Abstract
We present a protein–DNA docking benchmark containing 47 unbound–unbound test cases of which 13 are classified as easy, 22 as intermediate and 12 as difficult cases. The latter shows considerable structural rearrangement upon complex formation. DNA-specific modifications such as flipped out bases and base modifications are included. The benchmark covers all major groups of DNA-binding proteins according to the classification of Luscombe et al., except for the zipper-type group. The variety in test cases make this non-redundant benchmark a useful tool for comparison and development of protein–DNA docking methods. The benchmark is freely available as download from the internet.
INTRODUCTION
Biomolecular docking has become a mature discipline within structural biology (1). Docking aims at predicting the structure of a complex given the 3D structures of its components. The field of protein–protein docking in particular has seen extensive progress over the last decade as witnessed by recent CAPRI (Critical Assessment of Predicted Interactions) results, a community-wide blind docking experiment (2). For protein–DNA docking, however, progress lags behind. The scarcity of information for a proper identification of interaction surfaces on DNA and its inherent flexibility have hampered the development of effective docking methods. The field of protein–DNA docking is, however, receiving increased attention and efforts are put into the development of docking methods that address the above mentioned limitations (3). Considering the importance of biomolecular interactions in system biology, gaining insight into the biochemistry of recognition and gene expression is highly relevant (4). New developments in protein–DNA docking approaches are therefore expected.
A set of well-defined test cases that form a common ground for validating and comparing the different docking methods would facilitate the development of effective protein–DNA docking methods. Such a benchmark should contain the native structures of both protein and DNA in their unbound form together with the reference structure of the complex.
We have constructed a benchmark of 47 protein–DNA test cases in a similar manner as has been done for protein–protein docking (5). The benchmark covers all major groups of protein–DNA complexes according to the classification proposed by Luscombe et al. (6) except for the zipper-type group. It contains a variety of challenging systems in terms of size of the interaction interface, number of individual components present in the complex and conformational changes that the unbound components undergo upon complex formation. Its diversity makes it a comparison tool for different docking methods as their performance may vary depending on the type of complexes. This benchmark should benefit the entire docking community and offer a starting-point for the improvement of various algorithms.
MATERIALS AND METHODS
RCSB Protein Data Bank (PDB) query
A non-redundant benchmark was generated from structures deposited in the RCSB PDB (7). The PDB (as of September 2007) was queried for all entries containing X-ray crystallographic structures with a resolution better than 3.0 Å containing both protein and DNA. Complexes containing DNA structures with a sequence length smaller than 8 bp and protein structures containing mutations in the core and or interface region were removed.
For the resulting complexes, the PDB was queried for unbound protein entries. Structures resolved using NMR or X-ray crystallography with a resolution better than 3.0 Å were retrieved. Structures with a sequence similarity larger than or equal to 90% were removed. Structures were regarded as redundant if the raw alignment score is positive, >80% of their sequences are aligned and >60% of the sequences are identical. Sequence alignments were performed using the Needleman–Wunsch algorithm as implemented in the LSQMAN software package (8) with a gap penalty of 5.
Generation of unbound DNA models
Models for unbound DNA were generated using the DNA analysis and rebuilding program 3DNA (9) with the base-pair sequence of the DNA in the reference complex. The models were generated in canonical B-DNA conformation (fiber model 4) using the nucleotide building blocks as determined in the fiber diffraction studies of Chandrasekaran and Arnott (10). Structures with overhanging base-pairs were converted to all-paired structures by adding their Watson–Crick counterparts.
Structure post-processing
The residue numbering of the bound and unbound components was matched to allow for easy comparison. The DNA was assigned one chain identifier and renumbered. Structures of unbound proteins that contain more than one chain were assigned a single chain identifier instead of being separated into their individual components; residues were renumbered to avoid overlap in numbering. Atom and residue names were matched to the topallhdg5.3.pro (11) and dna-rna_allatom.top topology files (12) naming for direct use in HADDOCK (13).
Analysis
The size of the interaction interface between protein and DNA is expressed in terms of the buried surface area (BSA, Table 1) of the DNA in the complex. The BSA was calculated using NACCESS (Hubbard, S. J., Thornton, J. M. 1993) with a probe radius of 1.4 Å. The conformational changes between the unbound and the bound states are expressed in terms of the root mean square deviation (RMSD) calculated using ProFit (Martin, A.C.R., http://www.bioinf.org.uk/software/profit/). These were calculated in three different ways:
Conformational change of the protein–DNA interface was calculated by superimposition of all Cα and phosphate atoms at the interface. Residues belonging to the interface are identified as those having atoms within 5.0 Å intermolecular distance of one another (RMSD Inter., Table 1). The interface RMSD values were used to classify the test cases as ‘easy’, ‘intermediate’ or ‘difficult’ (see below).
As the conformational change in the DNA tends to affect the complete molecule, the RMSD of the DNA was calculated by superimposition of all phosphate atoms (RMSD DNA, Table 1).
Conformational changes in the protein, such as global domain reorientations and flexible segments not located at the interface are represented by means of the RMSD calculated over all Cα atoms of the protein (RMSD Prot, Table 1).
Table 1.
The protein–DNA benchmark
| Complex |
Protein |
DNA | RMSD |
||||||
|---|---|---|---|---|---|---|---|---|---|
| PDB ida | Cat.b | PDB ida | Description | Sequence 5′-3′c | Nr.d | BSAe | Inter.f | DNAg | Proth |
| ‘Easy’ targets | |||||||||
| 2c5r | 1 | 2bnkX | Phage PHI29 replication organizer protein P16.7 | TCCACCGG | 4 | 402 | 0.49 | 0.49 | 0.82 |
| 1pt3 (A:C:D) | 8 | 1m08X | Col-E7 nuclease domain | GCGATCGC | 2 | 730 | 1.35 | 2.09 | 1.36 |
| 1mnn | 1 | 1mn4X | Sporulation specific transcription factor NDT80 | TGCGACACAAAAACT | 2 | 1292 | 1.48 | 1.81 | 0.83 |
| 1fok | 1 | 2fokX | Restriction endonuclease FOKI | TCGGATGATAACGCTAGTCAT | 2 | 1920 | 1.53 | 2.51 | 1.09 |
| 1ksy (A:C:D:F) | 4 | 1f08X | Papillomavirus replication initiation domain E-1 | ATAATTGTTGTCAACAATTAT | 3 | 1020 | 1.58 | 2.56 | 0.52 |
| 3cro | 1 | 1zugN | Phage 434 CRO | AAGTACAAACTTTCTTGTAT | 3 | 1473 | 1.58 | 2.66 | 1.17 |
| 1emh | 8 | 1akzX | Human uracil-DNA glucosylase | TGT(P2U)ATCTTT | 2 | 869 | 1.62 | 4.53 | 1.46 |
| 1h9t | 1 | 1e2xX | FADR, fatty acid responsive transcription factor | CATCTGGTACGACCAGATC | 3 | 1622 | 1.68 | 3.88 | 0.77 |
| 1tro (A:C:I:J) | 1 | 3wrpX | TRP repressor | TGTACTAGTTAACTAGTACA | 3 | 1540 | 1.70 | 3.08 | 1.42 |
| 1by4 (A:B:E:F) | 2 | 1rxrN | Retinoid X receptor DNA binding domain | TAGGTCAAAGGTCAG | 3 | 1480 | 1.77 | 1.46 | 2.23 |
| 1hjc (A:B:C) | 5 | 1eanX | RUNX1 runt domain | GAACTCTGTGGTTGCGG | 2 | 634 | 1.80 | 2.88 | 0.97 |
| 1diz (A:E:F) | 8 | 1mpgX | E. coli 3-methyladenine DNA glycosylase II | TGACATGA(NRI)TGCCT | 2 | 805 | 1.82 | 5.80 | 0.46 |
| 1rpe | 1 | 1r63N | Phage 434 repressor | ACAAACAAGATACATTGTATA | 3 | 1430 | 1.87 | 2.97 | 0.94 |
| ‘Intermediate’ targets | |||||||||
| 1vrr | 8 | 1sdoX | Restriction endonuclease BSTYI | TTATAGATCTATAA | 3 | 2098 | 2.08 | 2.11 | 2.22 |
| 1f4k | 1 | 1bm9X | Replication terminator protein | CTATGAACATAATGTTCATAG | 3 | 1741 | 2.26 | 1.94 | 2.29 |
| 1k79 (A:B:C) | 1 | 1gvjX | ETS-1 DNA binding and autoinhibitory domain | TAGTGCCGGAAATGTG | 2 | 912 | 2.37 | 3.82 | 0.80 |
| 1kc6 (A:B:E:F) | 8 | 2audX | Restriction endonuclease HINCII | CCGGTCGACCGG | 3 | 2658 | 2.38 | 4.67 | 1.38 |
| 1ea4 (D:E:F:G:W:X) | 6 | 2cpgX | Transcription repressor COPG | TAACCGTGCACTCAATGCAATC | 3 | 1473 | 2.43 | 4.48 | 0.64 |
| 1z63 (A:C:D) | 8 | 1z6aX | Sulfolobus solfataricus SWI2/SNF2 ATPase core domain | ATTGCCGAAGACGAAAAAAA | 2 | 603 | 2.51 | 2.74 | 2.27 |
| 1r4o | 2 | 1gdcN | Glucocorticoid receptor | CCAGAACATCGATGTTCTGT | 3 | 1401 | 2.61 | 3.05 | 1.91 |
| 1azp | 6 | 1sapN | Hyperthermophile chromosomal protein SAC7D | GCGATCGC | 2 | 778 | 2.70 | 3.77 | 2.76 |
| 1w0t | 1 | 1ba5N | HTRF1 DNA-binding domain | CTGTTAGGGTTAGGGTTAGA | 3 | 1545 | 2.78 | 3.20 | 2.47 |
| 1cma | 6 | 1mjkX | Methionine repressor | TTAGACGTCT | 2 | 775 | 2.81 | 2.60 | 2.05 |
| 1jj4 | 4 | 1f9fX | Papillomavirus type 18 E2 | CAACCGAATTCGGTTG | 2 | 1169 | 2.83 | 3.32 | 2.25 |
| 1vas | 8 | 1eniX | T4 pyrimidine dimer specific excision repair | ATCGCGTTGCGCT | 2 | 1445 | 3.04 | 6.99 | 1.42 |
| 4ktq | 8 | 1ktqX | DNA polymerase I | GACCACGGCGC(DOC) | 2 | 1685 | 3.23 | 3.64 | 1.97 |
| 1z9c (A:C:D) | 1 | 1z91X | Organic hydroperoxide resistence transcription regulator | TACAATTTAATTGTATACAATT TAATTGTA | 3 | 2107 | 3.24 | 4.26 | 4.18 |
| 1ddn | 1 | 2tdxX | Diphtheria TOX repressor | ATATAATTAGGATAGCTTTACC TAATTATTTTAA | 5 | 2877 | 3.26 | 7.25 | 0.50 |
| 2irf | 1 | 1irgN | Interferon Regulatory Factor 2 | AAGTGAAAGUGA | 2 | 898 | 3.35 | 2.23 | 3.83 |
| 1jt0 | 1 | 1jusX | Multidrug binding transcription factor QACR | CTTATAGACCGATCGATCGG TCTATAAG | 2 | 2484 | 3.49 | 4.58 | 3.53 |
| 1g9z | 8 | 2o7mX | I-CreI endonuclease | GCAAAACGTCGTGAGACAGTTTCG | 2 | 3255 | 3.67 | 5.02 | 4.21 |
| 1a73 | 8 | 1evxX | Intron-encoded homing endonuclease I-PPOI | TTGACTCTCTTAAGAGAGTCA | 2 | 2076 | 4.26 | 8.22 | 1.20 |
| 2fio | 4 | 2fibX | Phage PHI29 transcription regulator P4 | AAAAACGTCAACATTTTATA AAAAAGTCTTGCAAAAAGT | 2 | 1114 | 4.41 | 8.03 | 0.67 |
| 1qne (A:C:D) | 5 | 1vokX | Adenovirus major late promotor TBP | GCTATAAAAGGGCA | 2 | 1487 | 4.57 | 8.54 | 0.89 |
| 1zs4 | 1 | 1zpqX | Phage lambda CII | CCTCGTTGCGTTTGTTTGCACGAAT | 2 | 1358 | 4.71 | 2.97 | 3.77 |
| ‘Difficult’ targets | |||||||||
| 1qrv | 4 | 1hmaN | High mobility group protein D | GCGATATCGC | 3 | 1204 | 5.19 | 7.68 | 3.91 |
| 1o3t | 1 | 1g6nX | CAP-CAMP | GCTTTTTACGCTAGATCTA GCGTAAAAAGCGC | 2 | 1277 | 5.20 | 10.6 | 2.55 |
| 1b3t | 4 | 1vhiX | Epstein-Barr virus nuclear antigen-1 | GGAAGCATATGCTTCCC | 2 | 2627 | 5.32 | 3.91 | 3.53 |
| 3bam | 8 | 1bamX | Restriction endonuclease BAMHI | TATGGATCCATA | 3 | 2208 | 5.55 | 2.19 | 4.50 |
| 1rva | 8 | 1rveX | Eco RV endonuclease | AAAGATATCTTT | 2 | 2350 | 5.68 | 9.78 | 3.88 |
| 1zme | 2 | 1ajyN | Proline utilization transcription activator PUT3 | ACGGGAAGCCAACTCCGT | 2 | 1362 | 5.76 | 4.68 | 8.64 |
| 1dfm | 8 | 1es8X | Restriction endonuclease BGLII | TATTATAGATCTATAAAT | 3 | 2735 | 6.31 | 3.04 | 4.68 |
| 1bdt | 6 | 1arqN | Phage P22 Arc gene regulating protein | TATAGTAGAGTGCTTCTATCATT | 3 | 2109 | 6.45 | 4.90 | 5.20 |
| 7mht | 8 | 2hmyX | HHAI methyltransferase | GTCAGCGCATGG | 2 | 1613 | 6.71 | 2.55 | 3.84 |
| 2fl3 | 8 | 1ynmX | Restriction endonuclease HINP1I | CCAGCGCTGG | 2 | 1670 | 6.71 | 2.95 | 4.37 |
| 1eyu | 8 | 1pvuX | PVUII endonuclease | TGACCAGCTGGTCA | 2 | 2068 | 6.82 | 4.49 | 6.36 |
| 2oaa | 8 | 2oa9X | Restriction endonuclease MVAI | GGTACCTGGATG | 2 | 2009 | 8.95 | 8.15 | 8.02 |
aThe RCSB PDB accession number for the structures used. Specific chains are in parenthesis. Structures for the unbound protein were either solved by X-ray crystallography (X) or NMR spectroscopy (N).
bThe classification of the protein–DNA complexes in eight different groups according to the scheme of Luscombe et al. (6).
cThe base sequence of the DNA in the bound complex also used for generating the unbound DNA structure. Some sequences contain modified bases. These are: DOC (2′,3′-dideoxycytidine-5′-monophosphate), NRI (phosphoric acid mono-(4-hydroxy-pyrrolidin-3-ylmethyl) ester) and P2U (2′-deoxy-pseudouridine-5′monophosphate).
dThe number of individual biomolecules that need to be docked to reconstruct the complex.
eBuried surface area of the DNA upon complex formation in Å2.
fThe RMSD (Å) from the bound form calculated over the interface Cα and phosphate atoms of the unbound protein structure after superposition onto the reference complex.
gThe RMSD (Å) from the bound form calculated over all phosphate atoms of the unbound DNA after superposition onto the reference complex.
hThe RMSD (Å) from the bound form calculated over Cα atoms of the unbound protein after superposition onto the reference complex.
COMPOSITION OF THE BENCHMARK
The protein–DNA benchmark version 1.0 (Table 1) contains 47 test cases. For all test cases, the unbound structures of both protein and DNA are available. In addition, the reference complexes have been separated into their DNA and protein bound forms. This should allow to evaluate the performance of a docking method for bound–bound, bound–unbound and unbound–unbound cases. Although the reference structure is always from X-ray crystallography, the unbound proteins contain both solution NMR and X-ray structures. The use of an ensemble of NMR structures as starting point for the docking provides an easy way for various docking algorithms to sample additional conformational space. The benchmark contains members of all major structural groups described by Luscombe et al. (6) apart from the zipper-type group. These are: 16 helix–turn–helix (group 1), three zinc-coordinating (group 2), five other α-helix (group 4), two β-sheet (group 5), four β-hairpin/ribbon (group 6) and 17 enzyme (group 8) complexes.
Each test case in the benchmark poses its own challenges for a docking algorithm. A common theme throughout the benchmark is ‘conformational changes’ either in the protein, the DNA or both. This benchmark differs from its protein–protein counterpart by the omnipresence of conformation changes. To provide some structure in the test cases, we classified them as ‘easy’, ‘intermediate’ or ‘difficult’. This classification is based on the interface RMSD values between the bound and unbound components of the complex:
‘easy’ test case: interface RMSD between 0.0 Å and 2.0 Å
‘intermediate’ test case: interface RMSD between 2.0 Å and 5.0 Å
‘difficult’ test case: interface RMSD above 5.0 Å.
An ‘easy’ test case
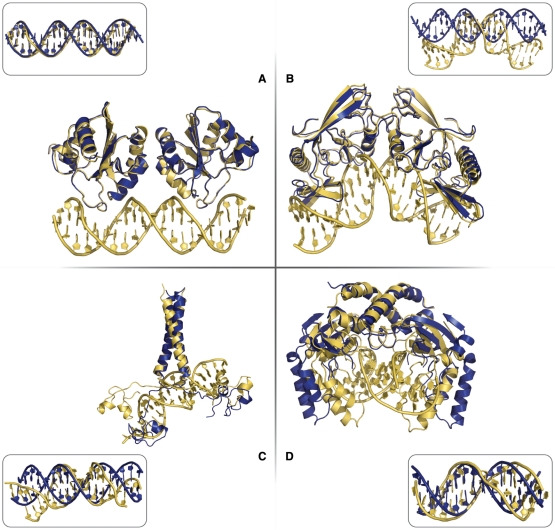
The individual components from this group of complexes do not change significantly the conformation of their interface upon binding. Conformational changes at the interface of the protein are mostly brought about by small flexible loop rearrangements. This does not mean that the components can always be regarded as rigid. Conformational changes at the interface of the DNA often cause the DNA to bend and twist in the interface region (see DNA RMSD values in Table 1). A representative example from this group is the Papillomavirus replication initiation domain E-1 (PDB entry 1ksy, Figure 1A).
Figure 1.
Illustration of ‘easy’ (interface RMSD < 2.0 Å), ‘intermediate’ (2.0 Å ≤ interface RMSD < 5.0 Å) and ‘difficult’ (interface RMSD ≥ 5.0 Å) test cases from the protein–DNA benchmark. ‘Easy’ test case: the Papillomavirus replication initiation domain E-1 (PDB id 1ksy) (interface RMSD = 1.6 Å) (A). ‘Intermediate’ test case: the intron-encoded homing endonuclease I-PPOI complex (PDB id 1a73) (interface RMSD = 4.3 Å) (B). ‘Difficult’ test cases: the proline utilization transcription activator (PDB id 1zme) (interface RMSD = 5.8 Å) (C) and the PVUII endonuclease complex (PDB id 1eyu) (interface RMSD = 6.8 Å) (D). The bound form of the complex is shown in yellow and the unbound protein in blue. The bound- and canonical B-form DNA structures are shown as insets to highlight the conformational changes in the DNA.
An ‘intermediate’ test case
Unbound components of this group undergo more pronounced structural rearrangements in their interface upon complex formation. The type of conformational changes involves global and local domain rearrangements in the protein and global conformational change in the DNA. An example is the intron-encoded homing endonuclease I-PPOI complex (PDB entry 1a73, Figure 1B), the protein shows little conformational change upon binding but the DNA is heavily kinked in its centre.
A ‘difficult’ test case
In the difficult cases, the extent of structural rearrangement upon complex formation increases even further. In addition to the conformational changes occurring in the ‘intermediate’ test cases, the ‘difficult’ group contains complexes with features like structural transitions and major domain reorientations in the protein. An example is the proline utilization transcription activator (PDB entry 1zme, Figure 1C), a protein that has two DNA interaction domains linked together by a long highly flexible loop; the dimerization interface connecting the two DNA interaction domains show a loop to sheet transition upon DNA binding. In the PVUII endonuclease complex (PDB entry 1eyu, Figure 1D), the individual protein chains do not show much conformational changes but a hinge point connecting them facilitates a ‘clamping’ motion upon binding. This results in a large RMSD between bound and unbound structures. This is an example of global domain motions upon binding.
The benchmark also contains several structures with special features such as strand breaks (PDB entries 1g9z, 1o3t and 3bam) and flipped out bases in the DNA (PDB entries 1diz, 1emh, 1vas and 7mht).
We constructed this benchmark as a test base to stimulate developments in the field of protein–DNA docking and will use it in particular for further developing our own protein–DNA docking approach (3). Ideally, the classification of ‘easy’, ‘intermediate’ or ‘difficult’ could have been based on docking results; at this stage, however, we chose to purely base it on conformational changes as measured by the RMSDs between bound and unbound form. Basing the classification on HADDOCK results would have introduced a bias not only toward the amount of conformational changes, but also toward our ability to predict protein–DNA interfaces since HADDOCK requires some kind of input to drive the docking process. We will of course proceed with evaluating our performance on this benchmark, but this is outside the scope of this article.
In conclusion, allowing for structural rearrangements in both protein and DNA during docking, while maintaining the helical character of DNA is a major challenge in protein–DNA docking. The large variety of protein–DNA complexes in the benchmark should provide a valuable test set to evaluate and improve docking algorithms. Version 1.0 of the benchmark is available from the web site: http://haddock.chem.uu.nl/dna/benchmark.html
ACKNOWLEDGEMENTS
Financial support for this research and the Open Access publication charges for this article was provided by the European Community (FP6 STREP project ‘ExtendNMR’, contract no. LSHG-CT-2005-018988, FP6 I3 project ‘EU-NMR’, contract no. RII3-026145 and FP7 I3 project ‘eNMR’, contract no. 213010-e-NMR) and from a VICI grant from the Netherlands Organization for Scientific Research (NWO) to A.M.J.J.B. (grant no. 700.96.442).
Conflict of interest statement. None declared.
REFERENCES
- 1.van Dijk AD, Boelens R, Bonvin AM. Data-driven docking for the study of biomolecular complexes. FEBS J. 2005;272:293–312. doi: 10.1111/j.1742-4658.2004.04473.x. [DOI] [PubMed] [Google Scholar]
- 2.Janin J. The targets of CAPRI rounds 6-12. Proteins. 2007;69:699–703. doi: 10.1002/prot.21689. [DOI] [PubMed] [Google Scholar]
- 3.van Dijk M, van Dijk AD, Hsu V, Boelens R, Bonvin AM. Information-driven protein-DNA docking using HADDOCK: it is a matter of flexibility. Nucleic Acids Res. 2006;34:3317–3325. doi: 10.1093/nar/gkl412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhodes D, Schwabe JW, Chapman L, Fairall L. Towards an understanding of protein-DNA recognition. Phil. Trans. Roy. Soc. Lond. 1996;351:501–509. doi: 10.1098/rstb.1996.0048. [DOI] [PubMed] [Google Scholar]
- 5.Mintseris J, Wiehe K, Pierce B, Anderson R, Chen R, Janin J, Weng Z. Protein-Protein Docking Benchmark 2.0: an update. Proteins. 2005;60:214–216. doi: 10.1002/prot.20560. [DOI] [PubMed] [Google Scholar]
- 6.Luscombe NM, Austin SE, Berman HM, Thornton JM. An overview of the structures of protein-DNA complexes. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-1-reviews001. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:301–303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sierk ML, Kleywegt GJ. Deja vu all over again: finding and analyzing protein structure similarities. Structure. 2004;12:2103–2111. doi: 10.1016/j.str.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 9.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandrasekaran RA, Arnott S. The structures of DNA and RNA helices in oriented fibers. In: Saenger W, editor. Landolt-Börnstein Numerical Data and Functional Relationships in Science and Technology. Vol. VII/1b. Springer, Berlin: 1989. pp. 31–170. [Google Scholar]
- 11.Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 12.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 13.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]