Abstract
It is generally believed that basic features of ribosomal functions are universally valid, but a systematic test still stands out for higher eukaryotic 80S ribosomes. Here we report: (i) differences in tRNA and mRNA binding capabilities of eukaryotic and bacterial ribosomes and their subunits. Eukaryotic 40S subunits bind mRNA exclusively in the presence of cognate tRNA, whereas bacterial 30S do bind mRNA already in the absence of tRNA. 80S ribosomes bind mRNA efficiently in the absence of tRNA. In contrast, bacterial 70S interact with mRNA more productively in the presence rather than in the absence of tRNA. (ii) States of initiation (Pi), pre-translocation (PRE) and post-translocation (POST) of the ribosome were checked and no significant functional differences to the prokaryotic counterpart were observed including the reciprocal linkage between A and E sites. (iii) Eukaryotic ribosomes bind tetracycline with an affinity 15 times lower than that of bacterial ribosomes (Kd 30 μM and 1–2 μM, respectively). The drug does not effect enzymatic A-site occupation of 80S ribosomes in contrast to non-enzymatic tRNA binding to the A-site. Both observations explain the relative resistance of eukaryotic ribosomes to this antibiotic.
INTRODUCTION
Translational decoding and peptide-bond formation are reactions of the ribosomal elongation cycle, the most conserved phase of ribosomal functions. On the contrary, the initiation, termination and recycling phases show significant differences between bacterial ribosomes on one hand and archaeal and eukaryotic ribosomes on the other indicated by specific factors (1–3). Factors of the elongation phase such as EF-Tu and EF-G in bacteria and EF1 and EF2 in archaea and eukaryotes have a common ancestor, respectively, and demonstrate the same mode of actions, namely delivery of aminoacyl-tRNA to the ribosomal A-site (EF-Tu and EF1) and promotion of translocation (EF-G and EF2). Only under defined growth conditions such as low pH and high salts bacteria require a third elongation factor LepA (EF4) that is also present in all bacteria related organelles, namely mitochondria and chloroplasts (4; Z. Karim, M. Pech, Y. Qin and K.H. Nierhaus, unpublished data).
A characteristic feature of ribosomes from all three domains is the presence of three tRNA-binding sites, namely the A, P and E sites (5). Moreover, it was shown for bacteria (6) and yeast (7) that A and E ribosomal sites are connected via reciprocal linkage that is occupation of the E-site induces a low affinity for binding an A-tRNA, and vice versa, the E-tRNA release is triggered by A-site occupation, precisely after decoding and before accommodation (8). This allostery between A and E-sites is an intrinsic feature of ribosomes, and lower eukaryotes such as yeast even need a unique elongation factor 3 (eEF3) to execute the allosteric interaction. eEF3 is an essential ATPase and it is required after translocation for opening the E site to allow E-tRNA release upon A-site occupation (7,9).
Regardless of the above-mentioned similarities between prokaryotic and eukaryotic elongation cycles first results of cryo-EM analysis indicate that the dynamical behavior of eukaryotic and prokaryotic ribosomes is different (10). Another conspicuous difference concerns the mRNA tunnel upstream the E site. In 80S ribosomes the mRNA of this region is surrounded by proteins that are not present in prokaryotes, whereas in 70S ribosomes both ribosomal proteins and rRNA are found (11,12). Undoubtedly, discovery and design of new antibacterial drugs need information about organization, functions and evolutionary conservation of mammalian elongation apparatus. Here we compare some basic functional features of ribosomes from higher eukaryotes (rabbit liver) with those from bacteria concerning binding of mRNA and tRNA and the effects of tetracycline, member of one of the most widely used antibiotic family in human, livestock and plant therapy.
MATERIALS AND METHODS
Materials
Re-associated 80S ribosomes from rabbit liver free of endogenous tRNAs and mRNAs were prepared according to (13) with slight modifications: in order to wash endogenous RNases from polysomes the post-mitochondrial supernatant was centrifuged through a 1 M sucrose cushion, which contained 0.5 M KCl. Re-association was confirmed by analytical centrifugation for 18 000 r.p.m., 17 h (Beckman SW40 rotor). The ribosome concentrations were calculated assuming the following ratios: 60 pmol/A260 for 40S subunits, 30 pmol/A260 for 60S subunits and 20 pmol/A260 for 80S ribosomes.
Heteropolymeric MFVK-mRNA 5′-GGGAAAAGAAAAGAAAAGAAA-AUG-UUC-GUU-AAA-GAAAAGAAAAGAAAU-3′ is 49 nucleotides long with four codons Met-Phe-Val-Lys in the middle was prepared with run-off transcription, according to (14). Note that the MFVK-mRNA does not contain a classical Shine-Dalgarno (SD) sequence unless the 5′-GGG triplet which can function as a weak SD in the absence of tRNA.
tRNAPhe and tRNAVal were purified from tRNAbulk derived from bovine liver by HPLC chromatography using ion-exchange DEAE and reversed phase Hypersil 5 C4 columns. The plasmids for human  and yeast
and yeast  were kindly provided by Dr M. Mirande (Gif-sur-Yvette, France) and Dr E. Sattlegger (Auckland, New Zealand), respectively. N-acetyl-[3H]Phe-tRNAPhe (Ac[3H]Phe-tRNAPhe), [14C]Val-tRNAVal and [14C]Lys-
were kindly provided by Dr M. Mirande (Gif-sur-Yvette, France) and Dr E. Sattlegger (Auckland, New Zealand), respectively. N-acetyl-[3H]Phe-tRNAPhe (Ac[3H]Phe-tRNAPhe), [14C]Val-tRNAVal and [14C]Lys-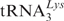 were purified by reversed-phase HPLC on a Nucleosil 300-5 C4 column using a methanol gradient as described (15). 3′-end 32P-labeling of tRNA was performed according to (16). 5′-end 32P-labeling of MFVK-mRNA was done according to (17).
were purified by reversed-phase HPLC on a Nucleosil 300-5 C4 column using a methanol gradient as described (15). 3′-end 32P-labeling of tRNA was performed according to (16). 5′-end 32P-labeling of MFVK-mRNA was done according to (17).
The elongation factors eEF1A and eEF2 from rabbit liver were isolated using combination of gel-filtration and ion-exchange chromatography as described (18,19).
All complexes were prepared in polyamine buffer A, which contained 20 mM HEPES-KOH (pH 7.5 at 0°C), 5 mM MgCl2, 100 mM KCl, 0.6 mM spermine, 0.8 mM spermidine and 6 mM 2-mercaptoethanol (20).
mRNA binding to ribosome and ribosomal subunits
Pre-incubation of 40S, 60S and 80S ribosomes with 32P-labeled heteropolymeric MFVK-mRNA (100 d.p.m./pmol) during 2 min at 37°C (molar ratio mRNA:40S/60S/80S = 10:1) was followed by an incubation for 15 min at 37°C in the presence or absence of deacylated tRNA (molar ratio tRNA:40S/60S/80S = 2:1). Then 40 pmol of each sample were loaded on 10–30% sucrose gradient (Beckman SW40 rotor) in buffer A and were centrifuged for 18 000 r.p.m., 17 h.
This technique was chosen instead of nitrocellulose filtration to decrease background level. A pre-treatment of filters with 0.5 M KOH (25°C, 25 min) (21) proposed as solution for this trouble for short oligonucleotides did not work in our case for heteropolymeric mRNA (49 nt).
The experiments with bacterial 70S and 30S were done in parallel in ionic conditions adjusted for an optimal bacterial translational system [20 mM HEPES-KOH (pH 7.5 at 0°C), 4.5 mM MgCl2, 6 mM 2-mercaptoethanol, 150 mM KCl, 0.05 mM spermine and 2 mM spermidine] (8,22).
Construction of the Pi complex
Re-associated 80S ribosomes (0.4 µM) were incubated with MFVK-mRNA for 2 min at 37°C. Then Ac[3H]Phe-tRNAPhe (2400 d.p.m./pmol) in indicated amounts was added and incubation continued for 20 min at 37°C. The nitrocellulose filtration (tRNA binding) and the puromycin reaction were performed as described elsewhere (23).
Construction of the PRE complex
80S ribosomes programmed with MFVK-mRNA and occupied by Ac[3H]Phe-tRNAPhe (described above) were incubated for 15 min at 37°C with pre-formed ternary complex (3 min 37°C; buffer A with 0.4 mM GTP, 2 mM phosphoenolpyruvate, 0.2 mg/ml pyruvate kinase; molar ratio ternary complex:80S = 2:1) containing eEF1A·GTP·[14C]Val-tRNAVal (580 dpm/pmol) for enzymatic A-site binding. The puromycin reaction yielded no product indicating that tRNAPhe was at the P site and di-peptide Phe-Val-tRNA at the A site. In experiments with non-hydrolysable analog of GTP, ternary complexes were formed and bound to 80S ribosomes in the presence of 0.4 mM GDPNP.
Monitoring di-peptide formation by HPLC
Aliquots of PRE complex containing 2 pmol of 80S were withdrawn and the binding and puromycin reactivity were assessed as described above. Another 8–10 pmol were analyzed for di-peptide synthesis by HPLC chromatography according to (24). The amount of formed di-peptide was normalized to the active fraction of ribosomes determined by the amount of bound Ac[3H]Phe-tRNAPhe.
Construction of the POST complex and subsequent ternary complex binding
Pi complexes programmed with MFVK-mRNA and Ac[3H]Val-tRNAVal (2500 d.p.m./pmol) at the P site were incubated with [32P]tRNAPhe (3000 d.p.m./pmol) to occupy the E site. The puromycin reaction indicated that about 70% of AcVal-tRNA remained at the P site. In such a manner a POST complex could be constructed without elongation factor 2 (eEF2) avoiding a purification (ultracentrifugation) step, which can be harmful for ribosomes. Increasing amounts of ternary complexes containing [14C]Lys-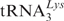 (680 dpm/pmol; AAA codon at the A site) were added. The fate of each tRNA could be monitored due to three different isotopic labels used. This set of experiments was carried out in the presence of 3 mM ATP.
(680 dpm/pmol; AAA codon at the A site) were added. The fate of each tRNA could be monitored due to three different isotopic labels used. This set of experiments was carried out in the presence of 3 mM ATP.
Susceptibility to tetracycline
Stock solution (15 mM) of tetracycline (USB Corp., USA) contained 25% ethanol and 10 mM Tris, the pH was adjusted to 7.8 and the solution was used immediately (25). Controls (minus tetracycline) were performed with assay mixtures containing corresponding concentrations of ethanol and Tris. In all cases parallel assays with 70S ribosomes were used as control and reference.
Poly(U)-directed poly(Phe) synthesis by E. coli ribosomes was performed as described in (22) in the presence of appropriate amounts of tetracycline (0.1 × 10–6 to 300 × 10–6 M) in a polyamine buffer containing 4.5 mM Mg2+, 3 mM ATP, 1.5 mM GTP, 5 mM acetyl-phosphate, 40 µg poly(U), 5 pmol 70S ribosomes, 140 µM [14C]Phe (∼60 dpm/pmol), 15 A260/ml tRNAbulk and S100 fraction. The amount of hot TCA precipitable radioactive material was determined after 60 min incubation at 37°C.
For the mammalian translation system optimal conditions were used according to ref. (26), namely 20 mM Hepes–KOH (pH 7.5 at 0°C), 2.5 mM Mg(acetate)2, 150 mM K(acetate), 3.5 mM spermidine, 4 mM β-mercaptoethanol, 9% glycerol, 21 mM phosphocreatine, 0.9 mM ATP, 0.1 mM GTP. The system was homogeneous: ribosomes and S-100 proteins were from rabbit liver and tRNAbulk from bovine liver. Overall procedure was the same as that used for E. coli translation system.
To test a possible influence of the drug on the P-site occupation MFVK-mRNA programmed ribosomes were incubated with increased amount of tetracycline (50 × 10–6 to 500 × 10–6 M) 10 min at 37°C and then Ac[14C]Phe-tRNA (1147 d.p.m./pmol) was added as described above.
A-site binding was tested as follows: ribosomes carrying deacylated tRNA at the P site were incubated with the drug (50 × 10–6 to 500 × 10–6 M), thereafter a two-molar excess of AcPhe-tRNA or ternary complex was added as substrate for the A site.
Affinity of tetracycline binding to free ribosomes was studied using [3H]tetracycline (NEN, Inc., 500 d.p.m./pmol).
RESULTS
mRNA and tRNA binding
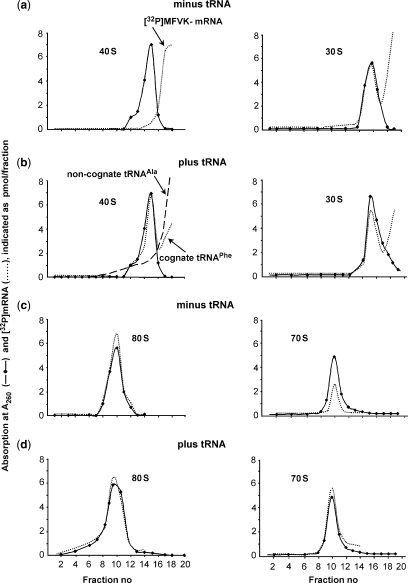
Binding of [32P]mRNA to pro- and eukaryotic ribosomes and their small subunits in the absence and presence of tRNA was compared by ultracentrifugation in 10–30% sucrose gradients (Figure 1). 40S subunits bind mRNA only in the presence of cognate tRNAPhe (Figure 1a and b, left panels). Replacing tRNAPhe by non-cognate tRNAAla abolished binding to background level (Figure 1b, dashed line). In contrast, 30S can interact with mRNA even in the absence of tRNA (Figure 1a and b, right panels). In the absence of template tRNA does not bind to 40S (Table 1) or 30S subunits (27,28). It follows that simultaneous presence of both mRNA and cognate tRNA ([32P] 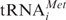 or [32P]tRNAPhe) is an absolute requirement for their binding to small eukaryotic ribosomal subunits allowing stoichiometric binding of both ligands. Note that we are analyzing non-enzymatic binding of deacylated tRNA to small subunits, whereas it is well documented that in vivo scanning 40S subunits do bind
or [32P]tRNAPhe) is an absolute requirement for their binding to small eukaryotic ribosomal subunits allowing stoichiometric binding of both ligands. Note that we are analyzing non-enzymatic binding of deacylated tRNA to small subunits, whereas it is well documented that in vivo scanning 40S subunits do bind 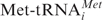 in presence of initiation factors (see ‘Discussion’ section).
in presence of initiation factors (see ‘Discussion’ section).
Figure 1.
Sucrose gradient profiles of small ribosomal subunits and ribosomes (A260) in the presence of [32P]mRNA (dotted lines) and—where indicated—tRNAPhe (dashed line in 1b represents experiment in the presence of non-cognate tRNAAla). Panels on the left and right sides show profiles obtained with eukaryotic and bacterial ribosomes, respectively.
Figure 2.
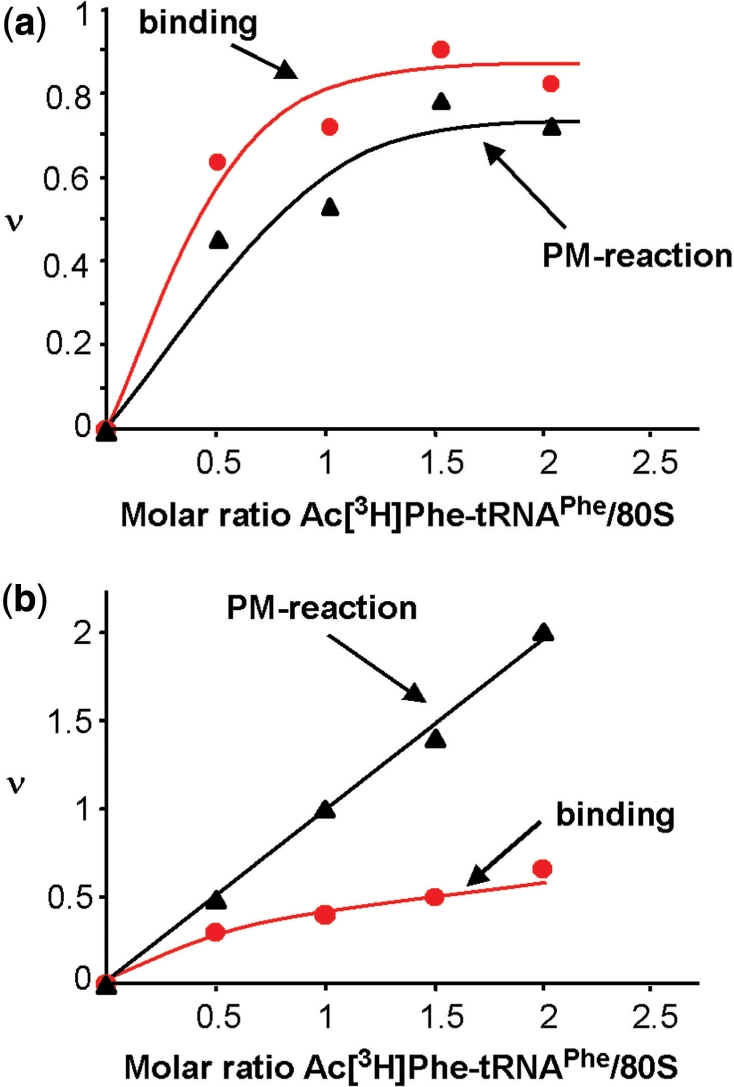
AcPhe-tRNA binding (red lines) and puromycin reaction (black lines) with 80S ribosomes in the presence (a) and absence (b) of MFVK-mRNA. ν, tRNA bound per ribosome in a binding assay or acylated tRNA per ribosome reacting with puromycin.
Table 1.
Binding of deacylated [32P]tRNAPhe to eukaryotic ribosomes and subunits
| 80S |
40Sa |
60S |
|||
|---|---|---|---|---|---|
| −mRNA | +mRNA | −mRNA | +mRNA | −mRNA | +mRNA |
| 0.77 ± 0.07 | 1 ± 0.01 | 0.1 ± 0.01 | 1 ± 0.03 | 0.56 ± 0.03 | 0.59 ± 0.06 |
ain experiments with 40S subunits bovine deacylated [32P]tRNAPhe or yeast [32P] were used yielding the same results.
were used yielding the same results.
The numbers give the bound tRNA per ribosome (ν), when two-molar excess was added over ribosomes. If indicated, MFVK-mRNA was used; at least three experiments were performed.
The large eukaryotic 60S subunit do not bind mRNA at all (profiles not shown), but can interact with deacylated tRNA even in the absence of mRNA (∼0.6 tRNA/60S, Table 1), in agreement with the observation that only one tRNA-binding site, the E site, is available on isolated bacterial 50S subunits (27).
mRNA comigrates with 80S ribosomes in the absence and presence of tRNA (Figure 1c and d, left panels) indicating a tRNA-independent mRNA binding to mammalian ribosomes. An analogous result was observed in filter-binding assays (0.77 tRNA/80S without mRNA, Table 1).
70S ribosomes bind one MFVK-mRNA per ribosome in the presence of tRNAPhe, without tRNA only half the value was observed (Figure 1c and d, right panels).
Characterization of Pi state
Programmed ribosomes containing only one tRNA at the P site represent the Pi state (i for initiation) (25). We have used AcPhe-tRNA, an analog of peptidyl-tRNA, for specific P site filling.
A saturation curve for AcPhe-tRNA leveled off at 0.8 tRNA per 80S in the presence of MFVK-mRNA containing only one codon for tRNAPhe, i.e. 80% of the ribosomes of our preparation participate in tRNA binding in the presence of template. In the absence of template up to 0.5 AcPhe-tRNA could be bound per 80S (Figure 2a and b, respectively).
The bound AcPhe-tRNA reacted quantitatively with puromycin in the presence of template indicating P-site location. Interestingly, in the absence of a template formation of AcPhe-puromycin exceeded by far the amount of bound AcPhe-tRNA and stopped only when the total input amount was consumed. For example, in Figure 2b two AcPhe-puromycin formed per ribosome versus 0.5 AcPhe-tRNA bound per ribosome. It follows that one ribosome can perform more than one puromycin reaction (in Figure 2b—four reactions) until the AcPhe-tRNA input has been used, but only in the absence of message (repetitive puromycin reaction, see ‘Discussion’ section). This observation has important consequences for a reliable interpretation of a puromycin reaction.
Characterization of PRE state
The PRE state (pretranslocational state) is characterized by a tRNA occupation of both A and P sites. After occupying the P-site with Ac[3H]Phe-tRNAPhe the A site was filled with a pre-formed ternary complex containing [14C]Val-tRNAVal corresponding to the GUU codon at the A site. Figure 3a shows that with increasing A-site occupation the puromycin reaction of the P-site bound Ac[3H]Phe-tRNA steadily decreased indicating formation of the di-peptide AcPhe-Val-tRNA at the A site. The formed di-peptide was directly demonstrated in a parallel experiment via HPLC analysis. The peak containing both radioactive amino acids (Ac[3H]Phe and [14C]Val) in stoichiometric amounts (Figure 3b, fractions 32–35) indicates di-peptides. Fractions 5–8 and 28–31 represent the excess of unbound [14C]Val-tRNAVal and Ac[3H]Phe-tRNAPhe, respectively.
Figure 3.
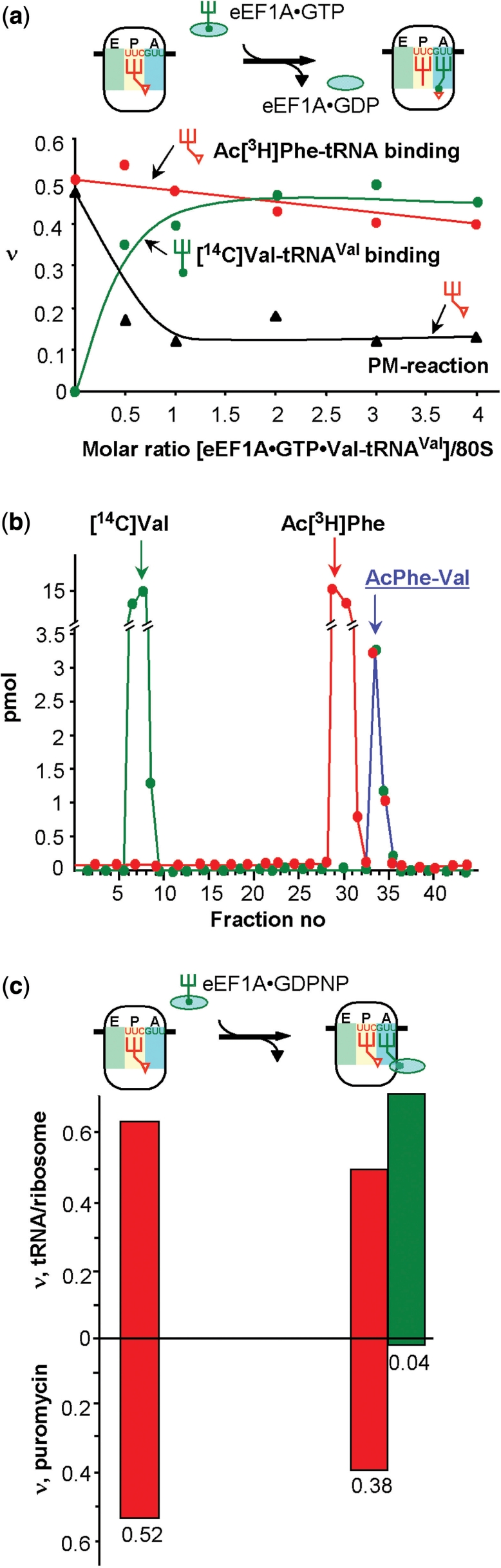
Addition of a ternary complexes (green) to a Pi state of 80S ribosomes. (a) Binding of the ternary complex eEF1A·GTP·Val-tRNA to 80S ribosomes in the Pi state carrying an AcPhe-tRNA (red line) at the P site in the presence of MFVK-mRNA. (b) Di-peptide formation in the assay shown in (a). (c) eEF1A·GDPNP·Val-tRNA (green bars) is added to Pi states carrying AcPhe-tRNA (red bars). ν, tRNA bound per ribosome in a binding assay or acylated tRNA per ribosome reacting with puromycin.
When the ternary complex was bound to the A site in the presence of the non-cleavable GTP analogue GDPNP, 75% of the AcPhe-tRNA still reacted with puromycin, whereas almost none of the Val-tRNA participated in reaction (Figure 3c). Thus, the ternary complex is fixed on the ribosome after the decoding process at the A site, but does not hamper the puromycin reaction with AcPhe-tRNA at the P site. It follows that Val-tRNA did not yet accommodate, i.e. the ternary complex is present in the A/T state as it is observed with bacterial ribosomes (8).
Allosteric interplay between A and E sites
The E-tRNA is released upon A-site occupation, indicating that the A site can influence the E site via a negative cooperativity. Also occupation of the E site influences negatively the tRNA affinity of the A site; the A site flips into a low-affinity state upon E site occupation as shown with ribosomes from bacteria and lower eukaryotes (6,7), preventing premature accommodation and peptide-bond formation of the aminoacyl-tRNA during or before decoding at the A/T state (29). This allosteric phenomenon of a reciprocal linkage between A and E sites is instrumental for an accurate protein synthesis (5).
To test whether this feature is also valid in higher eukaryotic 80S ribosomes, a posttranslocational complex was constructed with Ac[3H]Val-tRNAVal at the P site (MFVK-mRNA, GUU codon) and 32P-labeled deacylated tRNAPhe at the E site (UUC codon). According to the allosteric three-site model (6), the A-site occupation via addition of [14C]Lys- (AAA codon) together with eEF1A in the presence of GTP should trigger a corresponding release of deacylated tRNA from the E site. Note that eEF2 was not present in the reaction mixture and therefore a translocation cannot occur (see ‘Materials and Methods’ section). Three different radioactive labels (3H, 14C and 32P) present in one assay allow a simultaneous analysis of the tRNA fates at the three tRNA binding sites (Figure 4). Since about 65% of the ribosomes contained Ac[3H]Val-tRNAVal at the P-site, a maximum of 65% of ribosomes can have an occupied E site and thus can be present in the POST state. Addition of increasing amounts of A-site substrate (ternary complex containing [14C]Lys-
(AAA codon) together with eEF1A in the presence of GTP should trigger a corresponding release of deacylated tRNA from the E site. Note that eEF2 was not present in the reaction mixture and therefore a translocation cannot occur (see ‘Materials and Methods’ section). Three different radioactive labels (3H, 14C and 32P) present in one assay allow a simultaneous analysis of the tRNA fates at the three tRNA binding sites (Figure 4). Since about 65% of the ribosomes contained Ac[3H]Val-tRNAVal at the P-site, a maximum of 65% of ribosomes can have an occupied E site and thus can be present in the POST state. Addition of increasing amounts of A-site substrate (ternary complex containing [14C]Lys-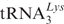 ) triggers the release of E-tRNA from 62% of the 80S ribosomes. The fact that the binding of Lys-tRNA saturates at about 90% obviously indicates that about 30% of this tRNA goes to ribosomes with an empty P site.
) triggers the release of E-tRNA from 62% of the 80S ribosomes. The fact that the binding of Lys-tRNA saturates at about 90% obviously indicates that about 30% of this tRNA goes to ribosomes with an empty P site.
Figure 4.

Binding of the ternary complex eEF1A·GTP·[14C]Lys-tRNA (green) to 80S ribosomes in the POST state carrying an Ac[3H]Val-tRNA (red) at the P and a [32P]tRNAPhe (orange) at the E site. The 100% binding corresponds to 0.4 tRNAPhe bound per ribosome (ν). For details see text.
Susceptibility to tetracycline
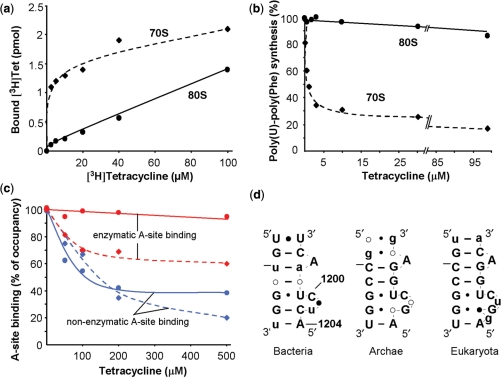
Tetracycline (Tet) binds to the 16S rRNA of 30S subunits in a sequence unspecific manner and blocks the binding of aminoacyl-tRNA to the A site (30–33). Titration of empty 70S and 80S with labelled [3H]Tet revealed an at least 15 times smaller apparent Kd with bacterial than with eukaryotic ribosomes (1–2 µM and ∼30 µM, respectively; Figure 5a). Binding was not yet saturated in the case of 80S ribosomes probably due to lower-affinity binding site(s), since more than one binding site per 70S ribosome has been described before (32–34).
Figure 5.
Analysis of tetracycline binding to 70S and 80S ribosomes. (a) Binding of [3H]Tet to empty 70S (diamonds, dashed line) and 80S (circles, solid line) ribosomes. (b) Poly(U)-poly(Phe) synthesis in bacterial (diamonds, dashed line) and mammalian (circles, solid line) systems in the presence of increasing amounts of tetracycline. The 100% corresponds to 60 and 15 incorporated Phe per ribosome in the bacterial and eukaryotic system, respectively. (c) Effect of tetracycline on non-enzymatic A-site binding with AcPhe-tRNA (blue) and enzymatic A-site binding with ternary complex (red). Deacylated tRNAfMet and 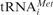 were used to occupy the P site in 70S (dashed line) and 80S systems (solid line), respectively. The 100% corresponds to a ν value of 0.7 for both 70S and 80S ribosomes for non-enzymatic A-site binding and 0.83 and 0.6 for 70S and 80S ribosomes, respectively, for enzymatic A-site binding. (d) Parts of helices 34 from phylogenetic conservation maps superimposed onto the bacterial (E. coli), archaeal (Methanococcus jannaschii) and eukaryotic (Saccharomyces cerevisiae) 16S-type secondary structures (http://www.rna.ccbb.utexas.edu/). ACGU, 98+% conserved positions; acgu, 90–98% conserved positions; filled circle, 80–90% conserved positions; open circle, less than 80% conserved positions. The nucleotides of the right strand are connected for the sake of clarity; number of sequences considered: Bacteria, 4214; Archaea, 174; Eukaryota, 1939.
were used to occupy the P site in 70S (dashed line) and 80S systems (solid line), respectively. The 100% corresponds to a ν value of 0.7 for both 70S and 80S ribosomes for non-enzymatic A-site binding and 0.83 and 0.6 for 70S and 80S ribosomes, respectively, for enzymatic A-site binding. (d) Parts of helices 34 from phylogenetic conservation maps superimposed onto the bacterial (E. coli), archaeal (Methanococcus jannaschii) and eukaryotic (Saccharomyces cerevisiae) 16S-type secondary structures (http://www.rna.ccbb.utexas.edu/). ACGU, 98+% conserved positions; acgu, 90–98% conserved positions; filled circle, 80–90% conserved positions; open circle, less than 80% conserved positions. The nucleotides of the right strand are connected for the sake of clarity; number of sequences considered: Bacteria, 4214; Archaea, 174; Eukaryota, 1939.
The drug slightly inhibits poly(Phe) synthesis of 80S (10% reduction of ribosomal activity in the presence of 300 µM), whereas inhibition was observed even below 5 µM in the bacterial system (70S, Figure 5b). Next, tRNA binding to P- and A-sites was studied in the presence of Tet. AcPhe-tRNA binding to the P site was not affected even by 500 µM in both systems (data not shown), whereas the A site binding of AcPhe-tRNA was sensitive to Tet in the bacterial and—interestingly—in the eukaryotic system as well (Figure 5c, blue lines). It follows that the specificity of the drug action seen in prevention of non-enzymatic tRNA binding to the A site was similar in both systems. However, when enzymatic A-site occupation with ternary complexes was studied, significant blockage of ternary complex binding was only observed in the bacterial rather than in the eukaryotic system (Figure 5c, dashed and solid red lines, respectively).
DISCUSSION
We analyzed basic functions of bacterial and eukaryotic ribosomes and their subunits and used the MFVK-mRNA with a length of 49 nt and a coding region in the middle for Met-Phe-Val-Lys lacking all translation initiation regulatory elements inherent to eukaryotic or prokaryotic mRNAs. Therefore it is a proper model-mRNA to compare mRNA and tRNA binding to ribosomes. The most conspicuous difference is the observation that the 40S subunits cannot bind either deacylated tRNA or mRNA alone, whereas 30S do bind efficiently mRNA in the absence of tRNA. mRNA binds only to 40S ribosomal subunits in the presence of cognate deacylated tRNAPhe rather then non-cognate tRNAAla (Figure 1a and b, left panels) indicating that tRNA fixes the reading frame via codon-anticodon interaction and stabilizes 40S–mRNA interactions. On the other hand, a 40S subunit binds the ternary complex eIF2·GTP·Met-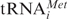 in the absence of mRNA during initiation (43S initiation complex) (2) before it interacts with the cap-region of mRNA in the presence of other initiation factors (48S initiation complex). Therefore, deacylated or acylated tRNA will not interfere with the initiation process, be it the scanning mechanism (35) or the tethering and clustering mechanisms (36).
in the absence of mRNA during initiation (43S initiation complex) (2) before it interacts with the cap-region of mRNA in the presence of other initiation factors (48S initiation complex). Therefore, deacylated or acylated tRNA will not interfere with the initiation process, be it the scanning mechanism (35) or the tethering and clustering mechanisms (36).
The specific 30S feature, namely interaction with mRNA in the absence of tRNA, reflects the initiation mode of Gram-negative bacteria, where ribosomal protein S1 in the absence of a Shine-Dalgarno (SD) sequence is a key recognition element in mRNA binding (37,38). It is shown that S1 is required for translation of most, if not all, natural mRNAs in E. coli, where only 57.1% of genes contain SD sequence (37,38). Together our data of ultracentrifugation and previously published results indicate that mRNA binding precedes that of tRNA on 30S (38,39). When both mRNA and cognate tRNA are present, the small ribosomal subunits bind both molecules efficiently and stoichiometrically, i.e. the binding of tRNA and mRNA to small subunits is highly cooperative.
80S ribosomes bind tRNA to the P site in the absence of mRNA (Table 1). This fact is long known for 70S ribosomes (40,41). Concerning mRNA binding in the absence of tRNA, 80S bind template in the absence of tRNA more efficient than 70S ribosomes (Figure 1c; only 50% of 70S bound mRNA under the same conditions). Presence of both tRNA and mRNA stabilizes the resulting ribosome complex as indicated by stoichiometric binding values with 70S and 80S ribosomes. Recently, the stabilization effect was directly measured with 70S ribosomes applying optical tweezers (42).
When the P sites of 80S ribosomes were titrated with AcPhe-tRNA in the presence of MFVK-mRNA, the binding curve leveled off at about 0.8 molecules per 80S as did the puromycin reaction (Figure 2a), whereas in the absence of mRNA the binding did not exceed 0.5 molecules per 80S (Figure 2b). Interestingly, the puromycin reaction surpassed the binding value by far in the absence of mRNA and went on until the input of AcPhe-tRNA was depleted. What happens is obviously a repetitive reaction, namely after the formation of AcPhe-puromycin the remaining deacylated tRNAPhe at the P site is chased by AcPhe-tRNA, so that the same ribosome can perform more then one puromycin reaction as long as the input AcPhe-tRNA is consumed. Under polyamine-conditions codon-anticodon interaction is able to fix deacylated tRNA at the P site thus preventing chasing, whereas non-programmed 80S ribosomes bind tRNA only weakly allowing chasing. A repetitive puromycin reaction is therefore an indication for the absence of mRNA or non-saturating mRNA conditions. The weak binding in the absence of mRNA can be explained by analogy with 70S ribosomes: a P-tRNA in non-programmed 70S ribosomes is losing its interactions with the small subunit and interacts exclusively with the large subunit (43). The important conclusion is that the puromycin reaction is only reliable in the presence of saturating amount of mRNA, otherwise it will be going on with time.
According to cryo-EM studies in bacteria the A-site occupation occurs in two structural clearly distinct steps, the decoding process and the accommodation (44). During decoding the aminoacyl-residue is >50 Å apart from the peptidyltransferase center (A/T state) giving room for free access of puromycin to the peptidyltransferase center. This state is visualized in the presence of the non-cleavable GTP analog GDPNP. Only after a successful decoding the aminoacyl-tRNA flips fully into the A site (accommodation). Therefore, the fact that in the presence of GDPNP AcPhe-tRNA interacts significantly with puromycin whereas the adjacent Val-tRNA does not (Figure 3c) indicates that also higher eukaryotic ribosomes run through the A/T state during A-site occupation, which is in accord with an analysis by cryo-electronmicroscopy (T. V. Budkevich, J. Giesebrecht, K. H. Nierhaus and C. T. M. Spahn, manuscript in preparation).
We have presented evidence that the reciprocal linkage between the A and E sites is of outmost importance for accurate protein synthesis (5), i.e. E-site occupation induces a low affinity mode for tRNA binding to the A site and vice versa, occupation of the A site triggers the release of the E-tRNA. This is also true for higher eukaryotic ribosomes, since occupation of the A site of POST ribosomes is accompanied by a corresponding release of the E-tRNA (Figure 4), the intervening P-tRNA is not affected serving for a welcome internal control. We conclude that the allosteric interaction of A and E sites is universal, since it has also been demonstrated in a bacterial system (6,45) as well as with yeast 80S ribosomes (7).
Tetracycline binds to the A site and prevents tRNA binding to this site (32,33,46,47). The primary binding site of this drug is nested in a universal rRNA fold of the sugar-phosphate backbone and not depending on a conserved sequence. Therefore, in principal, it should bind to all kinds of ribosomes, the 70S as well as the 80S type, with comparable affinity. Most contacts are formed with the irregular shallow/minor groove of h34 (1196–1200, 1053–1056; E. coli numbering), and additional contacts are found with h31 (964–967). For the first time, we compare drug binding to 70S and 80S ribosomes, and surprisingly, we observe binding of the antibiotic to empty 80S ribosomes with a 15-fold reduced affinity as compared with that to 70S ribosomes (Figure 5a). We speculate that this phenomenon might be due to structural differences of the primary tetracycline binding site between bacterial and eukaryotic rRNAs at helix h34 around residue 1200 (Figure 5d). Comparison of secondary structures of the 16S-type rRNA from the three phylogenetic domains (6326 sequences, http://www.rna.ccbb.utexas.edu/) revealed that in almost all eukaryotic sequences helix 34 contains one extra nucleotide before the universal A1204, which can cause a different tertiary organization of phosphate backbone in this area and is possibly responsible for the increase of the apparent Kd value. The observation of partial resistance of mammalian ribosomes to Tet was also confirmed in poly(U)-poly(Phe) synthesis (Figure 5b). Our data are in line with old results (48), where Tet at 400 µM concentration was more inhibitory (90%) in [14C]Leu incorporation into E. coli total protein than in a rat liver system (28%).
Tet blocks non-enzymatic A-site occupation with acetylated tRNAPhe (but not non-enzymatic P-site occupation; data not shown) in both systems to a comparable extent (Figure 5c, blue lines), which indicates that antibiotic binds to corresponding sites in 70S and 80S ribosomes. In striking contrast, interaction of eukaryotic ternary complex with 80S ribosomes was practically unaffected at up to 500 µM of the drug, whereas binding of bacterial analog was blocked for about 30% (Figure 5c, red lines). A possible reason is that in spite of the lower affinity of Tet for eukaryotic ribosomes mentioned above the affinity of tRNA to the A site in the absence of elongation factors (EF-Tu/eEF1A) is low enough to be blocked by Tet in both the bacterial and eukaryotic systems. Indeed, non-enzymatic binding to the A site of bacterial ribosomes occurs with an affinity which is about two orders of magnitudes smaller than that of enzymatic binding (49). Obviously, the relative resistance of eukaryotic systems to the effect of Tet—the prerequisite for its application in medicine—is caused by two factors: (i) the lower intrinsic affinity as shown by binding experiments with empty ribosomes (Figure 5a) and (ii) the stronger drug-chasing capability of eukaryotic ternary complexes (Figure 5c).
In vivo eukaryotic cells are essentially resistant against Tet in contrast to bacterial cells. It is generally believed that this is due to the fact that the drug can easier penetrate the bacterial rather than the eukaryotic cell membrane (50). Our results reveal additional and possibly decisive explanation of a preferential inhibition of bacterial rather than eukaryotic ribosomes.
ACKNOWLEDGEMENTS
We thank Dr. Oliver Vesper and Daniela Wittek for ribosomes and ribosomal subunits from E. coli strain CAN/20-12E (51) and also Edda Einfeldt for bacterial tRNA preparations. This work was funded by Ukrainian-German DFG project 436 UKR 113/64/1-1.
Conflict of interest statement. None declared.
REFERENCES
- 1.Londei P. Evolution of translational initiation: new insights from the archaea. FEMS Microbiol. Rev. 2005;29:185–200. doi: 10.1016/j.femsre.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Pestova TV, Hellen CU. Functions of eukaryotic factors in initiation of translation. Cold Spring Harb. Symp. Quant. Biol. 2001;66:389–396. doi: 10.1101/sqb.2001.66.389. [DOI] [PubMed] [Google Scholar]
- 3.Kisselev L. Polypeptide release factors in prokaryotes and eukaryotes: same function, different structure. Structure. 2002;10:8–9. doi: 10.1016/s0969-2126(01)00703-1. [DOI] [PubMed] [Google Scholar]
- 4.Qin Y, Polacek N, Vesper O, Staub E, Einfeldt E, Wilson DN, Nierhaus KH. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell. 2006;127:721–733. doi: 10.1016/j.cell.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 5.Wilson DN, Nierhaus KH. The E-site story: the importance of maintaining two tRNAs on the ribosome during protein synthesis. Cell Mol. Life Sci. 2006;63:2725–2737. doi: 10.1007/s00018-006-6125-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rheinberger H-J, Nierhaus KH. Allosteric interactions between the ribosomal transfer RNA-binding sites A and E. J. Biol. Chem. 1986;261:9133–9139. [PubMed] [Google Scholar]
- 7.Triana-Alonso FJ, Chakraburtty K, Nierhaus KH. The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. J. Biol. Chem. 1995;270:20473–20478. doi: 10.1074/jbc.270.35.20473. [DOI] [PubMed] [Google Scholar]
- 8.Dinos G, Kalpaxis DL, Wilson DN, Nierhaus KH. Deacylated tRNA is released from the E site upon A site occupation but before GTP is hydrolyzed by EF-Tu. Nucleic Acids Res. 2005;33:5291–5296. doi: 10.1093/nar/gki833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersen BF, Becker T, Blau M, Anand M, Halic M, Balar B, Mielke T, Boesen T, Pedersen JS, Spahn CMT, et al. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature. 2006;433:663–668. doi: 10.1038/nature05126. [DOI] [PubMed] [Google Scholar]
- 10.Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jørgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graifer DM, Karpova GG, Knorre DG. Location of template on the human ribosome as revealed from data on cross-linking with reactive mRNA analogs. Biochemistry. 2001;66:585–602. doi: 10.1023/a:1010220627612. [DOI] [PubMed] [Google Scholar]
- 12.Molotkov M, Graifer D, Demeshkina N, Repkova M, Ven'yaminova A, Karpova G. Arrangement of mRNA 3′ of the A site codon on the human 80S ribosome. RNA Biol. 2005;2:63–69. doi: 10.4161/rna.2.2.1756. [DOI] [PubMed] [Google Scholar]
- 13.Bommer U, Burkhardt N, Jünemann R, Spahn CMT, Triana-Alonso FJ, Nierhaus KH. Subcellular Fractionation. In: Graham J, Rickwoods D, editors. A Practical Approach. Oxford: IRL Press at Oxford University Press; 1996. pp. 271–301. [Google Scholar]
- 14.Triana-Alonso FJ, Dabrowski M, Wadzack J, Nierhaus KH. Self-coded 3′-extension of run-off transcripts produces aberrant products during in vitro transcription with T7 RNA polymerase. J. Biol. Chem. 1995;270:6298–6307. doi: 10.1074/jbc.270.11.6298. [DOI] [PubMed] [Google Scholar]
- 15.Triana F, Nierhaus K, Chakraburtty K. Transfer RNA binding to 80S ribosomes from yeast: evidence of three sites. Biochem. Mol. Biol. Int. 1994;33:909–915. [PubMed] [Google Scholar]
- 16.Silberklang M, Gillum AM, RajBhandary UL. The use of nuclease P1 in sequence analysis of end group labeled RNA. Nucleic Acids Res. 1977;4:4091–4108. doi: 10.1093/nar/4.12.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruce AG, Uhlenbeck OC. Reactions at the termini of tRNA with T4 RNA ligase. Nucleic Acids Res. 1978;5:3665–3677. doi: 10.1093/nar/5.10.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shalak VF, Budkevich TV, Negrutskii BS, El'skaya AV. A fast and effective method for purification of elongation factor 1 alpha from rabbit liver. Ukr. Biokhim. Zh. 1997;69:104–109. [PubMed] [Google Scholar]
- 19.Kemper WM, Merrick WC. Preparation of protein synthesis elongation factors from rabbit reticulocytes. Methods Enzymol. 1979;60:638–648. doi: 10.1016/s0076-6879(79)60060-5. [DOI] [PubMed] [Google Scholar]
- 20.El'skaya AV, Ovcharenko GV, Palchevskii SS, Petrushenko ZM, Triana-Alonso FJ, Nierhaus KH. Three tRNA binding sites in rabbit liver ribosomes and role of the intrinsic ATPase in 80S ribosomes from higher eukaryotes. Biochemistry. 1997;36:10492–10497. doi: 10.1021/bi970631e. [DOI] [PubMed] [Google Scholar]
- 21.Graifer DM, Malygin AA, Matasova NB, Mundus DA, Zenkova MA, Karpova GG. Studying functional significance of the sequence 980-1061 in the central domain of human 18S rRNA using complementary DNA probes. Biochim. Biophys. Acta. 1997;1350:335–344. doi: 10.1016/s0167-4781(96)00176-5. [DOI] [PubMed] [Google Scholar]
- 22.Szaflarski W, Vesper O, Teraoka Y, Plitta B, Wilson DN, Nierhaus KH. New features of the ribosome and ribosomal inhibitors: Non-enzymatic recycling, misreading and back-translocation. J. Mol. Biol. 2008;380:193–205. doi: 10.1016/j.jmb.2008.04.060. [DOI] [PubMed] [Google Scholar]
- 23.Rheinberger H-J, Geigenmüller U, Wedde M, Nierhaus KH. Parameters for the preparation of Escherichia coli ribosomes and ribosomal subunits active in tRNA binding. Methods Enzymol. 1988;164:658–670. doi: 10.1016/s0076-6879(88)64076-6. [DOI] [PubMed] [Google Scholar]
- 24.Márquez V, Wilson DN, Tate WP, Triana-Alonso F, Nierhaus KH. Maintaining the ribosomal reading frame: The influence of the E site during translational regulation of release factor 2. Cell. 2004;118:45–55. doi: 10.1016/j.cell.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 25.Hausner TP, Geigenmüller U, Nierhaus KH. The allosteric three site model for the ribosomal elongation cycle. New insights into the inhibition mechanisms of aminoglycosides, thiostrepton, and viomycin. J. Biol. Chem. 1988;263:13103–13111. [PubMed] [Google Scholar]
- 26.Ferreras AC, Bandeira E, Cayama E, Zambrano R, Avila H, Yépez A, Triana JL, Triana-Alonso FJ. Efficient and faithful in vitro translation of natural and synthetic mRNA with human ribosomes. Int. J. Mol. Med. 2004;13:527–536. [PubMed] [Google Scholar]
- 27.Gnirke A, Nierhaus KH. tRNA binding sites on the subunits of Escherichia coli ribosomes. J. Biol. Chem. 1986;261:14506–14514. [PubMed] [Google Scholar]
- 28.Kirillov SV, Makhno VI, Semenkov YP. Mechanism of codon-anticodon interaction in ribosomes. Direct functional evidence that isolated 30S subunits contain two codon-specific binding sites for transfer RNA. Nucleic Acids Res. 1980;8:183–196. doi: 10.1093/nar/8.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geigenmüller U, Nierhaus KH. Significance of the third tRNA binding site, the E site, on E. coli ribosomes for the accuracy of translation: an occupied E site prevents the binding of non-cognate aminoacyl-transfer RNA to the A site. EMBO J. 1990;9:4527–4533. doi: 10.1002/j.1460-2075.1990.tb07904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wurmbach P, Nierhaus KH. The inhibition pattern of antibiotics on the extent and accuracy of tRNA binding to the ribosome, and their effect on the subsequent steps in chain elongation. Eur. J. Biochem. 1983;130:9–12. doi: 10.1111/j.1432-1033.1983.tb07109.x. [DOI] [PubMed] [Google Scholar]
- 31.Geigenmüller U, Nierhaus KH. Tetracycline can inhibit tRNA binding to the ribosomal P site as well as to the A site. Eur. J. Biochem. 1986;161:723–726. doi: 10.1111/j.1432-1033.1986.tb10499.x. [DOI] [PubMed] [Google Scholar]
- 32.Brodersen DE, Clemons WM, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 33.Pioletti M, Schlünzen F, Harms J, Zarivach R, Glühmann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C, et al. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 2001;20:1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wirmer J, Westhof E. Molecular contacts between antibiotics and the 30S ribosomal particle. Methods Enzymol. 2006;415:180–202. doi: 10.1016/S0076-6879(06)15012-0. [DOI] [PubMed] [Google Scholar]
- 35.Kozak M. How do eukaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978;15:1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- 36.Chappell SA, Edelman GM, Mauro VP. Ribosomal tethering and clustering as mechanisms for translation initiation. Proc. Natl. Acad. Sci. USA. 2006;103:18077–18082. doi: 10.1073/pnas.0608212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tal M, Aviram M, Kanarek A, Weiss A. Polyuridylic acid binding and translating by Escherichia coli ribosomes: stimulation by protein I, inhibition by aurintricarboxylic acid. Biochim. Biophys. Acta. 1972;281:381–392. doi: 10.1016/0005-2787(72)90452-2. [DOI] [PubMed] [Google Scholar]
- 38.Sørensen MA, Fricke J, Pedersen S. Ribosomal protein S1 is required for translation of most, if not all, natural mRNAs in Escherichia coli in vivo. J. Mol. Biol. 1998;280:561–569. doi: 10.1006/jmbi.1998.1909. [DOI] [PubMed] [Google Scholar]
- 39.Ma J, Campbell A, Karlin S. Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. J. Bacteriol. 2002;184:5733–5745. doi: 10.1128/JB.184.20.5733-5745.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe S. Interaction of siomycin with the acceptor site of Escherichia coli ribosomes. J. Mol. Biol. 1972;67:443–457. doi: 10.1016/0022-2836(72)90462-7. [DOI] [PubMed] [Google Scholar]
- 41.Rheinberger H-J, Sternbach H, Nierhaus KH. Three tRNA binding sites on Escherichia coli ribosomes. Proc. Natl. Acad. Sci. USA. 1981;78:5310–5314. doi: 10.1073/pnas.78.9.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uemura S, Dorywalska M, Lee TH, Kim HD, Puglisi JD, Chu S. Peptide bond formation destabilizes Shine-Dalgarno interaction on the ribosome. Nature. 2007;446:454–457. doi: 10.1038/nature05625. [DOI] [PubMed] [Google Scholar]
- 43.Schäfer MA, Tastan AO, Patzke S, Blaha G, Spahn CM, Wilson DN, Nierhaus KH. Codon-anticodon interaction at the P site is a prerequisite for tRNA interaction with the small ribosomal subunit. J. Biol. Chem. 2002;277:19095–19105. doi: 10.1074/jbc.M108902200. [DOI] [PubMed] [Google Scholar]
- 44.Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat. Struct. Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- 45.Gnirke A, Geigenmüller U, Rheinberger H.-J, Nierhaus KH. The allosteric three-site model for the ribosomal elongation cycle. J. Biol. Chem. 1989;264:7291–7301. [PubMed] [Google Scholar]
- 46.Bodley JW, Zieve FJ. On the specificity of the two ribosomal binding sites: studies with tetracycline. Biochem. Biophys. Res. Commun. 1969;36:463–468. doi: 10.1016/0006-291x(69)90587-7. [DOI] [PubMed] [Google Scholar]
- 47.de Groot N, Panet A, Lapidot Y. The binding of purified Phe-tRNA and peptidyl-tRNAPhe to Escherichia coli ribosomes. Eur. J. Biochem. 1971;23:523–527. doi: 10.1111/j.1432-1033.1971.tb01649.x. [DOI] [PubMed] [Google Scholar]
- 48.Franklin TJ. The inhibition of incorporation of leucine into protein of cell-free systems from rat liver and Escherichia coli by chlortetracycline. Biochem. J. 1963;87:449–453. doi: 10.1042/bj0870449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schilling-Bartetzko S, Franceschi F, Sternbach H, Nierhaus KH. Apparent association constants of transfer RNAs for the ribosomal A-site, P-site, and E-site. J. Biol. Chem. 1992;267:4693–4702. [PubMed] [Google Scholar]
- 50.Vásquez D. Springer, Berlin, Heidelberg, New York: 1979. Inhibitors of Protein Synthesis; pp. 54–57. [Google Scholar]
- 51.Zaniewski R, Petkaites E, Deutscher MP. A multiple mutant of Escherichia coli lacking the exoribonucleases RNase II, RNase D, and RNase BN. J. Biol. Chem. 1984;259:11651–11653. [PubMed] [Google Scholar]