Abstract
Understanding the driving forces of gene expression variation within human populations will provide important insights into the molecular basis of human phenotypic variation. In the genome, the gene expression variability differs among genes, and at present, most research has focused on identifying the genetic variants responsible for the within population gene expression variation. However, little is known about whether microRNAs (miRNAs), which are small noncoding RNAs modulating expression of their target genes, could have impact on the variability of gene expression. Here we demonstrate that miRNAs likely lead to the difference of expression variability among genes. With the use of the genome-wide expression data in 193 human brain samples, we show that the increased variability of gene expression is concomitant with the increased number of the miRNA seeds interacting with the target genes, suggesting a direct influence of miRNA on gene expression variability. Compared with the non-miRNA-target genes, genes targeted by more than two miRNA seeds have increased expression variability, independent of the miRNA types. In addition, single-nucleotide polymorphisms (SNPs) located in the miRNA binding sites could further increase the gene expression variability of the target genes. We propose that miRNAs are one of the driving forces causing expression variability in the human genome.
INTRODUCTION
One of the major tasks for human genetics is to dissect the molecular basis of phenotypic variation. Over the past decades, studies have analyzed and unveiled the genetic variants in the human genome, such as single-nucleotide polymorphisms (SNPs) (1–3) and copy number variations (CNVs) (4), which contribute to gene expression variation, and eventually to phenotypic variation in human populations (5). At the same time, it has been shown that the variability of gene expression is highly different among genes (1,6). As an example, in lymphoblastoid cell lines, genes with chaperone regulatory activity show high expression variability whereas genes with extracellular function show low variability (6). However, few studies have focused on identifying genetic factors responsible for the observed difference of gene expression variability among genes.
In recent years, microRNAs (miRNAs) have been identified as a large class of small noncoding RNAs (∼22 nt), present in most metazoan and important for a diverse range of biological function (7–9). The human genome was predicted to encode nearly 1000 miRNAs which range from forming clusters to scattering across the genomic region (10,11). These miRNA genes act post-transcriptionally to target mRNA for translational repression, cleavage and destabilization (7,12,13), and different combinations of miRNAs could coordinately regulate a specific target gene in mammals (14,15). Due to the regulation of miRNAs, tens of thousands of mRNAs selectively maintain 7 nt sites in the 3′UTR region that match the sequences of the mature miRNAs. It seems that miRNAs make widespread impact on gene expression and 3′UTR evolution (16,17). Previous studies suggested that in mammal and fly species, miRNAs decreased the cross-species expression divergence and evolutionarily constrained gene expression variation (18). However, how miRNAs contribute to within species expression variability has not been tested yet. Recent studies (19–21) have generated genome-wide data for both miRNA expression and population level gene expression profiles in the brain, which can be used to address this question. In this study, with the use of the published expression data of human brain, we aim to exam whether miRNA is one of the forces driving gene expression variability in human populations.
MATERIALS AND METHODS
Gene expression data
We downloaded the cortical gene expression data of 193 filtered samples reported by Myers et al. (21). All samples were self-defined as ethnically European descent. All samples had an age at death from 65 to 100 years, and 46% of them were females. Transcripts that were detected in <5% of the samples were excluded. The expression profile were Rank-invariant normalized and log10 transformed as described before (21). We mapped the brain expressed probe set (14 078 probes) to the illumina probe annotation file to get the RefSeq IDs and excluded the RefSeq IDs without the unique locations in the human genome using UCSC human RefSeq transcript annotations. Finally, we got 12 277 RefSeq transcripts for further analysis.
When dividing the samples into groups by gender, collected brain regions or age, we also excluded the transcripts that were detected in <5% of each group.
Brain expressed miRNA selection
To maximize the collection of miRNAs preferentially expressed in the brain, two miRNA expression datasets were used. For the small RNA library dataset (20), mature miRNAs with equal to or more than six clones (∼0.4% relative cloning frequency) in the frontal-cortex of an adult were considered (28 miRNA selected). For another microarray dataset (19), we defined relatively high expression of miRNA with a threshold of 3000, which is six times higher than the background level (58 miRNAs selected).
Combining these two datasets, we obtained 64 distinct mature miRNAs, and merged them into 48 distinct miRNA seeds. To evaluate the conservation of each miRNA seed, we obtained the homologous copies of the 64 human precursor miRNAs in chimpanzee, rhesus monkey, mouse, rat and dog genomes using BLAST. A miRNA seed was considered conservative when at least one of the precursors belonging to a seed have totally identical sequences at position 1–8 in all the six species. Finally, we identified 42 conserved miRNA seeds.
3′UTR sequence alignment and miRNA binding site prediction
We downloaded the human RefSeq transcript annotations and the 17-way vertebrate multiZ alignments from the UCSC genome browser (22). To restrict the miRNA and 3′UTR to the same conservative criterion, we extracted genome-wide multiple alignments of six mammalian species from the multiZ alignments and removed insertions or deletions in the alignments caused by other species. The alignments were built from the following genome assemblies: Human March 2006 (hg18), Chimp November 2003 (panTro1), Macaque January 2006 (rheMac2), Mouse February 2006 (mm8), Rat November 2004 (rn4) and Dog May 2005 (canFam2). We used the RefSeq transcript annotations to the human genome to map multiple alignments of the 3′UTRs.
We used the term ‘miRNA site’ to refer a 7-mers in a 3′UTR with exact Watson–Crick complementary to bases 1–7 or 2–8 from the 5′end of the mature miRNA. We predicted conserved miRNA targets using the conserved 42 miRNA seeds by searching for such 7-mers completely conserved in the custom six-way multiZ alignments. This method is similar to the core PicTar algorithm, but it ignores the relatively small number of predicted imperfect binding sites (14,23).
For non-conserved miRNA target prediction, we searched for 7-mers for the 42 miRNA seeds using human RefSeq transcript 3′UTRs without conservation criteria. We defined the non-conserved transcript group by removing all genes which have conserved target sites.
A few studies suggested that part of non-conserved sites are functional when mRNA is co-expressed with miRNA (13,16,17,24). Because of the co-expression pattern of the 42 seeds and 12 277 Refseq transcripts, some non-conserved targets might also be subject to miRNA regulation. Thus we defined non-miRNA-target genes by removing both conserved and non-conserved targets. We focused on the analysis of the conserved targets because the inclusion of conservation criteria could substantially reduce the false positive rate and facilitate the signal-to-noise ratio evaluation (14,25).
We also used the TargetScan (25) and PITA (26) algorithms for target prediction. For TargetScan algorithm, compared with the method described above, it also considers an exact match to positions 2–7 or 2–8 of the mature miRNA with a downstream ‘A’ across from position 1 of the miRNA as a binding site. We used the data generated by UTRs and five-way UTR multiple sequence alignments with conserved and non-conserved sites corresponding to conserved miRNA families (TargetScan Release 4.1 http://www.targetscan.org). We downloaded the full dataset and divided the genes used for target prediction into three categories: conserved targets, non-conserved targets and non-miRNA-target genes of the brain expressed miRNAs. Finally, we mapped them to the 12 277 expressed Refseq transcripts. For PITA algorithm, we downloaded the data from: http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html. We considered the top predictions (having an 8-mer seed and a conservation score of 0.9 or higher) of the brain expressed miRNAs as the presumed targets. The non-miRNA-target genes were defined as the predicted targets of all miRNAs minus predicted targets of brain expressed miRNAs. Finally, we mapped them to the 12 277 expressed Refseq transcripts.
Signal-to-noise ratio calculation and coefficient of variation (CV) adjustment
Similar to previously described method (25), we generated two cohorts of randomized miRNA seeds. In the first protocol, for a given miRNA seed, we generated a series of random permutation with the same length and base composition as the seed, until a shuffled sequence was found to have approximately the same abundance of seed matches (±15%) in the human 3′UTRs compared with the original miRNA. In the second protocol, we shuffled the original seed with the precise dinucleotide composition using Altschul–Erikson dinucleotide shuffle algorithm (27), until the shuffled one had approximately the same abundance of seed matches (±15%) in the human 3′UTRs compared with the original miRNA. This protocol is not applicable to every seed for randomization while preserving the exact dinucleotide composition. Finally we got 23 shuffled ones of the 42 original seeds. The conclusions are consistent when we used the two different shuffling protocols. We only presented the results using the first protocol (25).
We defined the signal-to-noise ratio as the number of predicted conserved targets (the ‘signal’) dividing the number of conserved targets predicted by cohort of shuffled miRNAs (the ‘noise’). We adjusted the CVs (see Results section for definition) according to signal-to-noise ratio. For example, for a target gene group with 240 genes (grouped by seed number), if the signal-to-noise ratio is 2.4, it means that there are 140 genes above the noise. And for the remaining 100 genes, we could not distinguish them from the noise. Conservatively, we assumed that these 100 genes have the same CVs as the non-miRNA-target genes and corrected CVs by the observed signal-to-noise ratio. Finally, we adjusted the CVs using the following formula:
Cnon is the average CV of the non-miRNA-target genes. Ri is the signal-to-noise ratio of target genes recognized by i seeds. Xi is the adjusted average CV of group i. Ci is the original CV of group i. Because we cannot get the ratio for targets recognized by one seed, we considered the corrected CV as its original CV.
CV comparison between SNP-residing targets and non-SNP targets
We collected three SNP datasets for analysis. Firstly, we downloaded the genotype data of the 193 filtered samples reported by Myers et al. (21), which had 500 568 SNPs. We filtered the original data by PLINK analysis toolset (28) as described before (21): per sample call rate, ≥90%, per SNP call rate, ≥90%, per SNP minor allele frequency ≥1% and lack of significance (P < 0.05) for Hardy–Weinberg equilibrium tests. Finally, 366 140 filtered SNPs were chosen for further analysis. Secondly, we downloaded the validated SNPs in dbSNP127 version using Ensembl BioMart (5 752 639 SNPs). Thirdly, we downloaded genotype data from the Perlegen Sciences (1 586 383 SNPs) (29), and chose the SNPs which had per SNP minor allele frequency ≥1% in Americans of European ancestry. Finally, we combined the three SNP datasets and the SNPs with unique positions in the human genome were used. For the SNP-residing target analysis, if one target gene had a SNP located within the entire 22 nt sites in the 3′UTR complementary to the miRNA, we considered it as a SNP-residing target.
Target genes with more miRNA sites have an increased chance to obtain SNPs-residing binding site. Our data showed a positive correlation between CVs and miRNA seed numbers per target gene. To exclude the bias caused by seed numbers, we first counted the seed numbers of each gene in the 277 SNP-residing targets, then classified genes by seed numbers and got a list containing the information of the number of targets for a specific seed number. We also classified the genes by seed numbers in the non-SNP targets. Finally, we randomly extracted 277 genes in the non-SNP target dataset using the list inferred from SNPs-residing targets, i.e. for each class, we extracted the same number of genes according to the list. We repeated the extraction for 10 000 times, then compared the average CV of the SNP-residing targets with the normal distribution of the CVs of the randomly sampled non-SNP target dataset. We also performed the same analysis to correct the bias caused by the binding site numbers.
Data analysis
Because the distribution of 12 277 transcripts’ CVs does not fit a normal distribution, we used Mann–Whitney test when comparing the difference between gene groups. For the linear regression analysis, we transformed the CV values to approximately a normal distribution as described previously (30). In brief, we used a natural log transformation (CV' = In(CV + k)), where k was chosen to maximize the correlation with a linear fit at a normal probability plot. We further normalized CV′ to a mean of 0 and SD of 1. We also transformed the UTR lengths, which is highly skewed, to approximately a normal distribution using the log function.
A series of Perl scripts were written to perform target predictions and other data analysis. Gene expression analyses were performed by R (31) and bioconductor (32). Linear regression analyses with confounder (partial correlation) were performed by SPSS software.
RESULTS
Positive correlation between gene expression variability and miRNA seed number/binding site number per target gene
In general, multiple miRNAs could coordinately regulate one specific target gene (14,33). If miRNAs directly contribute to the gene expression variability, we would expect that for a target gene, the effect of miRNAs on gene expression becomes stronger when there are more miRNA targets in the 3′UTR region. To test this, we used several public datasets to study the miRNA-mediated gene regulation. We calculated the variability of individual gene using the data generated by Myers and colleagues (21), who carried out whole-genome expression analysis on 193 neuropathologically normal human cortex samples. For each gene, we defined the variability in gene expression levels among individuals as coefficient of variation (CV), i.e. the standard deviation divided by the mean, which is the most direct and unambiguous measurement of gene expression variability (34). In parallel, we selected miRNAs preferentially expressed in the brain by combining one microarray dataset (19) and one small RNA library dataset (20), and we identified 42 miRNA seeds [defined by position 1-8 of the mature miRNAs, critical for target recognition (35)] which are conserved in six mammalian species for analysis (Supplementary Table S1). We predicted the target genes using a widely accepted model in which miRNA-mRNA binding is nucleated by an exact Watson–Crick match to the first six to eight bases from the 5′end of miRNA [see Rajewsky (35)]. Using this approach, we identified 4358 conserved target genes and 1663 non-miRNA-target genes in Myers’ data (see Materials and Methods section).
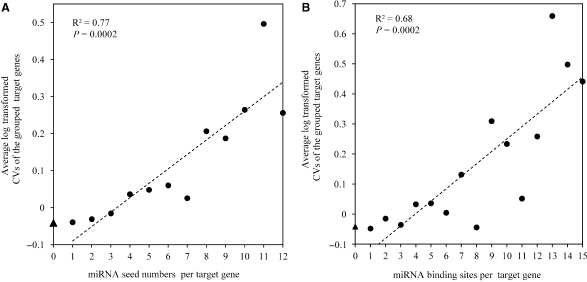
We first estimated the correlation between miRNA seed numbers per target gene and the CVs of target genes. Interestingly, the CV of a target gene significantly correlated with its targeted seed number (Spearman correlation ρ = 0.049, P = 0.001). Furthermore, when the target genes were grouped by targeted miRNA number, we found a positive correlation between the average log transformed CVs and the miRNA seed numbers (R2 = 0.77, P = 0.0002, Pearson correlation coefficient) (Figure 1A). These observations suggest that the miRNAs increase the CVs of the target genes, and the effect could be accumulative.
Figure 1.
Increased variability of gene expression is concomitant with the increased miRNA-mRNA interaction. (A) The correlation between miRNA seed numbers per target gene and the average CVs. The average CV of the non-miRNA-target genes is indicated in Y-axis by triangle. The dash line indicates the linear least square regression line. The R2 and P-value are indicated. The set of groups was restricted to those including at least 20 genes. (B) The correlation between miRNA binding sites per target gene and the average CVs.
Additionally, because one miRNA seed can have more than one binding sites in a specific gene, we also evaluated the relationship between the numbers of binding sites per gene and CVs. Again, we found a positive correlation between CV of a gene and its target binding site number (Spearman correlation ρ = 0.052, P = 0.001), and also a positive correlation between the average log transformed CVs and the miRNA binding site numbers (R2 = 0.68, P = 0.0002, Pearson correlation coefficient) (Figure 1B).
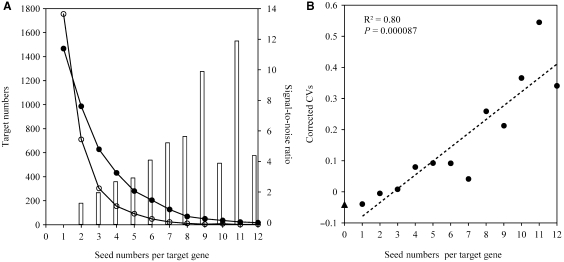
It was suggested that cooperation of miRNAs or multiplicity of target sites could boost the signal-to-noise ratio of prediction (14,36). To eliminate the potential effect of signal-to-noise ratio on the observed positive correlation between the miRNA seed numbers and the average CVs, we adjusted the CVs according to signal-to-noise ratio. Firstly, we estimated the signal-to-noise ratio using the ratio of the number of transcripts with n miRNA seeds for real versus random miRNA seeds. As expected, the cooperation of miRNAs does help to raise the signal-to-noise ratio (Figure 2A). We then adjusted the average CVs (see Materials and Methods section) and tested the relationship again. We found that although the adjusted CVs are larger than the original ones, the positive correlation still holds (R2 = 0.80, P = 0.000087, Pearson correlation coefficient) (Figure 2B). Because the above signal-to-noise ratio calculation method failed to obtain the signal-to-noise ratio of target genes recognized by one seed (Figure 2A), we also estimated the signal-to-noise ratio using the ratio of the number of transcripts with at least n miRNA seeds for real versus random miRNA seeds and corrected CVs using these ratios. Finally, we observed the same correlation (Supplementary Figure S1).
Figure 2.
Adjusting CVs by signal-to-noise ratio does not affect the positive correlation between miRNA seed numbers and CVs. (A) Plot of the number of target genes recognized by n seeds. The distribution for the real seeds (solid circles) is shown alongside the distribution for the random seeds (hollow circles). The line bar indicates the ratios of the number of transcripts with n seeds for real versus random seeds and the right Y-axis represents the signal-to-noise ratio. (B) The correlation between miRNA seed numbers per target gene and the adjusted CVs. The average CV of the non-miRNA-target genes is indicated in Y-axis by triangle. The dash line indicates the linear least square regression line. The R2 and P-value are indicated.
Similarly, we also estimated the signal-to-noise ratio using the ratio of the number of transcripts with n binding sites of real versus random miRNA seeds and adjusted the CVs using these ratios. We found that the multiplicity of targets helps to raise the signal-to-noise ratio and the positive correlation still holds (data not shown).
Length of 3′UTR and gene expression variability
Genes with longer 3′UTRs may have more miRNA target sites on average. As a result, the 3′UTR length could be a potential confounding factor. To exclude the effect of 3′UTR length, we tested the correlation between the average log transformed CVs and the miRNA seed numbers using a linear regression model with the average log transformed UTR length as the confounder. It is shown that the positive correlation still holds (R2 = 0.64, P = 0.0033), so does the positive correlation between the average log transformed CVs and the miRNA binding site numbers (R2 = 0.55, P = 0.0024).
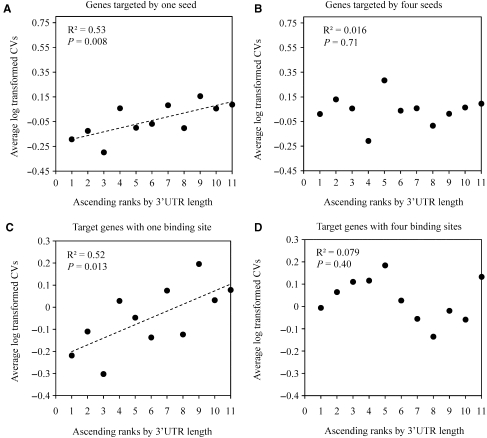
It is well known that besides of the miRNA target sites, the 3′UTRs also have other cis-acting elements or alternative polyadenylation signals affecting the mRNA expression levels (37–39). In addition, sequence variations [e.g. SNPs (40) and microsatellites (41)] in the 3′UTRs may affect cis-acting element efficiency, polyadenylation site selection and RNA secondary structure, and eventually influence mRNA stability (40). These factors may also contribute to gene expression variability in human populations, and therefore, the observed correlation between CVs and miRNA seed numbers/binding site numbers may be explained by the non-miRNA factors. To test the contribution of the non-miRNA factors, we used the 3′UTR length as the indicator because the longer the 3′UTR is, the more the regulators and genetic variants could reside in. Our data show that when the target genes are recognized by only one or two miRNA seeds, the CVs are positively related to the 3′UTR length (Figure 3A, Supplementary Table S2 for details). This observation suggests that the CVs of the target genes recognized by one or two seeds might be affected by the non-miRNA factors rather than miRNAs. In contrast, for target genes recognized by three or more seeds, the CVs are not related to the 3′UTR length (Figure 3B, Supplementary Table S2 for details), implying that miRNAs contribute to CVs of genes recognized by multiple miRNAs. Figure 3A and B show the examples of the relationship between the 3′UTR length and CVs. Similarly, for the target genes with one or two binding sites, the CVs are positively related to the 3′UTR length (Figure 3C, Supplementary Table S3 for details). In contrast, for genes with three or more binding sites, most of the CVs do not show correlation with the 3′UTR length (Figure 3D, Supplementary Table S3 for details).
Figure 3.
Examples of relationship between 3′UTR length and CVs. (A) Genes recognized by one seed were ascendingly ranked according to their 3′UTR lengths and incorporated into eleven groups with the same gene number. Spearman's rho was calculated between the 3′UTR ranks and the average CVs of each group. The positive correlation was identified. (B) For genes targeted by four seeds, using the same grouping approach, no correlation was identified between the 3′UTR ranks and the average CVs. (C) For genes with one miRNA binding site, using the same grouping approach, the positive correlation was identified. (D) For genes with four miRNA binding sites, using the same grouping approach, no correlation was identified.
Collectively, our data demonstrate that compared with the CVs of non-miRNA-target group, the CVs of target genes recognized by one or two miRNA seeds are similar (P = 0.77, two-tailed Mann–Whitney test), whereas the CVs of the target genes recognized by more than two miRNA seeds are significantly higher (P = 0.011, two–tailed Mann–Whitney test) (Table 1). When we divided the samples by gender or brain regions, the differences are still significant except for the temporal cortex (Table 2). In addition, the observed higher CVs of miRNA target genes do not depend on specific miRNAs (Supplementary Figure S2).
Table 1.
Comparison of gene expression variability between miRNA target genes and non-miRNA-target genes
| Seed region based method |
Experimentally verifieda |
|||
|---|---|---|---|---|
| Tar1-2/Non | Tar>2/Non | Targets/Non | ||
| Num | 2454/1663 | 1904/1663 | 109/1663 | |
| Median | 0.077/0.078 | 0.08/0.078 | 0.087/0.078 | |
| P-value# | 0.77 | 0.011* | 0.001* | |
Tar1-2, target genes recognized by 1–2 miRNA seeds; Tar>2, target genes recognized by three or more miRNA seeds; Non, non-miRNA-target genes; Num, gene numbers.
#Based on Two-tailed Mann–Whitney test.
*P < 0.05.
Table 2.
Comparison of gene expression variability between miRNA target genes (recognized by three or more miRNA seeds) and non-miRNA-target genes in sub-samples divided by gender or brain regions
| Male | Female | Frontal- cortex | Temporal- cortex | |
|---|---|---|---|---|
| Median (targets/non- targets) | 0.077/0.074 | 0.081/0.076 | 0.071/0.067 | 0.074/0.073 |
| P-value# | 0.028* | 0.00025* | 0.0079* | 0.17 |
#Based on Two-tailed Mann–Whitney test.
*P < 0.05.
As described above, miRNAs contribute to CVs of genes recognized by multiple miRNAs. To confirm this observation, we calculated the correlations between the CV of a target gene with more than two seeds/binding sites and its target seed/binding site number when considering the 3′UTR length as the confounding factor. Table 3 shows that the positive correlations still hold.
Table 3.
The relationship between CV of a gene and miRNA seed number/binding site number
| Seed region based method |
PITA method |
TargetScan method |
||
|---|---|---|---|---|
| CV/seed number | CV/binding site number | CV/seed number | CV/seed number | |
| r | 0.056 | 0.06 | 0.062 | 0.051 |
| P | 0.014* | 0.008* | 0.0032* | 0.048* |
The genes targeted by more than two miRNA seeds or with more than two binding sites were chosen. The r and P-values were calculated using log transformed 3′UTR length as the confounding factor.
To exclude the possibility that the pattern we observed was only restricted to a specific prediction algorithm, we performed the same data analysis using other two target prediction algorithms-TargetScan and PITA (25,26). The same positive correlation between expression variability of a gene and its targeted miRNA seed number was observed (TargetScan: ρ = 0.049, P = 0.002; PITA; ρ = 0.057, P = 0.002) (Table 3), as well as the same difference between miRNA target genes and non-miRNA-target genes (Supplementary Table S4). As the above analyses were based on the predicted target genes, we also calculated the average CV of the experimentally validated target genes of miR-124 (12) and found that the average CV of miR-124a's targets is significantly higher than that of the non-miRNA-target genes (Table 1).
SNPs located in the miRNA binding sites and gene expression variability
SNPs located in the miRNA binding sites are shown to affect the interaction between miRNAs and the target genes (42,43). Such variation in the binding sites is abundant in human (44), and may affect gene regulation and phenotypic variation. To evaluate the influence of such SNPs on the gene expression variability, we searched among 5 687 169 SNPs which were mapped to the unique positions in the human genome, and identified 277 genes with SNPs in the putative binding sites (defined as SNP-residing targets). We compared the average CV of SNP-residing targets with the average CV of non-SNP targets. The CV of SNP-residing targets is slightly higher than that of the randomly sampled non-SNP targets when corrected by seed numbers (Supplementary Figure S3, P = 0.047) or binding site numbers (P = 0.049) (refer to Materials and Methods seciton for details), suggesting that the SNPs might further increase the CVs within target gene groups. However, it should be noted that, because of the different sample sets of SNPs and the expression data, the above SNP based analysis needs to be confirmed using the same sample set in the future.
DISCUSSION
Previous study found that miRNA decreased the cross-species expression divergence, suggesting a role of miRNA in evolutionary constraints on gene expression variation (18). However, our analysis revealed an unexpected role of miRNAs on increasing within-species gene expression variability, independent of miRNA types or functional categories (see Supplementary Discussion). There are two possible explanations about this phenomenon. One possibility is that our result does reveal a general positive correlation pattern between the miRNA seed numbers per target gene and the CVs, and the expression variation of miRNAs themselves may lead to the increased CVs of the target genes. Most highly expressed miRNA sequences are conserved among species, suggesting functional constraints among species. However, the expression of miRNAs may fluctuate constantly at the population level during evolution to provide more options for adaptation. It is known that genes involved in basic cellular processes tend to avoid miRNA regulation due to short 3′UTRs that are specifically depleted of miRNA binding sites (16). Most of these genes are house-keeping or essential genes and the fluctuation of their expression is much lower compared with other genes (45,46). Consistent with this notion, our results show that the non-miRNA-target genes have lower expression variability compared with the target genes.
Alternatively, as the data analyzed in this study were from aged human subjects, our result may be explained by the increased fluctuation of miRNA expression during aging. To address this, we divided the 193 samples into three age groups (stage 1, 65- to 74-year-old; stage 2, 75- to 84-year-old; stage 3, 85 and older) (47,48), and compared the CVs between groups. Of the 11 958 genes surveyed, more than half of the genes showed decrease from stages 1 to 2 (6357 decreased genes and 5601 increased genes, P = 6.7 × 10−8, two-tailed Wilcoxon Signed Ranks Test) (Supplementary Figure S4). However, no significant increase or decrease was found between stages 2 and 3 (P = 0.61, two-tailed Wilcoxon Signed Ranks Test) (Supplementary Figure S4). This observation is consistent with a previous study (49), in which the individuals with the age ≤42 or ≥73-years-old showed more homogeneous pattern of gene expression in brain compared with individuals of 43- to 72-years-old. Interestingly, for target genes recognized by three or more miRNAs, there is no difference of CVs between stages 1 and 2 (Supplementary Figure S4) (P = 0.166, two-tailed Wilcoxon Signed Ranks Test), but we observed a significant increase from stages 2 to 3 (Supplementary Figure S4) (973 increased genes and 895 decreased ones, P = 0.001, two-tailed Wilcoxon Signed Ranks test). This observation suggests that aging might be another potential factor contributing to the expression fluctuation of miRNAs and eventually leads to increased target gene expression variability. Population level miRNA expression analysis of different developmental stages in the future will be informative to test this hypothesis.
In conclusion, we revealed that the increased expression variability of genes is concomitant with the increased number of miRNA seeds per target gene, suggesting direct influences of miRNA on gene expression variability, which may eventually contribute to phenotypic variation in human populations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This study was supported by grants from the National 973 project of China (2007CB947701, 2007CB815705), the Chinese Academy of Sciences (KSCX1-YW-R-34), the National Natural Science Foundation of China (30525028, 30630013 and 30623007), and the Natural Science Foundation of Yunnan Province of China.
We are grateful to Zhi-xiang Lu for his help in R programming. We also thank Leng Han, Ruo-lin Yang, Jin-kai Wang, Xia Zhang and Xin Li for their helpful discussions. Funding to pay the Open Access publication charges for this article was provided by the National 973 project of China (2007CB947701).
Conflict of interest statement: None declared.
REFERENCES
- 1.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, Burdick JT. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rockman MV, Kruglyak L. Genetics of global gene expression. Nat. Rev. Genet. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- 6.Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen KY, Morley M, Spielman RS. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat. Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 9.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 10.Megraw M, Sethupathy P, Corda B, Hatzigeorgiou AG. miRGen: a database for the study of animal microRNA genomic organization and function. Nucleic Acids Res. 2007;35:D149–D155. doi: 10.1093/nar/gkl904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li A, Mao L. Evolution of plant microRNA gene families. Cell Res. 2007;17:212–218. doi: 10.1038/sj.cr.7310113. [DOI] [PubMed] [Google Scholar]
- 12.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 13.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 14.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 15.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs Confer Robustness to Gene Expression and Have a Significant Impact on 3'UTR Evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 17.Farh KK-H, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The Widespread Impact of Mammalian MicroRNAs on mRNA Repression and Evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 18.Cui Q, Yu Z, Purisima EO, Wang E. MicroRNA regulation and interspecific variation of gene expression. Trends Genet. 2007;23:372–375. doi: 10.1016/j.tig.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A, Bentwich I, Einav U, Gilad S, Hurban P, Karov Y, et al. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A Mammalian microRNA Expression Atlas Based on Small RNA Library Sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, Kaleem M, Leung D, Bryden L, Nath P, et al. A survey of genetic human cortical gene expression. Nat. Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 22.Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat. Genet. 2006;38:1452–1456. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- 24.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 25.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 26.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 27.Altschul SF, Erickson BW. Significance of nucleotide sequence alignments: a method for random sequence permutation that preserves dinucleotide and codon usage. Mol. Biol. Evol. 1985;2:526–538. doi: 10.1093/oxfordjournals.molbev.a040370. [DOI] [PubMed] [Google Scholar]
- 28.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 30.Tirosh I, Weinberger A, Carmi M, Barkai N. A genetic signature of interspecies variations in gene expression. Nat. Genet. 2006;38:830–834. doi: 10.1038/ng1819. [DOI] [PubMed] [Google Scholar]
- 31.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2007. [Google Scholar]
- 32.Gentleman R, Carey V, Bates D, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hon LS, Zhang Z. The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biol. 2007;8:R166. doi: 10.1186/gb-2007-8-8-r166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 35.Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006;38(Suppl.):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 36.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross J. mRNA stability in mammalian cells. Microbiol. Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 39.Maret D, Boffa MB, Brien DF, Nesheim ME, Koschinsky ML. Role of mRNA transcript stability in modulation of expression of the gene encoding thrombin activable fibrinolysis inhibitor. J. Thromb. Haemost. 2004;2:1969–1979. doi: 10.1111/j.1538-7836.2004.00971.x. [DOI] [PubMed] [Google Scholar]
- 40.Boffa MB, Maret D, Hamill JD, Bastajian N, Crainich P, Jenny NS, Tang Z, Macy EM, Tracy RP, Franco RF, et al. Effect of single nucleotide polymorphisms on expression of the gene encoding thrombin-activatable fibrinolysis inhibitor: a functional analysis. Blood. 2008;111:183–189. doi: 10.1182/blood-2007-03-078543. [DOI] [PubMed] [Google Scholar]
- 41.Chen TM, Kuo PL, Hsu CH, Tsai SJ, Chen MJ, Lin CW, Sun HS. Microsatellite in the 3′ untranslated region of human fibroblast growth factor 9 (FGF9) gene exhibits pleiotropic effect on modulating FGF9 protein expression. Hum. Mutat. 2007;28:98. doi: 10.1002/humu.9471. [DOI] [PubMed] [Google Scholar]
- 42.Abelson JF, Kwan KY, O'Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 43.Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, Elton TS, Hatzigeorgiou AG, Antonarakis SE. Human microRNA-155 on Chromosome 21 Differentially Interacts with Its Polymorphic Target in the AGTR1 32 Untranslated Region: A Mechanism for Functional Single-Nucleotide Polymorphisms Related to Phenotypes. Am. J. Hum. Genet. 2007;81:405–413. doi: 10.1086/519979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, Caiment F, Elsen JM, Eychenne F, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat. Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 45.Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB. Noise minimization in eukaryotic gene expression. PLoS Biol. 2004;2:e137. doi: 10.1371/journal.pbio.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu H, Greenbaum D, Xin Lu H, Zhu X, Gerstein M. Genomic analysis of essentiality within protein networks. Trends Genet. 2004;20:227–231. doi: 10.1016/j.tig.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 47.Shumway-Cook A, Guralnik JM, Phillips CL, Coppin AK, Ciol MA, Bandinelli S, Ferrucci L. Age-associated declines in complex walking task performance: the Walking InCHIANTI toolkit. J. Am. Geriatr. Soc. 2007;55:58–65. doi: 10.1111/j.1532-5415.2006.00962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paloutzian RF, Park CL. Handbook of the Psychology of Religion and Spirituality. The Guilford Press, New York; 2005. [Google Scholar]
- 49.Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 50.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.