are tentative. One resonance in (B) remains unassigned. It resides in the spectral region of imino protons of mismatched or unpaired uridines. Its absence in (A) shows that it must be from a mismatched or unpaired U outside the segmentally labeled region. (C) Schematic representation of the labeling pattern in the uridine residues. (D) Schematic representation of the segmental labeling pattern in the full U9-ε-DHBV RNA. Brown uridine residues are 13C9/15N2/2H(1′, 3′, 4′, 5′, 5′′) -labeled, while the boxed uridine residues show imino resonances in the (1H imino,15N) HSQC NMR spectrum.
DISCUSSION
We have shown that multiple segmental labeling, as presented here, is an effective and straightforward method to specifically label one RNA segment, centrally positioned in a large RNA molecule, with the (combined) use of T4 DNA ligase and T4 RNA ligase 1. As demonstrated, the central segment of the 20 kDa 61-nt ε-DHBV RNA has been labeled with 13C9/15N2/2H(1′, 3′, 4′, 5′, 5′′)-labeled uridine residues. Resonance overlap is greatly reduced in the NMR spectrum compared to that of the fully selective U23-13C9/15N2/2H(1′, 3′, 4′, 5′, 5′′)-labeled sample (Figure 6A and B). As a result, the U28 imino resonance was revealed and could be assigned, a resonance that completely overlapped with the U58:G4 imino resonance in the fully selective uridine labeled sample, thereby providing direct evidence for the presence of the noncanonical U28:C37 base pair.
This multiple segmental labeling strategy using differentially stable isotope-labeled nucleotides can reduce spectral crowding significantly, by the ability to select and eliminate specific resonances and the reduction of line-widths resulting from deuteration. Potentially, any segmental labeling pattern can be applied to a large RNA, enabling studies of local structural and dynamical details within the natural conformational shape of the full RNA. However, the successful synthesis of multiple segmental labeled RNA requires special attention to segment design, ligation conditions and, if necessary, protection of termini.
Based on the DINAMelt folding predictions it was estimated that G1-A8, G9-U28 and G29-C61 was an effective segmentation pattern. In the first step of the two-step ligation, the possibility of G1-A8 RNA to form duplexes was not likely to interfere with the full base pairing of G1-A8 RNA with the DNA splint, which was predicted to hybridize with a higher stability (Table 1). In the second step of the two-step ligation, the intermediate G1-U28 RNA effectively hybridized with a 2-fold excess of G29-C61, canceling out duplex formation of G1-U28 and thereby the formation of side products.
In the one-pot ligation, segment G1-A8 was predicted to be correctly and completely base paired with segment G29-C61 with a ΔG value of −4.25 kcal/mol (Table 1). However, the G9-U28 segment was predicted to hybridize with the same part of G29-C61 with a stability of −7.4 kcal/mol (Table 1). This explains the formation of substantial amounts of G9-C61 RNA next to the full-length G1-C61 RNA, because G1-A8 RNA cannot efficiently be ligated to G9-U28 RNA if this segment is hybridized with G29-C61. When G1-A8 RNA is hybridized with G29-C61 RNA, it is possible that the G9-U28 RNA is correctly hybridized with the free part of the hybridized G29-C61 RNA, resulting in the resemblance of full G1-C61 RNA. G9-U28 RNA could also be present freely in the ligation mixture and undergo circularization or multimerization or hybridized G1-A8 RNA can be ligated to G29-61 RNA. The incorrect hybridization of G9-U28 to G29-C61 is assumed to partially interfere with the formation of the desired hybridized structure of G1-A8 + G29-C61 giving rise to the observed side products, although full ε-DHBV RNA remains the main product.
That the choice of segmentation sites is crucial is evident from ligation experiments that were carried out using a different segmentation pattern. Here, the G1-C61 ε-DHBV RNA was segmented into G1-C19, G20-U47 and G48-C61. Optimization ligation reactions with these three segments yielded no full-length G1-C61 product. T4 RNA ligase 1 only ligated the 3′-OH of G1-C19 to the 5′-PO4 of G48-C61 resulting in a 33 nt wrongly ligated product. Segment G20-U47 remained completely unreacted, clearly possessing inaccessible termini for ligation. Simulation with DINAMelt indicated that correct hybridization of G1-C19 and G48-C61 (ΔG = −9.6 kcal/mol) can indeed occur, whereas G20-U47 is predicted to fold as a stable hairpin (ΔG = −7.7 kcal/mol) and therefore not available for ligation. In conclusion, such interfering stable alternate structures should be avoided when designing the segmentation pattern. The hybridization predictions by DINAMelt are found to be reliable and to form a good framework for defining optimal ligation sites.
Synthesis of multiple segmental labeled ε-DHBV RNA
In the two-step and the one-pot labeling, protection of the 3′-end of segment G29-C61 RNA was essential to prevent the multimerization of segment G29-C61 and the formation of circular G29-C61.
The DNA splinted RNA ligation of the segments G1-A8 to G9-U28 resulted in only G1-U28 RNA as ligation product, even though the termini of the segments were not protected. The absence of side products is explained by the fact that T4 DNA ligase selectively ligates the nicked RNA–DNA hybrid. The subsequent ligation reaction of G1-U28 RNA to G29-C61 RNA required an excess of 3′-protected segment G29-C61, preventing the formation of circular G1-U28 and multimerized G1-U28. Thus, the conditions for the two-step ligation can be chosen in such way that side products are not formed and thereby eliminating the need for extensive protection of termini (i.e. the 5′-end and the 3′-end of the G1-U28 segment were not protected).
The formation of side products in the one-pot ligation cannot be prevented by protection of the involved termini, since their presence is required for the formation of the desired ligation product. The formation of these side products can however be minimized by optimizing the reaction conditions. Although wrongly ligated side products cannot be used, they do contain information about the hybridization interactions that occur in the one-pot ligation mixture. The formation of almost equimolar amounts of G9-C61 RNA next to G1-C61 RNA revealed that G9-U28 and G29-C61 are ligated into G9-C61 via the hybrid shown Figure 3C, which is nonnative, stable and does not allow subsequent hybridization with G1-A8. This ligation route also explains the formation of a substantial amount of 41 nt G1-A8 + G29-C61 RNA and circular G9-U28 RNA, because less stable, but correct, hybridization was predicted of G9-U28 RNA with the free part of the G29-C61 RNA when this is hybridized with G1-A8 RNA.
Correctly ligated side products lacking a segment can be purified and processed afterwards to yield additional full-length RNA. We found that, the 53-nt G9-C61 RNA side product could successfully be ligated to G1-A8 using T4 RNA ligase 1 or T4 RNA ligase 2 (yields 15 and 40%, respectively; results not shown). The higher ligation efficiency with T4 RNA ligase 2 suggests the presence of nicked dsRNA instead of gapped dsRNA, most likely because of the size heterogeneity in the segment G1-A8 isolate. Putatively, the G1-A8 (n + 1) is actually segment G1-A9, accidentally resulting in nicked dsRNA. Investigation of a comparative analytical denaturing PAGE (results not shown) revealed that in the DNA splinted RNA ligation the G1-A8 segment is used, whereas the putative G1-A9 segment is used in the one-pot ligation. The end product of the one-pot ligation thus contains one nucleotide extra in the lower stem that is now fully base paired as compared to the full-length RNA from the two-step ligation, which contains a bulged out uridine residue. This was ultimately evidenced by a slight difference in migration distance of both full-length products (results not shown). This shows that T4 RNA ligase 1, known to have a high preference for ligation of single stranded RNA, is also capable of joining RNA molecules in nicked RNA duplexes, albeit with a lower efficiency. This phenomenon has recently been described by Bullard and Bowater (30), who showed that nick joining activity of T4 RNA ligase 1 on duplex RNA is about 40 times lower than that of T4 RNA ligase 2. To prevent incorporation of segments of incorrect length in a one-pot ligation, it has to be assured that the size of the segments is homogeneous. If it is difficult to obtain single size RNA molecules, it is better to design ligation sites within a nicked RNA duplex and use T4 RNA ligase 2 for ligation or employ DNA splinted ligation with T4 DNA ligase.
We finally remark that the presented two-step ligation can easily be extended to more than three segments without loss in yield, e.g. by carrying out two DNA splinted ligations followed by ligation of the products using T4 RNA ligase 1. For four segments, the amount of side products in a one-pot ligation is likely to increase. Whether the yield will be sufficient for NMR studies depends on the extent to which segments can be designed to form stable native hybridizations.
CONCLUSIONS
Multiple segmental labeling of RNA with three segments has been demonstrated via two efficient and straightforward protocols, in which an RNA segment, selectively labeled with 13C9/15N2/2H(1′, 3′, 4′, 5′, 5′′)-labeled uridine residues, is introduced into the central position of the 20 kDa RNA encapsidation signal of DHBV. The multiple segmental labeled RNA molecules were obtained via a two-step and one-pot ligation reaction the latter giving somewhat lower yields and more intermediate and side products. The generally used protection of termini is minimized, which simplifies preparation and purification of segments. The choice of segmentation sites is crucial, to prevent trapping of segments into stable alternate structures. Our investigations with DINAMelt clearly demonstrate that ligation sites can reliably be predicted. The simplified NMR imino spectrum of the segmental labeled RNA provided direct evidence for the presence of a U28 H3-imino resonance, previously obscured in the spectrum of the fully labeled sample, and thus of the non-canonical base pair U28:C37. The described segmental labeling approaches can potentially be extended to more than three segments and to larger RNA molecules, allowing studies of structures and dynamics of key elements in large biological functional RNA molecules and RNA–protein complexes. Virtually, any segmental labeling pattern is achievable for a large RNA molecule. However, crucial parameters like thermodynamic stability of alternative structures of segments and accessibility for ligation should be simulated and carefully judged before initializing experiments.
ACKNOWLEDGEMENTS
Funding within context of the 6th framework program of the EU, STREP project FSG-V-RNA is gratefully acknowledged. We thank Dr Kirsten Ampt for critical reading of this article and Dr Frederic Girard for helpful discussions. Funding to pay the Open Access publication charges for this article was provided by EU / FP6 / STREP FSG-V-RNA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Batey RT, Battiste JL, Williamson JR. Preparation of isotopically enriched RNAs for heteronuclear NMR. Methods Enzymol. 1995;261:300–322. doi: 10.1016/s0076-6879(95)61015-4. [DOI] [PubMed] [Google Scholar]
- 2.Al-Hashimi HM. Dynamics-based amplification of RNA function and its characterization by using NMR spectroscopy. Chembiochem. 2005;6:1506–1519. doi: 10.1002/cbic.200500002. [DOI] [PubMed] [Google Scholar]
- 3.Cromsigt J, van Buuren B, Schleucher J, Wijmenga S. Resonance assignment and structure determination for RNA. Methods Enzymol. 2001;338:371–399. doi: 10.1016/s0076-6879(02)38229-6. [DOI] [PubMed] [Google Scholar]
- 4.Flinders J, Dieckmann T. NMR spectroscopy of ribonucleic acids. Prog. NMR Spect. 2006;48:137–159. [Google Scholar]
- 5.Furtig B, Richter C, Wohnert J, Schwalbe H. NMR spectroscopy of RNA. Chembiochem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- 6.Latham MR, Brown DJ, McCallum SA, Pardi A. NMR methods for studying the structure and dynamics of RNA. Chembiochem. 2005;6:1492–1505. doi: 10.1002/cbic.200500123. [DOI] [PubMed] [Google Scholar]
- 7.Lukavsky PJ, Puglisi JD. Structure determination of large biological RNAs. Methods Enzymol. 2005;394:399–416. doi: 10.1016/S0076-6879(05)94016-0. [DOI] [PubMed] [Google Scholar]
- 8.Tzakos AG, Grace CRR, Lukavsky PJ, Riek R. NMR techniques for very large proteins and RNAs in solution. Annu. Rev. Biophys. Biomol. Struct. 2006;35:319–342. doi: 10.1146/annurev.biophys.35.040405.102034. [DOI] [PubMed] [Google Scholar]
- 9.Wijmenga SS, van Buuren BNM. The use of NMR methods for conformational studies of nucleic acids. Prog. NMR Spect. 1998;32:287–387. [Google Scholar]
- 10.Wu HH, Finger LD, Feigon J. Structure determination of protein/RNA complexes by NMR. Methods Enzymol. 2005;394:525–545. doi: 10.1016/S0076-6879(05)94022-6. [DOI] [PubMed] [Google Scholar]
- 11.Al-Hashimi HM. Beyond static structures of RNA by NMR: folding, refolding, and dynamics at atomic resolution. Biopolymers. 2007;86:345–347. doi: 10.1002/bip.20754. [DOI] [PubMed] [Google Scholar]
- 12.Getz M, Sun XY, Casiano-Negroni A, Zhang Q, Al-Hashimi HM. NMR studies of RNA dynamics and structural plasticity using NMR residual dipolar couplings. Biopolymers. 2007;86:384–402. doi: 10.1002/bip.20765. [DOI] [PubMed] [Google Scholar]
- 13.Cromsigt J, Schleucher J, Gustafsson T, Kihlberg J, Wijmenga S. Preparation of partially H-2/C-13-labelled RNA for NMR studies. Stereo-specific deuteration of the H5′′ in nucleotides. Nucleic Acids Res. 2002;30:1639–1645. doi: 10.1093/nar/30.7.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cromsigt JAMTC, Schleucher J, Kidd-Ljunggren K, Wijmenga SS. Synthesis of specifically deuterated nucleotides for NMR studies on RNA. J. Biomol. Struct. Dyn. 2000;Sp. Iss. S2:211–219. doi: 10.1080/07391102.2000.10506624. [DOI] [PubMed] [Google Scholar]
- 15.Nikonowicz EP. Preparation and use of H-2-labeled RNA oligonucleotides in nuclear magnetic resonance studies. Methods Enzymol. 2001;338:320–341. doi: 10.1016/s0076-6879(02)38227-2. [DOI] [PubMed] [Google Scholar]
- 16.Tolbert TJ, Williamson JR. Preparation of specifically deuterated RNA for NMR studies using a combination of chemical and enzymatic synthesis. J. Am. Chem. Soc. 1996;118:7929–7940. [Google Scholar]
- 17.Tolbert TJ, Williamson JR. Preparation of specifically deuterated and C-13-labeled RNA for NMR studies using enzymatic synthesis. J. Am. Chem. Soc. 1997;119:12100–12108. [Google Scholar]
- 18.D'Souza V, Dey A, Habib D, Summers MF. NMR structure of the 101-nucleotide core encapsidation signal of the Moloney murine leukemia virus. J. Mol. Biol. 2004;337:427–442. doi: 10.1016/j.jmb.2004.01.037. [DOI] [PubMed] [Google Scholar]
- 19.D'Souza V, Summers MF. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature. 2004;431:586–590. doi: 10.1038/nature02944. [DOI] [PubMed] [Google Scholar]
- 20.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCVIRES domain II determined by NMR. Nat. Struct. Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 21.Kim I, Lukavsky PJ, Puglisi JD. NMR study of 100 kDa HCV IRES RNA using segmental isotope labeling. J. Am. Chem. Soc. 2002;124:9338–9339. doi: 10.1021/ja026647w. [DOI] [PubMed] [Google Scholar]
- 22.Ohtsuki T, Kawai G, Watanabe K. Stable isotope edited NMR analysis of Ascaris suum mitochondrial tRNA(Met) having a TV-replacement loop. J. Biochem. 1998;124:28–34. doi: 10.1093/oxfordjournals.jbchem.a022092. [DOI] [PubMed] [Google Scholar]
- 23.Ohtsuki T, Kawai G, Watanabe Y, Kita K, Nishikawa K, Watanabe K. Preparation of biologically active Ascaris suum mitochondrial tRNA(Met) with a TV-replacement loop by ligation of chemically synthesized RNA fragments. Nucleic Acids Res. 1996;24:662–667. doi: 10.1093/nar/24.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurata S, Ohtsuki T, Suzuki T, Watanabe K. Quick two-step RNA ligation employing periodate oxidation. Nucleic Acids Res. 2003;31:e145–e151. doi: 10.1093/nar/gng145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tzakos AG, Easton LE, Lukavsky PJ. Complementary segmental labeling of large RNAs: economic preparation and simplified NMR spectra for measurement of more RDCs. J. Am. Chem. Soc. 2006;128:13344–13345. doi: 10.1021/ja064807o. [DOI] [PubMed] [Google Scholar]
- 26.Xu J, Lapham J, Crothers DM. Determining RNA solution structure by segmental isotopic labeling and NMR: application to Caenorhabditis elegans spliced leader RNA 1. Proc. Natl Acad. Sci. USA. 1996;93:44–48. doi: 10.1073/pnas.93.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA-polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleppe K, Sande J.HVD, Khorana HG. Polynucleotide ligase-catalyzed joining of deoxyribo-oligonucleotides on ribopolynucleotide templates and ribo-oligonucleotides on deoxyribopolynucleotide templates. Proc. Natl Acad. Sci. USA. 1970;67:68–73. doi: 10.1073/pnas.67.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romaniuk PJ, Uhlenbeck OC. Joining of RNA molecules with RNA ligase. Methods Enzymol. 1983;100:52–59. doi: 10.1016/0076-6879(83)00045-2. [DOI] [PubMed] [Google Scholar]
- 30.Bullard DR, Bowater RP. Direct comparison of nick-joining activity of the nucleic acid ligases from bacteriophage T4. Biochem. J. 2006;398:135–144. doi: 10.1042/BJ20060313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marsh TL, Pace NR. Ribonuclease-P catalysis differs from ribosomal-RNA self-splicing. Science. 1985;229:79–81. doi: 10.1126/science.4012313. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 3rd. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 2001. 9.62–9.65. [Google Scholar]
- 33.Price SR, Ito N, Oubridge C, Avis JM, Nagai K. Crystallization of RNA-protein complexes. 1. Methods for the large-scale preparation of RNA suitable for crystallographic studies. J. Mol. Biol. 1995;249:398–408. doi: 10.1006/jmbi.1995.0305. [DOI] [PubMed] [Google Scholar]
- 34.Beck J, Nassal M. Formation of a functional hepatitis B virus replication initiation complex involves a major structural alteration in the RNA template. Mol. Cell. Biol. 1998;18:6265–6272. doi: 10.1128/mcb.18.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beck J, Nassal M. Reconstitution of a functional duck hepatitis B virus replication initiation complex from separate reverse transcriptase domains expressed in Escherichia coli. J. Virol. 2001;75:7410–7419. doi: 10.1128/JVI.75.16.7410-7419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beck J, Nassal M. Efficient hsp90-independent in vitro activation by Hsc70 and Hsp40 of duck hepatitis B virus reverse transcriptase, an assumed Hsp90 client protein. J. Biol. Chem. 2003;278:36128–36138. doi: 10.1074/jbc.M301069200. [DOI] [PubMed] [Google Scholar]
- 37.Beck J, Nassal M. Hepatitis B virus replication. World J. Gastroentero. 2007;13:48–64. doi: 10.3748/wjg.v13.i1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Girard FC, Ottink OM, Ampt KAM, Tessari M, Wijmenga SS. Thermodynamics and NMR studies on duck, heron and human HBV encapsidation signals. Nucleic Acids Res. 2007;35:2800–2811. doi: 10.1093/nar/gkm131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRpipe - a multidimensional spectral processing system based on unix pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]