Abstract
Expression and localization of myo-inositol-1-phosphate synthase (MIPS) in developing seeds of Arabidopsis thaliana was investigated. MIPS is an essential enzyme for production of inositol and inositol phosphates via its circularization of glucose-6-phosphate as the initial step. myo-inositol-6-phosphate (InsP6 or phytic acid) is the predominant form of phosphorus found in seeds and accumulates as a consequence of MIPS action. Three MIPS genes have been identified in Arabidopsis, all of which were expressed not only in siliques but in both leaves and roots. Immunoelectron microscopy using a MIPS antibody showed that MIPS localizes to the cytosol primarily in the endosperm during seed development and not in the embryo. This is consistent with results obtained using fluorescent microscopy and western blot analysis that showed a similar pattern of localization. However, InsP6, which is the final product of inositol phosphate metabolism, was present mainly in the embryo. This suggests that a complex interaction between the endosperm and embryo occurs during the synthesis and subsequent accumulation of InsP6 in developing seeds of Arabidopsis.
Keywords: Inositol, inositol phosphates, inositol-6-phosphates, myo-inositol-1-phosphate synthase, phosphate, phytate, vacuole
Introduction
myo-Inositol hexakisphosphate (InsP6; phytic acid) is the most abundant form of phosphorus found in plant seeds. InsP6 in seed is bound to minerals (K, Ca, Mg, Zn, and Mn) to form inclusion particles (globoids) which are commonly located in protein storage vacuoles (Lott et al., 1979). In germinating seeds, InsP6 is hydrolysed into myo-inositol and inorganic phosphates (Pi) by phytase (myo-inositol hexakisphosphate phosphatase) and used for subsequent seedling growth (Loewus and Murthy, 2000; Raboy, 2003). In yeast, InsP6 is also suggested to be involved in mRNA export from the nucleus (Miller et al., 2004) and chromatin remodelling (Shen et al., 2003). More recently, InsP6 has been found to act as a co-factor for auxin binding to the TIR1 plant hormone auxin receptor (Tan et al., 2007). Thus, InsP6 is attracting increased attention not only as a seed storage component but also as a regulator of cellular functions.
From a nutritional perspective, high InsP6 in grain is considered to be undesirable due to its binding of essential micronutrient metals. Considerable effort has therefore been undertaken to isolate low InsP6 mutants from various plant species, including barley (Hatzack et al., 2000; Dorsch et al., 2003), maize (Raboy et al., 2000), rice (Larson et al., 2000; Kuwano et al., 2006), and soybean (Wilcox et al., 2000). One such mutant in soybean showed 50% reduction of InsP6 and an ∼10-fold increase in the Pi content of seeds, which was associated with lower expression of a MIPS [Ins(3)P1 synthase] gene (Hitz et al., 2002). Similarly, RNA interference (RNAi)-mediated silencing of MIPS gene expression in transgenic soybean resulted in a 95% reduction of InsP6 in seeds (Nunes et al., 2006). However, the transgenic lines showed impaired seed development, and antisense lines of MIPS genes in potato plants similarly have altered plant morphology (Keller et al., 1998). These may be caused by important roles in the cellular functions of InsP6.
MIPS mediates the conversion of Glc-6-P to Ins(3)P1, and its activity is essential for the subsequent synthesis of InsP6. myo-inositol can also be derived from Ins(3)P1 via dephosphorylation by myo-inositol-1-phosphate phosphatase (Loewus and Murthy, 2000). Inositol is known to play important roles in plant development. Various derivatives of myo-inositol (e.g. muco-inositol, pinitol) also function in environmental stress tolerance. MIPS is therefore an important enzyme for inositol and inositol phosphate metabolism in plants.
InsP6 synthesis requires either the sequential phosphorylation of Ins(3)P1 or the phosphorylation of phosphatidyl inositol intermediates via myo-inositol. A low phytic acid mutant in maize (lpa3) has a mutation in the gene encoding a myo-inositol kinase (MIK) that is normally expressed in developing embryos (Shi et al., 2005). Another maize low phytic acid mutant (lpa2) has altered expression of an Ins(1,3,4)P3 5/6 kinase (PPK) gene (Shi et al., 2003), both of which caused a significant reduction in the InsP6 content in seeds with a concomitant increase in Pi. Ins(1,4,5)P3 kinase (AtIPK2) and Ins 2-kinase (AtIPK1) have similarly both been shown to be responsible for InsP6 synthesis in Arabidopsis (Stevenson-Paulik et al., 2005). These results indicate that InsP6 synthesis is complex, involving at least two alternative pathways. As part of this complexity, it is likely that enzyme reactions, such as MIPS action and sequential phosphorylation, are separated in different cells or tissues. In addition, it has been shown that addition of an excess of Pi to Catharanthus roseus suspension-cultured cells induced a large accumulation of InsP6 in the vacuole (Mitsuhashi et al., 2005). Although excess Pi increased InsP6 synthesis, the MIPS and Ins(1,4,5)P3 kinase immunocontents did not change, suggesting complex regulation of InsP6 synthesis, by not only enzymes but also substrate levels.
In the present study, the focus was on the role of MIPS in InsP6 synthesis in developing seeds of Arabidopsis. Yoshida et al. (1999) have shown that expression of a rice MIPS gene (RINO1) was detected between 4 d and 7 d after anthesis in the scutellum and aleurone layer, coinciding with the appearance of InsP6. Treatment of rice cultured cells with abscisic acid (ABA) and sucrose together also resulted in higher levels of MIPS transcript accumulation, suggesting a synergistic induction of the MIPS gene in developing rice seeds (Yoshida et al., 2002). However, the subcellular localization of MIPS was not reported. More recently, Chiera and Grabau (2007) have shown in soybean seed that MIPS (Gm MIPS1) is localized in non-embryo tissues and appeared to be associated subcellularly with plastid-like structures that stained with MIPS antibody. MIPS activity has been reported to be widely distributed in intracellular compartments including membrane-bound organelles and cell walls, as well as cytoplasm (Lackey et al., 2003). Despite this, the site(s) of InsP6 synthesis in seed cells is poorly understood. Most InsP6 is accumulated in the vacuole, which becomes globoid in seeds, but it is not known how InsP6 is transported to the vacuole. Using electron microscopy, Otegui et al. (2002) observed that in developing seeds of Arabidopsis, globoids with Mg2+, K+, and Ca2+ were present within the embryo and that the endosperm of chalazal cells transiently contain Mn- and/or Zn-phytate. More recently, Shi et al. (2007) reported that the maize lpa1 gene encodes an MRP-type ABC transporter and, since it is known that the maize LPA1 mutant has less InsP6, this ABC transporter may be associated with InsP6 storage.
To clarify InsP6 synthesis during Arabidopsis seed maturation, MIPS localization was examined using immunohistology, and it is shown here that MIPS proteins localize to the cytosol of the Arabidopsis seed endosperm. Moreover, the presence of InsP6 within both embryo and endosperm tissues suggests an interaction between the tissues in the synthesis and subsequent storage of InsP6 in developing seeds of Arabidopsis.
Materials and methods
Plant materials
Seeds of Arabidopsis thaliana (Columbia accession) were surface sterilized with 95% ethanol and then sown onto 0.2% gellan gum (Wako, Tokyo, Japan) in 1/2 MS medium (Wako) with 3 mg l−1 thiamine-HCl, 0.5 mg l−1 pyridoxine, and 5 mg l−1 nicotinic acid. After incubation at 4 °C for 4 d to break dormancy, the seeds were germinated and grown at 23 °C under continuous light. After ∼14 d the seedlings were transferred into vermiculite medium for subsequent growth.
Developing seeds were harvested from Arabidopsis plants having 10–12 siliques. Seeds harvested from the sixth to eighth siliques were separated into seed coat and embryo using tweezers under a binocular (SZX16, Olympus). The seed coat and embryo were washed with RNase-free water three times to remove fragile endosperm tissues.
RT-PCR and real-time RT-PCR
Total RNA was extracted from tissue using the RNeasy Plant Mini Kit (QIAGEN Inc., Valencia, CA, USA) according to protocols provided by the manufacturer. First-strand cDNA was generated by reverse transcription with reverse transcriptase XL (AMV) (Takara Bio Inc., Shiga, Japan) using oligo(dT primer), 5′- CTGATCTAGAGGTACCGGATCCTTTTTTTTTTTTTTTTTTTT. Real-time PCR amplification was performed using the SYBR® Premix Ex Taq™ (TaKaRa Bio Inc.,) and a real-time PCR detector (TaKaRa Smart Cycler II system). PCR was performed using gene-specific oligonucleotide primer pairs based on unique sequences for each MIPS gene and an Actin-2 (control) gene. The primer sequences used were: for AtMIPS1 (At2g22240), 5′-GCGGGATCCCATGGAGTACAAGTGAAGGATGAG-3′ and 5′-GCGGAATTCGAAAATCCATATTCATAGATCATAAG-3′; AtMIPS2 (At4g39800), 5′-GCGGAATTCAAGTGAACATGAAGAAGCATGAAC-3′ and 5′-GCGATCGATGGAACCAAAACCATGATTATATATCTC-3′; AtMIPS3 (At5g10170), 5′-GCGATCGATTCTCGAGTACAAGTGATCAAAGAGAC-3′ and 5′-GCGCTCGAGCCCAAATATATATTATAGTTTGAAATG-3′; and for Actin-2 (At3g18780), 5′-TTTGTTCCAGCCCTCGTTTGT-3′ and 5′-TCATGCTGCTTGGTGCAAGT-3′. In both PCR methods, the same primers sets were used for each gene.
Preparation of antibodies against MIPS
MIPS antibody was prepared according to Mitsuhashi et al. (2005). An expressed sequence tag (EST) clone (accession no. AV525103) for the Arabidopsis MIPS-2 gene (At4g39800) was provided by Kazusa DNA Research Institute. Oligonucletide primers 5′-GAATTCATGTTTATTGAGAGCTTCAAAGTT-3′ and 5′-CTCGAGCTTGAACTCCATGATCATGTTGTT-3′ were designed on the basis of N- and C-terminal sequences of the MIPS-2 gene, respectively. The amplified DNA was digested with XhoI and EcoRI, and inserted into the XhoI and EcoRI sites of the pET21a vector (EMD Biosciences, San Diego, CA, USA). The pET21::MIPS2 plasmid was introduced into Escherichia coli strain BL21(DE3) (EMD Biosciences). The recombinant protein was purified via a 6×His tag by using a HiTrap Chelating HP Column (Amersham Biosciences, Piscataway, NJ, USA) and used as antigen. Specific antisera raised in rabbit were provided by Shibayagi Co., Ltd (Gunma, Japan).
Preparation of thin sections
Developing Arabidopsis seeds with torpedo-shaped embryos were vacuum infiltrated for 1 h with a fixative that consisted of 4% paraformaldehyde, 1% glutaraldehyde, and 0.06 M sucrose in 0.05 M cacodylate buffer, pH 7.4. The tissues were cut into slices of <1 mm in thickness with a razor blade and treated for another 2 h with freshly prepared fixative.
Immunoelectron microscopy
Immunogold labelling procedures were essentially the same as described previously (Hara-Nishimura et al., 1993), except for the use of anti-AtMIPS2 antibodies (Mitsuhashi et al., 2005). Post-fixation was omitted for immunoelectron microscopy. The samples were dehydrated in a graded dimethylformamide series at –20 °C and embedded in London Resin White (London Resin Co., Basingstoke, Hampshire, UK). Blocks were polymerized under a UV lamp and sectioned with a Reichert ultramicrotome (Leica, Heidelberg, Germany). Ultrathin sections were incubated with 1% (w/v) bovine serum albumin (BSA) in phosphate-buffered saline (PBS) for 1 h at room temperature and then incubated with anti-AtMIPS2 antibodies in blocking solution overnight at 4 °C. The sections were washed with PBS and then incubated for 30 min at room temperature with a solution of protein A-gold (10 nm; GE Healthcare Biosciences, Tokyo, Japan) in the blocking solution. The sections were washed with distilled water and then were stained with uranyl acetate and lead citrate. After staining, all sections were examined with a transmission electron microscope (model 1200EX; JEOL, Tokyo, Japan) at 80 kV.
Immunofluorescence analysis
Developing seeds of Arabidopsis were fixed for 40 min in 7.2% (w/v) formaldehyde, 0.1% (v/v) Nonidet P-40, 10% (v/v) dimethylsulphoxide, and 50 mM Na-phosphate buffer, pH 7.2. Seeds were then washed twice with Tris-buffered saline–Tween (TBS-T) for 5 min, incubated in TBS-T containing 5% (w/v) Cellulase Onozuka R-10 (Yakult, Tokyo, Japan) and 2% (w/v) Pectolyase Y-23 (Kikkoman, Tokyo, Japan) for 20 min at 30°C, washed twice with TBS-T, incubated in blocking buffer [2% (w/v) BSA and TBS-T] for 30 min, and then incubated with anti-AtMIPS2 or pre-immune antibodies in the blocking buffer for 40 min. After this the seeds were washed three times for 5 min each, incubated for 1 h with goat anti-rabbit IgG antibodies conjugated with Alexa Fluor 488 (absorbance, 495 nm; emission, 519 nm; Molecular Probes, Eugene, OR), washed three times for 5 min with TBS-T, and mounted.
Immunoblot analysis
Seed tissues of Arabidopsis were homogenized in 10 mM Tris-HCl, pH 7.5, before centrifugation to collect soluble proteins. Proteins (5 μg per lane) were subjected to SDS–PAGE and were transferred electrophoretically to a polyvinylidene difluoride membrane according to the manufacturer's protocol (Biocraft, Tokyo, Japan). The membrane was incubated with the antibodies against AtMIPS2. Horseradish peroxidase-conjugated donkey antibodies directed against rabbit IgG (GE Healthcare Biosciences) were used as the secondary antibodies. The antibody-labelled proteins were visualized with an enhanced chemiluminescence kit (ECL system; GE Healthcare Biosciences). Antibodies against Arabidopsis 2S albumin and 12S globulin were also used as controls (Shimada et al., 2003).
Measurement of inositol phosphates
Inositol phosphates and Pi were measured using a DX-500 ion chromatography system (Dionex, Osaka, Japan) consisting of a gradient pump, a 25 μl sample loop, and a conductivity detector as described previously (Baluyot and Hartford, 1996; Sekiguchi et al., 2000; Mitsuhashi et al., 2005). Dionex IonPac AS11 (2 mm ID×250 mm) and IonPac AG11 (2 mm ID×50 mm) columns packed with anion-exchange resin were used as the separation columns. The Dionex ASRS-Ultra anion self-regenerating suppressor was operated in the external water mode at 100 mA. The Dionex PeakNet workstation was used for data processing. A Dionex EG40 eluent generator equipped with an EGC-KOH cartridge was used. A Dionex IonPac ATC-1 (4 mm ID×35 mm), a high-capacity anion-exchange column, was placed at the pump outlet to remove the small amount of dissolved carbon dioxide and carbonate in the deionized water. This procedure allowed InsP1 to InsP6 to be resolved in a single gradient elution. A 25 μl aliquot of the filtered samples was automatically injected by an autosampler AS-50 (Dionex). The flow-rate was 0.35 ml min−1 at 35 °C. The concentration gradient (5–80 mM KOH) was generated by EG40. The detection limit (S/N = 3) for InsP6 was 50 nM. Mitsuhashi et al. (2005) showed chromatograms of all 64 isomers of InsPn. They finally separated three of six InsP1 isomers, three of 15 InsP2 isomers, eight of 20 InsP3 isomers, eight of 15 InsP4 isomers, and three of six InsP5 isomers independently, although enantiomers were not separated. InsP6 was purchased from Sigma (St Louis, MO, USA).
Results
Investigation of MIPS gene expression in Arabidopsis
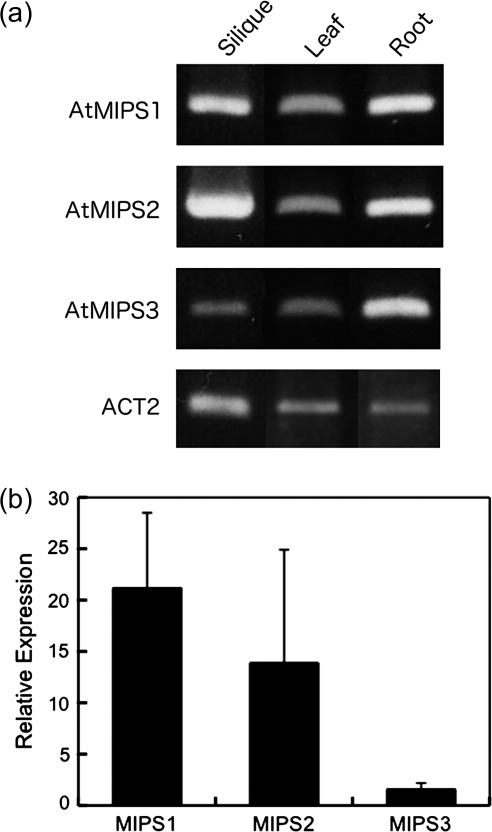
The objective of the present work is to clarify the localization of InsP6 synthesis during seed maturation in Arabidopsis. Conversion of Glc-6-P to Ins(3)P1 is mediated by MIPS which is an initial key step in InsP6 synthesis. In the Arabidopsis genome, there are three isoforms of the MIPS gene, i.e. AtMIPS1 (At2g22240), AtMIPS2 (At4g39800), and AtMIPS3 (At5g10170). To elucidate whether any one of these genes is responsible for InsP6 synthesis in developing seeds, the expression pattern of the MIPS isoforms was investigated using RT-PCR. As the nucleotide sequences of the three genes is highly conserved (>79%), specific PCR primers were designed for the C-terminus-encoding and 3′-untranslated region (UTR) sequences, respectively. All three AtMIPS genes were expressed in developing siliques, leaves, and roots (Fig. 1a), suggesting that the AtMIPS1, AtMIPS2, and AtMIPS3 genes may also all be expressed in developing seeds. Real-time PCR analysis using the same primer sets revealed that the relative expression of AtMIPS1 and AtMIPS2 in developing siliques was higher than that of AtMIPS3.
Fig. 1.
Expression of MIPS genes in Arabidopsis. (a) Organ specificity of MIPS gene expression. Shown is RT-PCR of total RNA extracted from green siliques (Silique), mature leaves (Leaf), and roots (Root) using gene-specific primers for the Arabidopsis AtMIPS1 (At2g22240), AtMIPS2 (At4g39800), AtMIPS3 (At5g10170), and ACTIN2 (ACT2; At3g18780) genes. (b) Relative expression levels of the three MIPS genes relative to the ACT2 gene in green siliques measured with real-time PCR. Values are expressed with mean ± SE (n = 3).
Immunolocalization of MIPS proteins in developing seeds of Arabidopsis
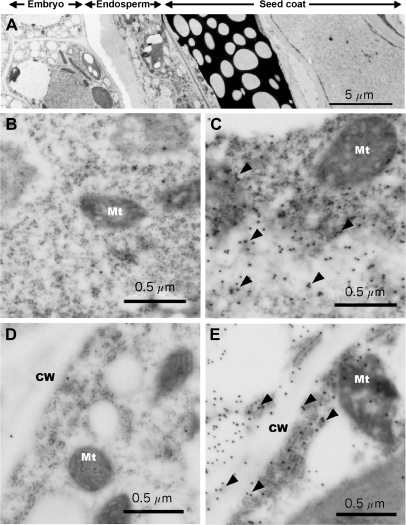
In Arabidopsis, it is not clear when InsP6 accumulation is initiated during seed development. Two different stages (torpedo and mature) of developing seeds were therefore investigated for localization of AtMIPS proteins with immunoelectron microscopy using a polyclonal antibody raised against AtMIPS2. However, this antibody does not distinguish between the three isoforms of MIPS due to high amino acid sequence homology (>89%) across the proteins. Indeed, MIPS proteins of another species (Catharanthus roseus) were also detected with this antibody (Mitsuhashi et al., 2005). The immunolocalization data obtained therefore are likely to represent the additive signals of the three AtMIPS proteins which were all expressed in developing siliques. The density of immunogold particles showed a similar frequency between cells in torpedo and mature stages (Fig. 2). The subcellular localization indicated the presence of immunogold particles mainly in the cytosol of the endosperm (Fig. 2C, E; arrowheads), with negligible presence in other organelles, cell walls, and membrane structures. Both torpedo and mature stage embryonic cells showed only low background levels of gold particles (Fig. 2), indicating that AtMIPS proteins appear to be specifically located within the endosperm cytosol.
Fig. 2.
Immunoelectron microscopy of MIPS protein in Arabidopsis seeds. (A) Transverse section of whole mature seeds. (B–D) Thin sections of torpedo-stage seeds (B and C) and mature-stage seeds (D and E). B and D show embryo tissue, and C and E show endosperm. CW, cell wall; Mt, mitochondria. Arrowheads show gold particles with anti-AtMIPS2 antibody.
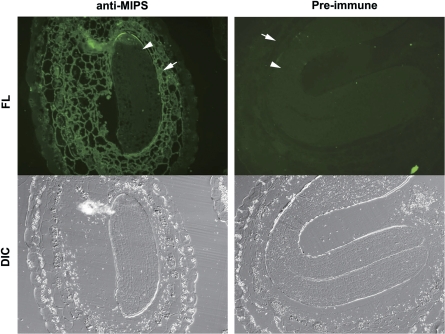
Localization of AtMIPS proteins was also investigated using immunofluorescent photomicroscopy. Mature stage seed sections were exposed to anti-AtMIPS2 serum or pre-immune control serum, and then to a secondary antibody with a fluorescent conjugate (Alexa Fluor 488). Consistent with the immunolocalization results, the fluorescent signal of the anti-AtMIPS antibody was also detected in the endosperm only and not in the embryo (Fig. 3). Control serum showed no positive signals in any cells.
Fig. 3.
Immunofluorescence microscopy of MIPS protein in mature seed sections. Sections were treated with either anti-AtMIPS2 serum (left) or pre-immune serum (right) followed by a secondary anti-rabbit antibody with a fluorescent conjugate (Alexa Fluor 488). DIC, differential interference contrast images; FL, fluorescence images. The arrowhead shows embryo and the arrow shows endsperm tissue.
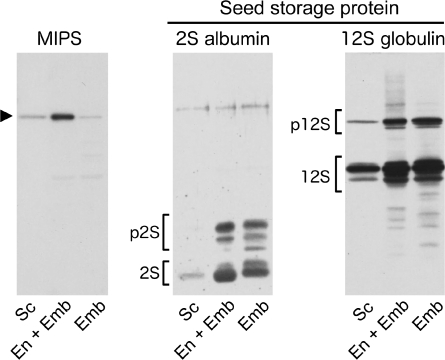
In addition to immunohistochemical analysis, the tissue-specific location of MIPS was also investigated by western blot analysis. Developing seeds were collected from Arabidopsis plants with between 10 and 12 siliques. Individual seeds were then dissected to separate the seed contents, containing the embryo and its surrounding endosperm, from the seed coat. In some cases, the endosperm was also removed from the embryo by washing with Tris buffer, and protein distributions were then assayed using MIPS, 2S albumin, and 12S globulin antibodies. AtMIPS protein (60 kDa band) showed low abundance in the seed coat and was detected primarily in non-washed tissues only, with a marked reduction when the endosperm was removed (Fig. 4). In contrast, the 2S albumin and 12S globulin seed storage proteins showed high abundance in both unwashed and washed tissues, suggesting either an embryo-specific location or a presence in both embryo and endosperm tissues.
Fig. 4.
Tissue specificity of MIPS and seed storage proteins in developing seeds of Arabidopsis as determined by immunoblot analysis with MIPS2, 2S albumin, and 12S globulin antibodies. Developing seeds (torpedo to mature stages) were separated into seed coat (Sc) and seed contents which were either left intact as endosperm and embryo (En+Emb), or washed to remove the endosperm (Emb). p2S and 2S are pro- and mature forms of 2S albumin, and p12S and 12S are pro- and mature forms of 12S globulin, respectively. The arrowhead at 60 kDa indicates MIPS.
Localization of InsP6 in Arabidopsis seeds
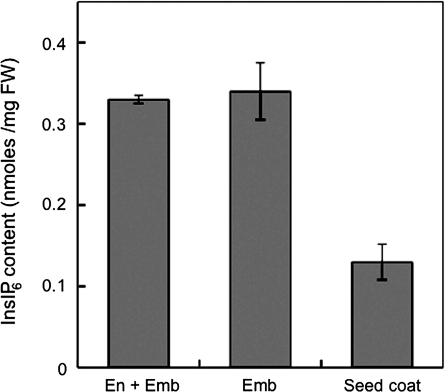
The presence of InsP6 was similarly measured in embryo and embryo+endosperm tissues using ion chromatography. InsP6 was clearly detected in the embryo and was not reduced by removal of the endosperm by washing (Fig. 5 and Supplementary Fig. S1 available at JXB online). This suggests that InsP6 accumulates primarily within the embryo despite the AtMIPS proteins being predominantly localized to the endosperm, as determined by the immunohistochemical analyses. Therefore, it is evident that the site of initial synthesis of inositol phosphates in Arabidopsis seeds (through the action of MIPS genes) may differ from the site of eventual accumulation of InsP6 and that there is interaction between the endosperm and embryo in the transport of InsP6 (or lower order derivatives) across the different tissues.
Fig. 5.
InsP6 content in endosperm, embryo tissue, and seed coat. InsP6 was measured using ion chromatography. Values are expressed with mean ± SE (n = 3–6).
Discussion
Arabidopsis contains three isoforms of the MIPS gene which were ubiquitously expressed in the various organs examined (i.e. developing siliques, leaves, and roots). MIPS proteins may be involved not only in InsP6 synthesis in developing seeds, but also in other physiological processes, such as raffinose metabolism (Hitz et al., 2002), salt tolerance (Ishitani et al., 1996; RayChaudhuri and Majumder, 1996), and ABA responses (Flores and Smart, 2000; Yoshida et al., 2002), via inositol or phosphatidylinositol synthesis. In Arabidopsis, AtMIPS gene isoforms may similarly have broad functionality in different tissues. However, differences of tissue specificity and the isoform(s) that is responsible for InsP6 synthesis in developing seeds remain unclear.
Although it was evident that all three isoforms were expressed in developing siliques (Fig. 1), InsP6 synthesis requires many enzymatic steps after MIPS reaction. It is reasonable to suppose that the complex pathways for InsPn synthesis exist separately in different tissues or different cellular compartments. In the present study, it was found that MIPS proteins localize in endosperm, but InsP6 was mainly accumulated in the embryo. The relationship between MIPS reaction and InsP6 synthesis is not direct. However, it is also known that MIPS mutation decreases InsP6 accumulation (Hitz et al., 2002). Thus, identification of MIPS localization would help to clarify the mechanism of InsP6 synthesis during seed maturation.
Localization of AtMIPS proteins in developing seeds
Immunoelectron microscopy and immunofluorescent photomicroscopy of developing seeds revealed that AtMIPS proteins in Arabidopsis were predominantly localized to the cytosol within the seed endosperm. The presence of MIPS within the endosperm was further verified by immunolocalization on western blots. In Figs 2–4, a small amount of MIPS proteins was also detectable in the embryo. Although it cannot be concluded that this resulted from contamination, the largest amounts of MIPS exist in the endosperm. Unfortunately, the antibody used could not distinguish between the three MIPS isoforms, so it was not possible to determine the relative contribution of each of the proteins to the localization that was observed, and thus the specific function of the different isoforms cannot yet be elucidated. Also, the antibody could not detect post-translational modifications, such as phosphorylation. Thus, the present data may not be related to the real activity of MIPS proteins. It is important to measure the enzymatic activity of MIPS in the embryo and endosperm. This is a possible subject of future research.
In monocots, InsP6 has been shown to accumulate in both the embryo and aleurone layer, whereas in dicots it accumulates only within the cotyledons of the embryo (Reddy and Sathe, 2002). In rice, InsP6-containing globoids have been shown to appear within the scutellum and aleurone layers of developing seeds, which coincides with the localization of the RINO1 (a rice MIPS gene) transcript (Yoshida et al., 1999). In contrast, for Arabidopsis it is shown that AtMIPS proteins localized to the endosperm, whilst InsP6 accumulated predominantly within the embryo. This raises an interesting question as to why the localization of MIPS proteins does not appear to coincide with that of accumulation of InsP6, and suggests some degree of cooperativity between the two tissues.
Conversion from Glc-6-P to Ins(3)P1 is considered to be one of the rate-limiting steps for InsP6 synthesis (Hitz et al., 2002; Kuwano et al., 2006; Nunes et al., 2006). Based on the present observations, Arabidopsis embryos did not contain a significant amount of MIPS relative to endosperm, and therefore are unlikely to be the site for initial synthesis of InsP6. It is more likely that synthesis of InsP6 is dependent on precursors provided by the endosperm, which is consistent with that recently reported for soybean (Chiera and Grabau, 2007). In one of the pathways of InsP6 synthesis, Ins(3)P1 synthesized by MIPS activity is first broken down to Ins and then rephosphorylated to Ins(3)P1. This represents a futile cycle if it occurs within one cell. Perhaps these steps are separated in different cells or tissues. However, further work to establish tissue-dependent metabolism of InsPn metabolism in developing seeds is required.
Translocation from endosperm to embryo
During seed development, InsP6 and minerals (Mn and Zn) have been shown to be transiently stored in charazal endosperm of Arabidopsis (Otegui et al., 2002) or small particles of castor bean (Greenwood and Bewley, 1984). In Arabidopsis seed, the endosperm serves to support the growth and development of the embryo. As for most dicot seeds (including Arabidopsis) the endosperm is ephemeral, and stored nutrients are absorbed by the embryo as the seed matures (Berger, 1999, 2003). However, as symplasmic connections are not present between the endosperm and embryo (Patrick and Offler, 2001), nutrients must be transferred through membrane transporters or via specialized cells, such as the transfer cells of the cotyledons or other contributing parts (Thompson et al., 2001). If the initial step of MIPS-mediated synthesis of InsP6 occurs in the endosperm, and InsP6 subsequently accumulates in the embryo, then there is a need for transport of InsP6 or its precursors to the embryo. Whilst the presence of such transporters for transfer of phosphorylated inositols has not been reported, a recent study by Shi et al. (2007) has identified a role for an ABC transporter in InsP6 accumulation in corn and it may function in the transport of phosphorylated inositols. Alternatively, it is possible that Ins(3)P1 is dephosphorylated by inositol monophosphatase within the endosperm and transported as myo-inositol to the embryo via an inositol transporter. Translocated myo-inositol may then be phosphorylated sequentially to InsP6 by inositol kinases. In preliminary experiments, the expression of both inositol transporter genes (four isoforms) and several InsP3 kinase genes has been detected in siliques from Arabidopsis (Supplementary Fig. S2 at JXB online). An attempt has also been made to detect the expression differences of MIPS and those genes between the endosperm and embryo, but the expression of all genes in both tissues was always detected, because mRNA contamination could not be removed from each tissue. Clearly, there is need for a better understanding of the role of specific genes in inositol phosphate metabolism in developing seeds, and of the relationship between endosperm and embryo tissues for InsP6 synthesis.
In the present study, it was shown that three isoforms of MIPS are expressed in developing siliques and that the MIPS protein localizes to the cytosol of the seed endosperm. Such an observation has important implications for the synthesis of InsP6, which was found to be present predominantly within the embryo of developing and mature Arabidopsis seeds.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Examples of a chromatogram of InsP6 measurements with ion chromatography. (a) InsP6 in endosperm and embryo. (b) InsP6 in embryo after washing out with pure water. (c) InsP6 in seed coat.
Fig. S2. Expression of inositol transporters (a) and several InsP3 kinase genes (b) in silique, leaf, and root tissue. PPK, inositol 1,3,4-trisphosphate 5/6-kinase; ITR, inositol transporter; IPK2, Ins(1,4,5)P3 kinase. The following primer sets were used for RT-PCR, AtITR1 (At4g16480) 5′-GCATGTCTTATCATCTTAGCCACG-3′, 5′-GCCCCAAAAACACTATAGCTAAGA-3′, AtITR2 (At2g35740) 5′-ACTTGCCTTGTCATTTTGGCT-3′ and 5′-AAGGAAGACAATGGCCAGGT-3′, AtITR3 (At2g43330) 5′-GATAGTCCAAATGGCTGGATTTC-3′ and 5′-CTAAGCCAAGCACAGCGAG-3′, AtITR4 (At1g30220) 5′-CATTATCTCGCTAGGAATACTAACCG-3′ and 5′-GTCCCAATCCAAGAAGAGCA-3′, PPK1 (At5g16760) 5′-TCGAACACTCAAGGCAACGA-3′ and 5′-TCCGGGACACCAAATCTCTC-3′, PPK2 (At4g08170) 5′-CCGGAGGCTGTCAATAATGC-3′ and 5′-CACAAGAGACCCGTGGGAAG-3′, PPK3 (At4g33770) 5′-GCAGACTTGGACCCTCGTGT-3′ and 5′-TTTGACCTGCGCCAGATTTT-3′, AtIPK2α (At5g07370) 5′-TAAAGGGAAATGATGATGATGCTA-3′ and 5′-CTAAGAATCTGCAGACTCATCTGG-3′, AtIPK2β (At5g61760)5′-CAATCTTGATGCAAGGAGGAG-3′ and 5′-CTAGCGCCCGTTCTCAAGT-3′.
Supplementary Material
Acknowledgments
We are very grateful to Ms Kazumi Tsutsui for her excellent technical assistance and Dr Emmanuel Delhaize (CSIRO) for his kind discussion and correction of this manuscript. An EST clone (accession no. AV525103) was kindly provided by Kazusa DNA Research Institute. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (16085204) by the Japanese Ministry of Education, Culture, Sports, Science and Technology, and CREST of JST to TM. NM was supported by a JSPS research fellowship for young scientists.
Glossary
Abbreviations
- Glc-6-P
glucose-6-phosphate
- InsP6
myo-inositol hexakisphosphate
- MIPS
myo-inositol-1-phosphate synthase
- Pi
inorganic phosphate
References
- Baluyot ES, Hartford CG. Comparison of polyphosphate analysis by ion chromatography and by modified end-group titration. Journal of Chromatography A. 1996;739:217–222. [Google Scholar]
- Berger F. Endosperm development. Current Opinion in Plant Biology. 1999;2:28–32. doi: 10.1016/s1369-5266(99)80006-5. [DOI] [PubMed] [Google Scholar]
- Berger F. Endosperm: the crossroad of seed development. Current Opinion in Plant Biology. 2003;6:42–50. doi: 10.1016/s1369526602000043. [DOI] [PubMed] [Google Scholar]
- Chiera J, Grabau E. Localization of myo-inositol phosphate synthase (GmMIPS-1) during the early stages of soybean seed development. Journal of Experimental Botany. 2007;58:2261–2268. doi: 10.1093/jxb/erm101. [DOI] [PubMed] [Google Scholar]
- Dorsch JA, Cook A, Young KA, Anderson JM, Bauman AT, Volkmann CJ, Murthy PPN, Raboy V. Seed phosphorus and inositol phosphate phenotype of barley low phytic acid genotypes. Phytochemistry. 2003;62:691–706. doi: 10.1016/s0031-9422(02)00610-6. [DOI] [PubMed] [Google Scholar]
- Flores S, Smart CC. Abscisic acid-induced changes in inositol metabolism in Spirodela polyrrhiza. Planta. 2000;211:823–832. doi: 10.1007/s004250000348. [DOI] [PubMed] [Google Scholar]
- Greenwood JS, Bewley JD. Subcellular distribution of phytin in the endosperm of developing castor bean: a possibility for its synthesis in the cytoplasm prior to deposition within protein bodies. Planta. 1984;160:113–120. doi: 10.1007/BF00392859. [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura I, Takeuchi Y, Nishimura M. Molecular characterization of a vacuolar processing enzyme related to a putative cysteine proteinase of Schistosoma mansoni. The Plant Cell. 1993;5:1651–1659. doi: 10.1105/tpc.5.11.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzack F, Johansen KS, Rasmussen SK. Nutritionally relevant parameters in low-phytate barley (Hordeum vulgare L.) grain mutants. Journal of Agriculture and Food Chemistry. 2000;48:6074–6080. doi: 10.1021/jf000669p. [DOI] [PubMed] [Google Scholar]
- Hitz WD, Carlson TJ, Kerr PS, Sebastian SA. Biochemical and molecular characterization of a mutation that confers a decreased raffinosaccharide and phytic acid phenotype on soybean seeds. Plant Physiology. 2002;128:650–660. doi: 10.1104/pp.010585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Majumder AL, Bornhouser A, Michalowski CB, Jensen RG, Bohnert HJ. Coordinate transcriptional induction of myo-inositol metabolism during environmental stress. The Plant Journal. 1996;9:537–548. doi: 10.1046/j.1365-313x.1996.09040537.x. [DOI] [PubMed] [Google Scholar]
- Keller R, Brearley CA, Trethewey RN, Müller-Röber B. Reduced inositol content and altered morphology in transgenic potato plants inhibited for 1D-myo-inositol 3-phosphate synthase. The Plant Journal. 1998;16:403–410. [Google Scholar]
- Kuwano M, Ohyama A, Tanaka Y, Mimura T, Takaiwa F, Yoshida KT. Molecular breeding for transgenic rice with low-phytic-acid phenotype through manipulating myo-inositol 3-phosphate synthase gene. Molecular Breeding. 2006;18:263–272. [Google Scholar]
- Lackey KH, Pope PM, Johnson MD. Expression of 1L-myoinositol-1-phosphate synthase in organelles. Plant Physiology. 2003;132:2240–2247. doi: 10.1104/pp.103.020610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson SR, Rutger JN, Young KA, Raboy V. Isolation and genetic mapping of a non-lethal rice (Oryza sativa L.) low phytic acid 1 mutation. Crop Science. 2000;40:1397–1405. [Google Scholar]
- Loewus FA, Murthy PPN. myo-Inositol metabolism in plants. Plant Science. 2000;150:1–19. [Google Scholar]
- Lott JNA, Spitzer E, Vollmer CM. Calcium distribution in globoid crystals of Cucurbita cotyledon protein bodies. Plant Physiology. 1979;63:847–851. doi: 10.1104/pp.63.5.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AL, Suntharalingam M, Johnson SL, Audhya A, Emr SD, Wente SR. Cytoplasmic inositol hexakisphosphate production is sufficient for mediating the Gle1-mRNA export pathway. Journal of Biological Chemistry. 2004;279:51022–51032. doi: 10.1074/jbc.M409394200. [DOI] [PubMed] [Google Scholar]
- Mitsuhashi N, Ohnishi M, Sekiguchi Y, Kwon Y-U, Chang Y-T, Chung S-K, Inoue Y, Reid RJ, Yagisawa H, Mimura T. Phytic acid synthesis and vacuolar accumulation in suspension-cultured cells of Catharanthus roseus induced by high concentration of inorganic phosphate and cation. Plant Physiology. 2005;138:1607–1614. doi: 10.1104/pp.105.060269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes AC, Vianna GR, Cuneo F, Amaya-Farfán J, de Capdeville G, Rech EL, Aragão FJ. RNAi-mediated silencing of the myo-inositol-1-phosphate synthase gene (GmMIPS1) in transgenic soybean inhibited seed development and reduced phytate content. Planta. 2006;224:125–132. doi: 10.1007/s00425-005-0201-0. [DOI] [PubMed] [Google Scholar]
- Otegui MS, Capp R, Staehelin LA. Developing seeds of Arabidopsis store different minerals in two types of vacuoles and in the endoplasmic reticulum. The Plant Cell. 2002;14:1311–1327. doi: 10.1105/tpc.010486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick JW, Offler CE. Compartmentation of transport and transfer events in developing seeds. Journal of Experimental Botany. 2001;52:551–564. [PubMed] [Google Scholar]
- Raboy V. myo-Inositol-1,2,3,4,5,6-hexakisphosphate. Phytochemistry. 2003;64:1033–1043. doi: 10.1016/s0031-9422(03)00446-1. [DOI] [PubMed] [Google Scholar]
- Raboy V, Gerbasi PF, Young KA, Stoneberg SD, Pickett SG, Bauman AT, Murthy PPN, Sheridan WF, Ertl DS. Origin and seed phenotype of maize low phytic acid 1-1 and low phytic acid 2-1. Plant Physiology. 2000;124:355–368. doi: 10.1104/pp.124.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RayChaudhuri A, Majumder AL. Salinity-induced enhancement of 1-myo-inositol 1-phosphate synthase in rice (Oryza sativa L.) Plant, Cell and Environment. 1996;19:1437–1442. [Google Scholar]
- Reddy NR, Sathe SK. Food phytates. Boca Raton, FL: CRC Press; 2002. [Google Scholar]
- Sekiguchi Y, Matsunaga A, Yamamoto A, Inoue Y. Analysis of condensed phosphates in food products by ion chromatography with an on-line hydroxide eluent generator. Journal of Chromatography A. 2000;881:639–644. doi: 10.1016/s0021-9673(99)01278-9. [DOI] [PubMed] [Google Scholar]
- Shen X, Xiao H, Ranallo R, Wu W-H, Wu C. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003;299:112–114. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- Shi J, Wang H, Hazebroek J, Ertl DS, Harp T. The maize low-phytic acid 3 encodes a myo-inositol kinase that plays a role in phytic acid biosynthesis in developing seeds. The Plant Journal. 2005;42:708–719. doi: 10.1111/j.1365-313X.2005.02412.x. [DOI] [PubMed] [Google Scholar]
- Shi J, Wang H, Schellin K, Li B, Faller M, Stoop JM, Meeley RB, Ertl DS, Ranch JP, Glassman K. Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nature Biotechnology. 2007;25:930–937. doi: 10.1038/nbt1322. [DOI] [PubMed] [Google Scholar]
- Shi J, Wang H, Wu Y, Hazebroek J, Meeley RB, Ertl DS. The maize low-phytic acid mutant lpa2 is caused by mutation in an inositol phosphate kinase gene. Plant Physiology. 2003;131:507–515. doi: 10.1104/pp.014258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Fuji K, Tamura K, Kondo M, Nishimura M, Hara-Nishimura I. Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 2003;100:16095–16100. doi: 10.1073/pnas.2530568100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson-Paulik J, Bastidas RJ, Chiou S-T, Frye RA, York JD. Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proceedings of the National Academy of Sciences, USA. 2005;102:12612–12617. doi: 10.1073/pnas.0504172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- Thompson RD, Hueros G, Becker H-A, Maitz M. Development and fuctions of seed transfer cells. Plant Science. 2001;160:775–783. doi: 10.1016/s0168-9452(01)00345-4. [DOI] [PubMed] [Google Scholar]
- Wilcox JR, Premachandra GS, Young KA, Raboy V. Isolation of high seed inorganic P, low-phytate soybean mutants. Crop Science. 2000;40:1601–1605. [Google Scholar]
- Yoshida KT, Fujiwara T, Naito S. The synergistic effects of sugar and abscisic acid on myo-inositol-1-phosphate synthase expression. Physiologia Plantarum. 2002;114:581–587. doi: 10.1034/j.1399-3054.2002.1140411.x. [DOI] [PubMed] [Google Scholar]
- Yoshida KT, Wada T, Koyama H, Mizobuchi-Fukuda R, Naito S. Temporal and spatial patterns of accumulation of the transcript of myo-inositol-1-phosphate synthase and phytin-containing particles during seed development in rice. Plant Physiology. 1999;119:65–72. doi: 10.1104/pp.119.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.