Abstract
Gene targeting via homologous recombination (HR) is an important application in biotechnology and medicine. However, in mammalian cells HR is much less efficient than random integration. Triplex-forming oligonucleotides (TFOs) linked to DNA damaging agents (e.g. psoralen) can stimulate HR, providing the potential to improve gene therapy applications. To elucidate factors affecting TFO-directed psoralen interstrand crosslink (ICL)-induced recombination, we constructed a series of plasmids with duplicated supF reporter genes, each containing an inactivating deletion, to measure HR frequencies in mammalian cells. Our results indicated that TFO-directed ICL-induced recombination frequencies were higher in the plasmids with larger distances between duplicated supF genes than with a smaller separation distance. However, the position of the ICL relative to the reporter genes did not affect HR frequencies. Recombination spectra were altered by the distance between supF copies. Although single-strand annealing (SSA) recombinants were predominant in all plasmid substrates, the plasmid with the shortest interval (60 bp) revealed a significant proportion of gene conversions (GCs). GCs occurred exclusively in the gene containing the shortest deletion, regardless of the distance between supF genes, ICL position or deletion orientation. Our analyses indicated that SSA is the predominant mechanism of ICL processing of these substrates in mammalian cells.
INTRODUCTION
Homologous recombination (HR) provides an effective approach to targeted genome modification. Gene targeting is not only useful for creating animal models of human disease, but may provide a strategy for treating genetic disorders in humans (1,2). A limitation to the current technology is that the rate of HR in mammalian cells is extremely low (10−8–10−5 events per cell per generation), while nonhomologous recombination or random integration occurs with a frequency ∼100- to 1000-fold higher than HR in mammalian cells (3). Efforts have been made to stimulate HR frequencies using a variety of strategies (4). For example, the introduction of a DNA double-strand break (DSB) at a specific site in the genome with the sequence-specific endonuclease I-SceI has been shown to enhance the frequency of HR by several orders of magnitude (5–8). However, this strategy is not amenable for therapeutic applications, due to the requirement of pre-introduction of the I-SceI recognition sequence at the target site in the genome.
An alternative strategy is the use of triplex technology to target DNA damage in a site-specific fashion to stimulate HR at the targeted site. Triplex-forming oligonucleotides (TFOs) can recognize and bind duplex target DNA in a sequence-specific fashion via Hoogsteen hydrogen bonding between the TFO and the purine-rich strand of the target duplex DNA (9). Triplex formation itself can result in increased frequencies of HR and mutation, likely due to its recognition and processing by DNA repair proteins (10–14). Furthermore, TFOs can be conjugated to DNA damaging agents (e.g. psoralen), which have been shown to stimulate HR and/or mutagenesis in a site-specific fashion (15). The DNA damaging agent psoralen, which is activated by UVA irradiation to form DNA interstrand crosslinks (ICLs), has been conjugated to TFOs to induce HR (4,16–22). Psoralens are naturally occurring planar tri-heterocyclic compounds consisting of a furan ring and a pyrone ring. Psoralen intercalates in DNA, and upon absorption of photons at 365 nm forms covalent crosslinks, preferentially at 5′-dTpA-3′ sequences (23,24).
While psoralen-conjugated TFOs have been used in mammalian systems to enhance the frequency of HR, many of the factors influencing ICL-induced HR in these systems, such as the distance between the tandemly duplicated reporter genes and the position of the ICL relative to reporter genes, have not been studied in detail. To address these questions, we designed a series of plasmid substrates with increasing distances (60bp, 350bp, 700 bp and 1.4 kb) between the two supF reporter genes, and varying TFO-psoralen ICL positions relative to the supF genes nearer to the upstream copy, centrally located between the two supF genes, or nearer to the downstream copy of the supF gene. Using these constructs, we determined ICL-induced HR frequencies and recombinant spectra in mammalian cells. Our results demonstrated that in all plasmid substrates tested, the TFO-directed psoralen ICL enhanced the HR frequency relative to control treatment groups. The increase in HR frequency was greater in plasmids containing more than 60 bp of intervening DNA sequence between the reporter supF genes, although the position of the ICL relative to the reporter genes did not have a significant effect. An intriguing result was that gene conversion (GC) recombinants were only observed in substantial proportions in the construct with the shortest sequence length separating the supF duplications. Further characterization of GC recombinants revealed a polarity, with all the conversion events occurring in the supF gene containing the shorter deletion, regardless of the position of the deletion relative to the TFO binding site.
MATERIALS AND METHODS
Plasmid substrate construction
The pSupFG1 plasmid is a shuttle vector, which can replicate in human cells due to the SV40 origin of replication and the large T antigen sequence, and can also replicate in Escherichia coli due to a ColE1-derived origin of replication. The supF gene in pSupFG1 encodes a suppressor tRNA, which can suppress amber mutations in the lacZ gene in the indicator E. coli strain MBM7070. The E. coli strain, MBM7070 has the genotype: F-lacZ (am)CA7020, lacY1, hsdR−, hsdM+, araD139 Δ(araABC-leu)7679, galU, galK, rpsL, thi (25,26), has been used extensively in studies of recombination events in mammalian cells (21,27–29). Background events that might have occurred in the bacteria (and not in the mammalian cells) were controlled in several ways. First, background frequencies of spontaneous recombination events in the bacterial cells were measured by transfecting the plasmid substrates directly into the MBM7070 indicator bacterial cells; HR frequencies in the bacterial cells did not exceed 0.8 × 10−4 for any of the reporter plasmids, which were substantially lower than those found after replication in the mammalian cells. In addition, the plasmids were treated with DpnI restriction enzyme following recovery from the mammalian cells (and prior to transfection in the bacterial cells), to remove plasmids that had not replicated in the mammalian cells from further analysis; therefore, only those plasmids that had processed the crosslink (leading to stimulation in HR) in the mammalian cells were subjected to the blue–white screen in the bacteria. The pSupFG1 shuttle vector was modified by incorporating two tandem mutant supF genes containing different deletions; the upstream supF gene contains an 8 bp deletion at the 3′-end of the reporter gene (at original position 82–90 bp), and the downstream supF gene contains a 24 bp deletion at the 5′-end of the reporter gene (at original position 17–41 bp). Between these two mutated supF genes, a 60 bp intervening spacer DNA sequence was inserted, which contains a unique TFO binding site for pTFO1 (psoralen-5′-TGTGGTGGGGGGTTTGGGG-3′) and a psoralen crosslinking site, to produce the parental plasmid, pTFOLK (Figure 1A). To study the effect of distance between the supF reporter genes and the location of the psoralen ICL on HR frequency, we inserted a series of DNA sequences of varying lengths from the human GAPDH gene to construct the following plasmids; p350 (containing a 350 bp DNA insertion between the two supF genes), p700 (containing a 700 bp insertion) and p1.4K (containing a 1.4 kb insertion). To exclude the possibility of DNA sequence effects on HR, we designed a series of primers to insert DNA sequences from the same region of the human GAPDH gene for the construction of the plasmids. To determine the effect of ICL position on HR, we constructed a series of plasmids with varying distances between the supF genes and also with different ICL positions: nearer to the upstream copy (pTFO350, pTFO700 and pTFO1.4K), nearer to the downstream copy (p350TFO, p700TFO and p1.4KTFO) or centrally located between the two reporter copies (Figure 1B and C; p350Mid, p700Mid and p1.4KMid).
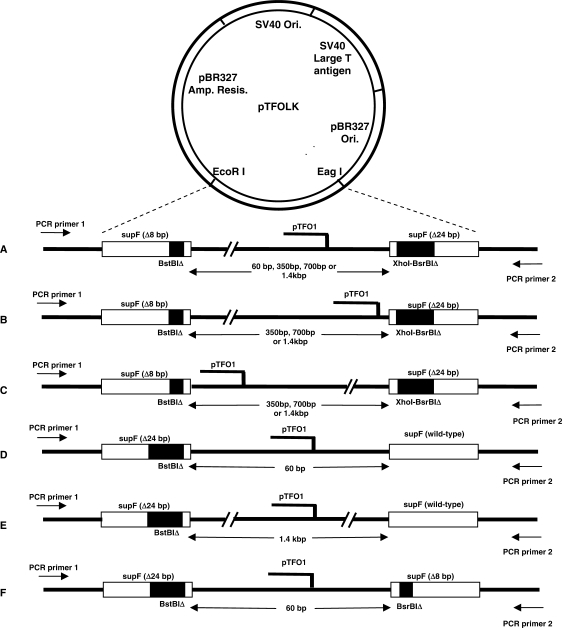
Figure 1.
Plasmid substrates. (A) Map of the parental plasmid substrate, pTFOLK. pTFOLK contains tandemly arranged supF genes each containing a different deletion at a different location, separated by 60 bp and a TFO1 binding site adjacent to a psoralen crosslinking site. The black bars indicate the deletions. The 8 bp deletion in the upstream supF gene is located at the BstBI site, and the 24 bp deletion in the downstream supF gene spans the XhoI to BsrBI sites. The binding site for the psoralen-conjugated TFO (pTFO1) was either centrally located between the supF copies (A), was nearer to the downstream supF gene (B) or was placed nearer to the upstream supF gene (C). (D and E) Plasmid substrates used to confirm that the distance between the supF promoter and the supF genes did not affect the function of the genes. The plasmid in 1D (p24BstB-TFOLK) is similar to pTFOLK plasmid, except that the upstream supF gene contains a larger deletion (24 bp) at the BstBI site, and the downstream supF gene is wild-type. The plasmid in 1E (p24BstB–1.4K) is similar to plasmid p24BstB–TFOLK, the only difference being the distance between the supF genes is 1.4 kb rather than 60 bp. (F) Plasmid p24BstB/TFOLK/BsrB was designed to determine the effect of deletion size on gene conversion events. This plasmid is similar to pTFOLK, except that the deletion positions were inverted; the larger 24 bp deletion at the BstBI site is in the upstream supF gene, while the smaller 8 bp deletion at the BsrBI site is in the downstream supF gene.
To exclude the possibility that the GC events occurring in the downstream supF gene were masked by the distance from the promoter, we constructed the plasmids shown in Figure 1D and E. These plasmids contain a 24-bp deletion at the BstBI restriction site in the upstream supF gene and a wild-type, functional supF gene downstream. Between these two genes, the shortest and longest (60 bp and 1.4 kb, respectively), intervening DNA sequences were inserted.
To characterize the GC events, another plasmid was generated, which is similar to pTFOLK, except that the deletion positions were reversed, with the larger deletion (24 bp) in the upstream supF gene, and the shorter deletion (8 bp) in the downstream copy (Figure 1F). All plasmid sequences were confirmed by DNA sequencing.
Substrate preparation
Duplex plasmid substrates are the original plasmids without any treatment. UVA-treated plasmid substrates are plasmids irradiated with 1.8 J/cm2 of UVA (365 nm). Triplex-modified plasmids (3 μg) were incubated with pTFO1 oligonucleotide (1 μl of TFO at 1.2 × 10−4 M) in triplex binding buffer [10 mM Tris–HCl, pH 7.6, 10 mM MgCl2 and 10% (vol/vol) glycerol] in a total volume of 50 μl at 37°C overnight. The ICL-modified plasmid substrates were incubated with pTFO1 under the same conditions as triplex-modified plasmids, except that the plasmids were also irradiated with 1.8 J/cm2 UVA light (365 nm). The UVA- and ICL-treated plasmid substrates were covered by Mylar film, which effectively eliminates UV with wavelengths <320 nm (30).
Recombination assays
HeLa cells were seeded in T-25 flasks and expanded to ∼80–90% confluence. Then cells were transfected with 4 μg plasmid using Geneporter reagent according to the manufacturer's instructions (Genlantis, San Diego, CA, USA). On the next day, cells were washed twice with PBS and incubated in 5 ml 10% FBS EMEM culture medium at 37°C, 5% CO2 for another 24 h. Forty-eight hours after transfection, cells were harvested with trypsin–EDTA followed by centrifugation at 1000 rpm at 4°C for 5 min. Cell pellets were lysed and plasmid DNA was isolated using a Qiagen miniprep kit (Qiagen, Valencia, CA, USA). The plasmid DNA preparation was extracted with phenol/chloroform, ethanol-precipitated, and then subjected to DpnI digestion.
Competent E. coli MBM7070 indicator cells were electroporated with plasmid DNA isolated from HeLa cells. After electroporation, the transfected MBM7070 cells were suspended in SOC media and incubated in a 37°C shaker for 45 min. Finally, the bacterial cells were spread on to LB plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal), isopropyl-β-d-thiogalactoside (IPTG) and Ampicillin selection medium (XIA medium) for blue-white colony screening. PCR was performed on DNA isolated from bacterial colonies for genotype analysis of the recombinants. The primer sequences used were: 5′-TTGCCGGGAAGCTAGAGTAA-3′ (PCR primer 1, Figure 1), and 5′-TTTTTGTGATGCTCGTCAGG-3′ (PCR primer 2, Figure 1). PCR reaction conditions included denaturing at 90°C, annealing at 54.7°C and elongation at 72°C. After 35 cycles, the PCR products were run on a 1% agarose gel.
Statistical analyses
Data from the recombination assays were analyzed by one-way ANOVA to compare the recombination frequencies among all groups. To compare the recombination frequencies among the different ICL treated plasmids with different intervening spacer lengths or different ICL positions, the Tukey Honestly Significant Difference (HSD) method was used. Analysis of GC frequencies was conducted by the two independent proportions test.
RESULTS
TFO-directed psoralen ICLs enhance the frequency of HR
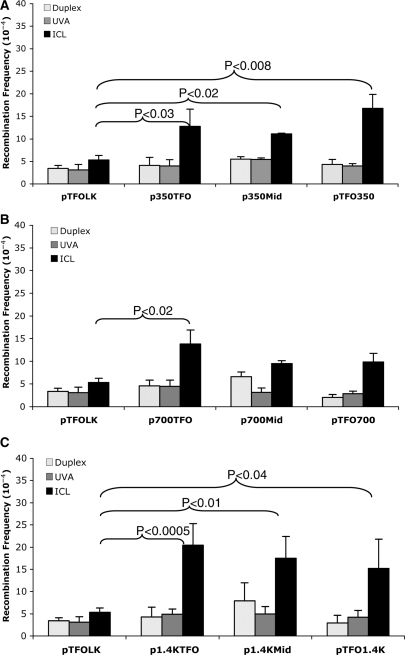
Using the plasmids shown in Figure 1, we performed a series of recombination assays in HeLa cells. Plasmids with no treatment (duplex), with only UVA treatment (UVA), and with triplex structures only (in the absence of UVA, triplex) served as controls for the pTFO1 + UVA-treated plasmid (ICL). Crosslinked plasmid substrates were prepared in vitro prior to transfection into HeLa cells to ensure the same efficiency of ICL formation in all plasmid substrates. Plasmids were isolated from HeLa cells 48 h after transfection to allow for replication and repair to occur prior to determining HR frequencies. The results (Figure 2A–C) demonstrated that the TFO-directed ICL HR up to ∼4-fold (∼20 × 10−4) over background frequency (∼5 × 10−4 in the case of p1.4KTFO). The controls showed no increase in HR frequency over background, as expected.
Figure 2.
HR frequencies induced by TFO-directed psoralen ICLs on plasmids in HeLa cells. Plasmids containing 350 bp (A), 700 bp (B) or 1.4 kb (C) between supF copies were treated as described in the Materials and Methods section: white bars, plasmids with no treatment; gray bars, plasmids treated with UVA only; black bars, plasmids treated with the psoralen-conjugated TFO1 + UVA irradiation. Plasmids were isolated from HeLa cells 48 h after transfection and HR frequencies determined by blue-white screening of the colonies. All results shown are from at least three independent experiments (error bars represent the SDs), P-values were derived from one-way ANOVA.
The effects of distance between the two reporter genes and psoralen ICL position on the frequency of HR
In order to determine if the distance between the reporter genes affected the frequency of HR, we performed experiments utilizing the plasmid substrates shown in Figure 1A–C. Interestingly, for the plasmid with the shortest distance between the reporter genes (60 bp), pTFOLK, the TFO-directed ICL induced HR frequency was <2-fold above the background levels (Figure 2A), and no significant differences were observed between ICL-containing and duplex plasmids. However, when the distance between the supF genes was increased to 350 bp, the TFO-directed psoralen ICL had a significant effect on the recombination frequency. In the presence of a TFO-directed psoralen ICL, all plasmids containing 350 bp between reporter genes (p350TFO, p350Mid and pTFO350) showed a ≥2-fold stimulation in recombination frequencies compared to pTFOLK (Figure 2A). In Figure 2B recombination frequencies are shown for the p700 series of plasmids. While 700 bp between supF genes also led to an increase in recombination frequencies compared to those from pTFOLK, there were no significant differences in the frequencies between the plasmids containing 350 versus 700 bp between reporter genes (cf. Figure 2A and B). When the distance between reporter genes was increased to 1.4 kb, the TFO-directed psoralen ICL resulted in a 4-fold stimulation of recombination over that found in pTFOLK, which is similar to the increases detected in the plasmids containing 350 or 700 bp between reporter genes (Figure 2C). Another interesting finding was that the recombination frequencies induced by TFO-psoralen is independent of the position of the ICL relative to the reporter genes (Figure 2A–C). That is, in the plasmids with the same insertion distance between two supF genes, the recombination frequencies are similar among the plasmid substrates regardless whether the plasmids with the ICL position were nearer to the upstream copy of the supF gene, nearer to the downstream copy or centrally located (Figure 1A–C).
To determine if triplex formation itself was contributing to the TFO-directed psoralen ICL-stimulated recombination, we performed a set of experiments using the TFO only in the absence of the psoralen ICL, and measured the frequencies of HR following incubation in HeLa cells. Triplex formation itself does not stimulate recombination under the conditions of our assay (data not shown). Again, in the presence of the TFO-directed psoralen ICL, the plasmid containing the greatest distance between reporter genes (1.4 kb), showed the greatest stimulation in recombination. This result demonstrated that recombination was enhanced by the TFO-directed psoralen ICL, and not by the triplex structure itself.
Recombinant spectra are affected by the distance between the reporter genes, but not by the ICL position
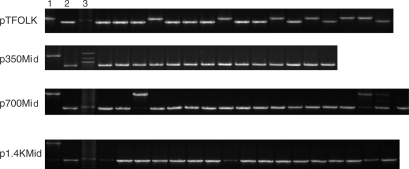
Because we found that the frequency of HR induced by ICLs was only affected by the distance between the reporter copies, but not by ICL position, we wanted to determine if the types of recombination events were altered by distance and/or ICL position. In our experimental system, recombinants (i.e. blue colonies) can result from two different mechanisms, single-strand annealing (SSA) or GC. SSA occurs between tandemly arranged homologous sequences, resulting in deletion of the intervening DNA sequences between the regions of homology, thus leaving only one copy of the homologous gene. GCs, on the other hand, represent a nonreciprocal exchange of sequence information from one of the two homologous genes to the other, retaining both copies. To characterize the recombinants generated in HeLa cells, we performed colony PCR using the primer set shown in Figure 1A. Recombinants derived from a SSA event will contain only one copy of the supF gene, revealing a faster migrating product by agarose gel analysis, while those derived from GC events will contain both copies (with one copy corrected to the wild-type sequence, and one copy containing the original deletion), such that the product will migrate similarly to that of the parental plasmid as visualized by agarose gel analysis (Figure 3). The proportions of GC events occurring on the plasmids in HeLa cells are shown in Table 1. Plasmid pTFOLK, containing the shortest distance (60 bp) between the two supF genes, showed a substantial proportion of GCs following ICL treatment (∼21%), in marked contrast to the other plasmids, which contained lengths from 350 bp to 1.4 kb between supF duplications, and generated <4% GC events.
Figure 3.
Characterization of recombinants. Lane 1 contains the products generated from a colony PCR reaction with the untreated parental plasmid, subjected to gel electrophoresis on a 1% agarose gel. Lane 2 contains PCR products generated from pSupFG1 plasmid containing only one copy of the supF gene. Lane 3 contains 100 bp DNA ladder. The slower migrating product indicates a gene conversion event, i.e. two copies of the supF gene and the intervening DNA sequence. The faster migrating product runs at the same position as the single copy pSupFG1 plasmid, indicating a recombinant generated by SSA. All recombinant samples shown are from ICL treatment groups.
Table 1.
Gene conversion events in recombinant colonies
| pTFOLK | p350TFO | p350Mid | pTFO350 | p700TFO | p700Mid | pTFO700 | p1.4KTFO | p1.4KMid | pTFO1.4K | p24BstB/ TFOLK/BsrB | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Duplex | 10/77 (13%) | 3/53 (5.7%) | 1/80 (1.3%) | 2/56 (3.6%) | 2/31 (6.5%) | 7/63 (11.1%) | 3/75 (4%) | 0/53 (0%) | 1/78 (1.3%) | 0/41 (0%) | 1/81 (1.2%) |
| UVA | 2/35 (5.7%) | 0/73 (0%) | 4/53 (7.5%) | 1/68 (1.5%) | 2/53 (3.8%) | 3/55 (5.5%) | 2/54 (3.7%) | 0/53 (0%) | 2/43 (4.7%) | 0/36 (0%) | ND |
| ICL | 14/66 (21.2%) | 1/47 (2.1%) | 1/128 (0.8%) | 2/71 (2.8%) | 0/46 (0%) | 3/85 (3.5%) | 2/73 (2.7%) | 0/42 (0%) | 1/113 (0.9%) | 0/69 (0%) | 1/98 (1%) |
ND, not determined.
Gene conversions occur exclusively in the supF copy containing the shorter deletion
To further characterize the GC events, a total of 36 recombinant plasmids derived from GC events in HeLa cells were sequenced (data not shown). Strikingly, the sequencing results demonstrated that in all cases, correction to wild-type had occurred in the upstream supF gene containing the shorter deletion, while the downstream supF gene containing the larger deletion remained in mutant form. To exclude the possibility that potential GC events occurring in the downstream copy were masked by the increased distance from its promoter, we constructed two additional plasmids (Figure 1D and E). These plasmids each contain a 24 bp deletion at the BstBI site in the upstream supF gene, and a wild-type, functional supF gene downstream. The only difference between the plasmids is that the intervening DNA length between the supF genes is 60 bp in p24BstB–TFOLK and 1.4 kb in p24BstB–1.4K. Both plasmids produced blue colonies when grown on XIA selection medium, suggesting that the distance from the promoter in the p1.4 plasmids still allows for detection of a functional supF gene. This result further supports the observation that the GC events occurred exclusively in the gene containing the shorter deletion.
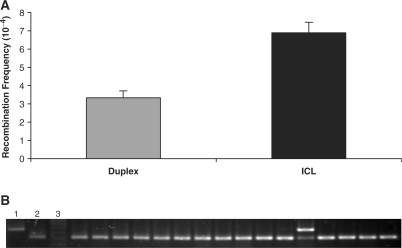
To better understand the polarity of the GC events (i.e. if the deletion size and/or gene position affected the outcome), we constructed plasmid p24BstB/TFOLK/BsrB, which has the 24 bp deletion in the upstream supF gene and the 8 bp deletion in the downstream copy. Thus, the deletion positions were inverted and separated by the same 60 bp sequence as in pTFOLK (Figure 1F). Forty-eight hours after transfection of the psoralen-crosslinked plasmid into HeLa cells, DNA was isolated and the HR frequency and spectrum were determined. We found that while the frequency of HR was similar to that found in pTFOLK, the frequency of GC events was substantially reduced (Figure 4A and B). However, the GC event that was detected now occurred in the downstream supF gene containing the shorter deletion. These results suggest that GC in our system might depend on both the position and size of the deletion. Larger deletions nearer to the promoter appear to reduce the GC frequency, under the conditions of our assay.
Figure 4.
HR frequencies induced by TFO-directed ICLs on plasmid p24BstB/TFOLK/BsrB in HeLa cells. (A) Recombination frequencies were measured 48 h after plasmid p24BstB/TFOLK/BsrB alone (duplex), or treated with pTFO1 and UVA irradiation (ICL), were transfected into HeLa cells. Error bars represent SDs. (B) Representative colony PCR analysis of recombinants (as described in Figure 3). Lanes 1–3 contain the same samples as described in Figure 3, lanes 1–3.
DISCUSSION
This study was undertaken to investigate the influence of some of the variables contributing to the stimulation of HR in mammalian cells by TFO-directed ICL formation in recombination-reporter plasmid substrates. The variables studied included the effects of the distance between homologous supF reporter gene duplications, and the placement of the TFO-directed ICL in the intervening sequence separating the supF genes, on induced HR frequencies and recombinant spectra. Our results confirm that triplex technology is a useful means to induce site-specific HR in mammalian cells, consistent with other studies of triplex-mediated enhancement of HR (16–22). However, in contrast to several published studies (16,17,20), we found that the triplex structure itself was not sufficient to induce HR in the absence of the TFO-directed psoralen ICL (data not shown). This discrepancy may be explained by the differences in HR reporter systems. Our plasmid reporter system does not allow for mutation detection, as does the reporter system used by Faruqi et al. (20), and triplex structures are known to induce mutagenesis, even in the absence of DNA damage (10,31,32). In addition, our recombination reporter system has a minimum of 60 bp between homologous reporter genes, with the TFO-binding site contained within this intervening sequence, whereas the reporter plasmid used by Faruqi et al. (17,20) contained only 9 bp between homologous reporter genes, and positioned the TFO binding site within the upstream copy of the supF gene.
Effects of distance between supF reporter genes on HR frequencies
Our results clearly demonstrate that the distance between the two tandemly arranged supF genes has a significant effect on TFO-directed psoralen ICL-induced HR frequencies, with distances longer than 60 bp (i.e. 350 bp, 700 bp and 1.4 kb) between the reporter genes resulting in much higher levels of ICL-induced HR (Figure 2). While the mechanism underlying this distance-dependent increase in HR is not clear, there are several possible explanations. For example, during ICL uncoupling, incisions made in the vicinity of the ICL may require further processing into a single-stranded gap to provoke efficient strand invasion, as has been demonstrated in bacterial cells during ICL-induced RecA-mediated strand exchange (33). Supportive evidence for this notion is also provided from in vitro studies by Bessho and Sancar (34) suggesting that a 22–28 nt single-stranded region was removed only from the 5′ end of an ICL. Thus, gap formation in pTFOLK, with the shortest intervening length, may reduce the extent of homology in the supF gene(s), resulting in lower levels of HR. However, in the plasmids containing longer intervening sequence distances (p350, p700 and p1.4K), gap formation is not likely to result in substantial reduction in supF homology, and therefore would not result in suppression of HR. In addition, as the length of the intervening sequence increases, so does the probability of nicks being induced within this region, thereby facilitating HR.
From results using a similar plasmid substrate containing a site-specific, but non-TFO-directed ICL, Zheng et al. (29) proposed a model for ICL processing that includes the generation of two single-stranded gaps on complementary strands 5′ to the ICL. After annealing of homologous sequences between the two tandem supF genes, the flap containing the ICL is removed by a second round of excision. In our study, it is possible that increasing the distance between homologous sequences can increase the efficiency of the annealing step, resulting in higher levels of induced SSA (29).
Effect of distance between homologous reporter genes on induced recombinant spectra
In mammalian cells, mitotic recombination between tandemly duplicated sequences generates primarily GCs (35–37). However, nonconservative products, which can arise by single reciprocal exchange or SSA are also generated; these recombinants are characterized by the loss of one of the gene duplications and all intervening sequences. Results of a number of studies support SSA as the most likely mechanism for generating these nonconservative recombination products in mitotic mammalian cells (35,38–40). In intrachromosomal recombination-reporter systems, which utilize I-SceI-induced DSBs to initiate recombination, products arising from SSA increase significantly (40,41). In plasmid substrates such as those used in the present study, SSA recombinants are the predominant product (42–45). Our results from the present study confirm this observation for ICL-initiated recombination; however, although GC recombinants did not predominate, their presence indicated that there are multiple recombination mechanisms involved in processing TFO-directed psoralen ICLs.
Table 1 presents the distributions of GC recombinants observed in the recombination-reporter plasmids used in this study. TFO-directed psoralen ICL-induced recombination in the pTFOLK plasmid generated ∼20% GC products; this ratio of GC:SSA recombinants (1:4) approximates the inverse of the ratio of GC:SSA recombinants observed in intrachromosomal tandemly repeated recombination-reporter substrates (36,37). However, as the distance between the supF reporter genes was increased, ICL-induced GCs were reduced to near background levels and SSA products constituted almost all the recombinants generated. One implication of this result is that conversion tracts may be somewhat restricted for ICL processing compared to DSBs, since another study has shown increased conversion frequencies with increasing intervening sequence length in I-SceI-induced recombination between NEO repeats in mammalian cells (8); however, this study was performed using an intrachromosomal recombination-reporter substrate in which the I-SceI site was located within the upstream NEO copy, and so may not be strictly comparable. There is evidence supporting the role of a DSB intermediate in the processing of ICLs (46,47); however, DSBs are more recombinogenic than ICLs (29), thus our results suggest that processing of a TFO-directed ICL may not always lead to a DSB intermediate.
Another possible mechanistic difference between HR processing of DSBs and ICLs is that DSB-induced recombination events utilize functional mismatch repair (MMR) to correct heterologous intermediates (48), whereas the recombination events induced by TFO-directed ICLs may not require a functional MMR pathway (17). However, our sequencing analyses of GC products suggests that processing of the psoralen ICLs was error free, since no point mutations were identified in the vicinity of the ICL site. Because the TFO binds to its target site duplex via Hoogsteen hydrogen bonds, the cell may recognize the triplex structure as a type of mismatch and perhaps some elements of the MMR machinery are recruited and participate in ICL processing. In support of this idea, published work from our laboratory indicates that MSH2 participates in an error-free pathway of TFO-directed ICL processing (14). In addition, MMR has been in implicated in recombination to ensure fidelity (49,50). Processing of ICLs thus seem to involve MSH2, a protein from the MMR pathway, but this appears to be a separate and unique function of this protein outside of its role in MMR per se (14,51).
Effect of ICL placement on induced HR frequencies and recombinant spectra
The position of the TFO-directed psoralen ICL relative to the duplicated supF reporter genes had no effect on either induced HR frequencies (Figure 2A–C) or recombinant spectra (Table 1). In studies using yeast (52,53), DSBs between duplicated sequences resulted in predominantly SSA recombinants, whereas a DSB in one gene duplicate resulted in conversions (54,55). In studies using mammalian cells (44,56), the efficiency of SSA was modulated by the position of the DSB, with positioning close to the middle provoking the highest level of SSA. A possible explanation may be that the simultaneous exposure of complementary single strands in the repeated genes resulted in the most efficient annealing of complementary homologous single-stranded DNA after exonucleolytic resection of the ends (44,56). Since we do not observe any effect with TFO-directed ICLs in our experimental system, our results are consistent with the notion that not all ICLs are processed through DSB intermediates.
Polarity of gene conversions
A striking observation from our experiments was that GCs occurred exclusively in the upstream copy of the supF gene containing the smaller deletion. A similar finding was observed by Glazer and colleagues (20), although the HR substrate used in their study contained point mutations in the duplicated reporter genes rather than deletions. We also observed that the preference for GC events in the upstream copy was independent of the TFO-directed ICL, occurring spontaneously even in untreated plasmids. We excluded the possibility that GCs occurred in the downstream copy but were potentially undetectable due to increased distance from the promoter, inhibiting transcription of the supF gene (Figure 1D and E). It has been reported that transcription can stimulate recombination (57–59), and because the upstream copy of the supF gene is in closer proximity to the promoter than the downstream copy, this could have influenced copy choice for GCs. To determine if the size of the deletion was involved in copy selection for conversions, or the relative positions of the genes to the promoter, we modified the substrate to contain the 8 bp deletion in the downstream supF gene, with the larger (24 bp) in the upstream copy. Our results demonstrated that the HR spectra were largely dependent on the relative positions of the deletions (Figure 1F), such that the GC events resulted in correction of the smaller deletion regardless of its location relative to the promoter.
Weiss and Wilson (60) reported that GC events were unaffected by mismatched loop size in an SV40-based system, although this system is quite different from the one used in our study. A study by Taghian and Nickoloff (7) measured conversion tracts around I-SceI-induced DSBs in intrachromosomal recombination substrates in CHO cells and found that conversion tracts were larger and bidirectional as compared to yeast, where they were primarily unidirectional and smaller. In yeast, nonconservative (SSA or crossover) recombinant products are less likely to occur if conversion tracts are smaller (61). In our system, if conversion tracts are restricted after the initial processing of a TFO-directed ICL, the efficiency of GC of the recombination intermediate might depend on the size of the deletion to be repaired by HR, with smaller deletions having a higher probability of conversion. Clearly, further experimentation is warranted to determine the mechanism of copy choice in the GC events and why the smaller deletion is preferred for correction.
ACKNOWLEDGEMENTS
We thank Ms Sarah Henninger for assistance in preparation of the article. We also appreciate suggestions on plasmid construction from Dr Guliang Wang in our laboratory, and assistance with statistical analyses from Kevin Lin. This work was supported by National Institutes of Health/NCI grant P01 CA097175 (K.M.V.) and an NIEHS Center Grant ES007784. Funding to pay the Open Access publication charges for this article was provided by NIH/NCI grant P01 CA097175.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vijayaraj P, Sohl G, Magin TM. Keratin transgenic and knockout mice: functional analysis and validation of disease-causing mutations. Methods Mol. Biol. 2007;360:203–251. doi: 10.1385/1-59745-165-7:203. [DOI] [PubMed] [Google Scholar]
- 2.Fischer A, Hacein-Bey-Abina S, Cavazzana-Calvo M. Gene therapy of metabolic diseases. J. Inherit. Metab. Dis. 2006;29:409–412. doi: 10.1007/s10545-006-0270-7. [DOI] [PubMed] [Google Scholar]
- 3.Bollag RJ, Waldman AS, Liskay RM. Homologous recombination in mammalian cells. Annu. Rev. Genet. 1989;23:199–225. doi: 10.1146/annurev.ge.23.120189.001215. [DOI] [PubMed] [Google Scholar]
- 4.Vasquez KM, Marburger K, Intody Z, Wilson JH. Manipulating the mammalian genome by homologous recombination. Proc. Natl Acad. Sci. USA. 2001;98:8403–8410. doi: 10.1073/pnas.111009698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenneman M, Gimble FS, Wilson JH. Stimulation of intrachromosomal homologous recombination in human cells by electroporation with site-specific endonucleases. Proc. Natl Acad. Sci. USA. 1996;93:3608–3612. doi: 10.1073/pnas.93.8.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl Acad. Sci. USA. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taghian DG, Nickoloff JA. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol. Cell. Biol. 1997;17:6386–6393. doi: 10.1128/mcb.17.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schildkraut E, Miller CA, Nickoloff JA. Gene conversion and deletion frequencies during double-strand break repair in human cells are controlled by the distance between direct repeats. Nucleic Acids Res. 2005;33:1574–1580. doi: 10.1093/nar/gki295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vasquez KM, Wilson JH. Triplex-directed modification of genes and gene activity. Trends Biochem. Sci. 1998;23:4–9. doi: 10.1016/s0968-0004(97)01158-4. [DOI] [PubMed] [Google Scholar]
- 10.Wang G, Seidman MM, Glazer PM. Mutagenesis in mammalian cells induced by triple helix formation and transcription-coupled repair. Science. 1996;271:802–805. doi: 10.1126/science.271.5250.802. [DOI] [PubMed] [Google Scholar]
- 11.Vasquez KM, Christensen J, Li L, Finch RA, Glazer PM. Human XPA and RPA DNA repair proteins participate in specific recognition of triplex-induced helical distortions. Proc. Natl Acad. Sci. USA. 2002;99:5848–5853. doi: 10.1073/pnas.082193799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thoma BS, Wakasugi M, Christensen J, Reddy MC, Vasquez KM. Human XPC-hHR23B interacts with XPA-RPA in the recognition of triplex-directed psoralen DNA interstrand crosslinks. Nucleic Acids Res. 2005;33:2993–3001. doi: 10.1093/nar/gki610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reddy MC, Christensen J, Vasquez KM. Interplay between human high mobility group protein 1 and replication protein A on psoralen-cross-linked DNA. Biochemistry. 2005;44:4188–4195. doi: 10.1021/bi047902n. [DOI] [PubMed] [Google Scholar]
- 14.Wu Q, Christensen LA, Legerski RJ, Vasquez KM. Mismatch repair participates in error-free processing of DNA interstrand crosslinks in human cells. EMBO Rep. 2005;6:551–557. doi: 10.1038/sj.embor.7400418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain A, Wang G, Vasquez KM. Biochimie. 2008. DNA triple helices: Biological consequences and therapeutic potential. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo Z, Macris MA, Faruqi AF, Glazer PM. High-frequency intrachromosomal gene conversion induced by triplex-forming oligonucleotides microinjected into mouse cells. Proc. Natl Acad. Sci. USA. 2000;97:9003–9008. doi: 10.1073/pnas.160004997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faruqi AF, Datta HJ, Carroll D, Seidman MM, Glazer PM. Triple-helix formation induces recombination in mammalian cells via a nucleotide excision repair-dependent pathway. Mol. Cell. Biol. 2000;20:990–1000. doi: 10.1128/mcb.20.3.990-1000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan PP, Lin M, Faruqi AF, Powell J, Seidman MM, Glazer PM. Targeted correction of an episomal gene in mammalian cells by a short DNA fragment tethered to a triplex-forming oligonucleotide. J. Biol. Chem. 1999;274:11541–11548. doi: 10.1074/jbc.274.17.11541. [DOI] [PubMed] [Google Scholar]
- 19.Datta HJ, Chan PP, Vasquez KM, Gupta RC, Glazer PM. Triplex-induced recombination in human cell-free extracts. Dependence on XPA and HsRad51. J. Biol. Chem. 2001;276:18018–18023. doi: 10.1074/jbc.M011646200. [DOI] [PubMed] [Google Scholar]
- 20.Faruqi AF, Seidman MM, Segal DJ, Carroll D, Glazer PM. Recombination induced by triple-helix-targeted DNA damage in mammalian cells. Mol. Cell. Biol. 1996;16:6820–6828. doi: 10.1128/mcb.16.12.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalish JM, Seidman MM, Weeks DL, Glazer PM. Triplex-induced recombination and repair in the pyrimidine motif. Nucleic Acids Res. 2005;33:3492–3502. doi: 10.1093/nar/gki659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandor Z, Bredberg A. Triple helix directed psoralen adducts induce a low frequency of recombination in an SV40 shuttle vector. Biochim. Biophys. Acta. 1995;1263:235–240. doi: 10.1016/0167-4781(95)00109-t. [DOI] [PubMed] [Google Scholar]
- 23.Cimino GD, Gamper HB, Isaacs ST, Hearst JE. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Ann. Rev. Biochem. 1985;54:1151–1193. doi: 10.1146/annurev.bi.54.070185.005443. [DOI] [PubMed] [Google Scholar]
- 24.Moor AC, Gasparro FP. Biochemical aspects of psoralen photochemotherapy. Clin. Dermatol. 1996;14:353–365. doi: 10.1016/0738-081x(96)00065-x. [DOI] [PubMed] [Google Scholar]
- 25.Seidman MM, Dixon K, Razzaque A, Zagurski RJ, Berman ML. A shuttle vector plasmid for studying carcinogen-induced point mutations in mammalian cells. Gene. 1985;38:233–237. doi: 10.1016/0378-1119(85)90222-7. [DOI] [PubMed] [Google Scholar]
- 26.Protic-Sabljic M, Tuteja N, Munson PJ, Hauser J, Kraemer KH, Dixon K. UV light-induced cyclobutane pyrimidine dimers are mutagenic in mammalian cells. Mol. Cell. Biol. 1986;6:3349–3356. doi: 10.1128/mcb.6.10.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faruqi AF, Seidman MM, Segal DJ, Carroll D, Glazer PM. Recombination induced by triple helix-targeted DNA damage in mammalian cells. Mol. Cell. Biol. 1996;16:6820–6828. doi: 10.1128/mcb.16.12.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faruqi AF, Datta HJ, Carroll D, Seidman MM, Glazer PM. Triple-helix formation induces recombination in mammalian cells via a nucleotide excision repair-dependent pathway. Mol. Cell. Biol. 2000;20:990–1000. doi: 10.1128/mcb.20.3.990-1000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng H, Wang X, Legerski RJ, Glazer PM, Li L. Repair of DNA interstrand cross-links: interactions between homology-dependent and homology-independent pathways. DNA Repair. 2006;5:566–574. doi: 10.1016/j.dnarep.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 30.White AL, Jahnke LS. Removing UV-A and UV-C radiation from UV-B fluorescent lamp emissions. Differences in the inhibition of photosynthesis in the marine alga Dunaliella tertiolecta using chromate versus cellulose acetate-polyester filters. Photochem. Photobiol. 2004;80:340–345. doi: 10.1562/2003-12-31-RA-040. [DOI] [PubMed] [Google Scholar]
- 31.Vasquez KM, Wang G, Havre PA, Glazer PM. Chromosomal mutations induced by triplex-forming oligonucleotides in mammalian cells. Nucleic Acids Res. 1999;27:1176–1181. doi: 10.1093/nar/27.4.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasquez KM, Narayanan L, Glazer PM. Specific mutations induced by triplex-forming oligonucleotides in mice. Science. 2000;290:530–533. doi: 10.1126/science.290.5491.530. [DOI] [PubMed] [Google Scholar]
- 33.Sladek FM, Munn MM, Rupp WD, Howard-Flanders P. In vitro repair of psoralen-DNA cross-links by RecA, UvrABC, and the 5′-exonuclease of DNA polymerase I. J. Biol. Chem. 1989;264:6755–6765. [PubMed] [Google Scholar]
- 34.Bessho T, Mu D, Sancar A. Initiation of DNA interstrand cross-link repair in humans: the nucleotide excision repair system makes dual incisions 5′ to the cross-linked base and removes a 22- to 28-nucleotide-long damage-free strand. Mol. Cell. Biol. 1997;17:6822–6830. doi: 10.1128/mcb.17.12.6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bollag RJ, Liskay RM. Direct-repeat analysis of chromatid interactions during intrachromosomal recombination in mouse cells. Mol. Cell. Biol. 1991;11:4839–4845. doi: 10.1128/mcb.11.10.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sargent RG, Rolig RL, Kilburn AE, Adair GM, Wilson JH, Nairn RS. Recombination-dependent deletion formation in mammalian cells deficient in the nucleotide excision repair gene ERCC1. Proc. Natl Acad. Sci. USA. 1997;94:13122–13127. doi: 10.1073/pnas.94.24.13122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 39.Tutt A, Bertwistle D, Valentine J, Gabriel A, Swift S, Ross G, Griffin C, Thacker J, Ashworth A. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20:4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al-Minawi AZ, Saleh-Gohari N, Helleday T. The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells. Nucleic Acids Res. 2008;36:1–9. doi: 10.1093/nar/gkm888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sargent RG, Meservy JL, Perkins BD, Kilburn AE, Intody Z, Adair GM, Nairn RS, Wilson JH. Role of the nucleotide excision repair gene ERCC1 in formation of recombination-dependent rearrangements in mammalian cells. Nucleic Acids Res. 2000;28:3771–3778. doi: 10.1093/nar/28.19.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakrabarti S, Seidman MM. Intramolecular recombination between transfected repeated sequences in mammalian cells is nonconservative. Mol. Cell. Biol. 1986;6:2520–2526. doi: 10.1128/mcb.6.7.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng WP, Nickoloff JA. Mismatch repair of heteroduplex DNA intermediates of extrachromosomal recombination in mammalian cells. Mol. Cell. Biol. 1994;14:400–406. doi: 10.1128/mcb.14.1.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin FL, Sperle K, Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol. Cell. Biol. 1984;4:1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seidman MM. Intermolecular homologous recombination between transfected sequences in mammalian cells is primarily nonconservative. Mol. Cell. Biol. 1987;7:3561–3565. doi: 10.1128/mcb.7.10.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McHugh PJ, Gill RD, Waters R, Hartley JA. Excision repair of nitrogen mustard-DNA adducts in Saccharomyces cerevisiae. Nucleic Acids Res. 1999;27:3259–3266. doi: 10.1093/nar/27.16.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol. Cell. Biol. 2000;20:7980–7990. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller EM, Hough HL, Cho JW, Nickoloff JA. Mismatch repair by efficient nick-directed, and less efficient mismatch-specific, mechanisms in homologous recombination intermediates in Chinese hamster ovary cells. Genetics. 1997;147:743–753. doi: 10.1093/genetics/147.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alani E, Reenan RA, Kolodner RD. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics. 1994;137:19–39. doi: 10.1093/genetics/137.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Worth L, Jr, Clark S, Radman M, Modrich P. Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. Proc. Natl Acad. Sci. USA. 1994;91:3238–3241. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang N, Lu X, Zhang X, Peterson CA, Legerski RJ. hMutSbeta is required for the recognition and uncoupling of psoralen interstrand cross-links in vitro. Mol. Cell. Biol. 2002;22:2388–2397. doi: 10.1128/MCB.22.7.2388-2397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fishman-Lobell J, Rudin N, Haber JE. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol. Cell. Biol. 1992;12:1292–1303. doi: 10.1128/mcb.12.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sugawara N, Haber JE. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol. Cell. Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ray A, Siddiqi I, Kolodkin AL, Stahl FW. Intra-chromosomal gene conversion induced by a DNA double-strand break in Saccharomyces cerevisiae. J. Mol. Biol. 1988;201:247–260. doi: 10.1016/0022-2836(88)90136-2. [DOI] [PubMed] [Google Scholar]
- 56.Lin FL, Sperle KM, Sternberg NL. Extrachromosomal recombination in mammalian cells as studied with single- and double-stranded DNA substrates. Mol. Cell. Biol. 1987;7:129–140. doi: 10.1128/mcb.7.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bratty J, Ferbeyre G, Molinaro C, Cedergren R. Stimulation of mitotic recombination upon transcription from the yeast GAL1 promoter but not from other RNA polymerase I, II and III promoters. Curr. Genet. 1996;30:381–388. doi: 10.1007/s002940050146. [DOI] [PubMed] [Google Scholar]
- 58.Takeuchi Y, Horiuchi T, Kobayashi T. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 2003;17:1497–1506. doi: 10.1101/gad.1085403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ercan S, Reese JC, Workman JL, Simpson RT. Yeast recombination enhancer is stimulated by transcription activation. Mol. Cell. Biol. 2005;25:7976–7987. doi: 10.1128/MCB.25.18.7976-7987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weiss U, Wilson JH. Repair of single-stranded loops in heteroduplex DNA transfected into mammalian cells. Proc. Natl Acad. Sci. USA. 1987;84:1619–1623. doi: 10.1073/pnas.84.6.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aguilera A, Klein HL. Yeast intrachromosomal recombination: long gene conversion tracts are preferentially associated with reciprocal exchange and require the RAD1 and RAD3 gene products. Genetics. 1989;123:683–694. doi: 10.1093/genetics/123.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]