Abstract
Plants can survive a limiting nitrogen (N) supply by developing a set of N limitation adaptive responses. However, the Arabidopsis nla (nitrogen limitation adaptation) mutant fails to produce such responses, and cannot adapt to N limitation. In this study, the nla mutant was utilized to understand further the effect of NLA on Arabidopsis adaptation to N limitation. Grown with limiting N, the nla mutant could not accumulate anthocyanins and instead produced an N limitation-induced early senescence phenotype. In contrast, when supplied with limiting N and limiting phosphorus (Pi), the nla mutants accumulated abundant anthocyanins and did not show the N limitation-induced early senescence phenotype. These results support the hypothesis that Arabidopsis has a specific pathway to control N limitation-induced anthocyanin synthesis, and the nla mutation disrupts this pathway. However, the nla mutation does not affect the Pi limitation-induced anthocyanin synthesis pathway. Therefore, Pi limitation induced the nla mutant to accumulate anthocyanins under N limitation and allowed this mutant to adapt to N limitation. Under N limitation, the nla mutant had a significantly down-regulated expression of many genes functioning in anthocyanin synthesis, and an enhanced expression of genes involved in lignin production. Correspondingly, the nla mutant grown with limiting N showed a significantly lower production of anthocyanins (particularly cyanidins) and an increase in lignin contents compared with wild-type plants. These data suggest that NLA controls Arabidopsis adaptability to N limitation by channelling the phenylpropanoid metabolic flux to the induced anthocyanin synthesis, which is important for Arabidopsis to adapt to N limitation.
Keywords: Anthocyanin, Arabidopsis, adaptation, lignin, N limitation, nla mutant, phosphorus limitation
Introduction
Plant growth and development requires abundant nitrogen (N) nutrient (Marschner, 1995) since N constitutes 1.5–2% of plant dry matter and is the key component of many important macromolecules such as proteins, nucleic acids, and chlorophyll. Nevertheless, plants also have the ability to adapt to a limiting N supply growth condition, and such adaptability is an essential survival strategy for plants to finish their life cycles successfully and produce offspring under N limitation. In agriculture, enhancing crop adaptability to N limitation can maintain or increase crop yields with reduced N fertilizer application (Duvick, 1997; Tollenaar and Wu, 1999; Ding et al., 2005). To acclimate to N limitation, plants such as Arabidopsis have evolved a set of adaptive responses. These responses include the reduction of growth and photosynthesis, the remobilization of N from old, mature organs to actively growing ones, and the accumulation of abundant anthocyanins (Khamis et al., 1990; Bongue-Bartelsman and Phillips, 1995; Chalker-Scott, 1999; Ding et al., 2005; Diaz et al., 2006; Peng et al, 2007a, b). The Arabidopsis mutant nla (nitrogen limitation adaptation), which was identified in a previous study (Peng et al, 2007a), fails to adapt to N limitation, but instead produces an N limitation-induced early senescence phenotype. Physiological and biochemical analysis revealed that the nla mutant's phenotype was caused by the failure to develop the essential adaptive responses to N limitation, and one of the most obvious and constant defects caused by the nla mutation was the lack of N limitation-induced anthocyanin accumulation (Peng et al., 2007a).
Anthocyanins are a group of colourful secondary metabolites and have diverse functions in plants. In flower petals, fruit skins, and seed coats, anthocyanins provide various colours such as red and purple that are essential for plants to recruit pollinators and attract animals to disperse seeds (Winkel-Shirley, 2001). Acting as a ‘sunscreen’, anthocyanins shield leaves, especially senescing leaves, from excess light, and thus protect the photosynthetic apparatus from photodamage and facilitate the recovery of nutrients, especially N, from senescing leaves (Smillie and Hetherington, 1999; Hoch et al, 2003). The biochemical and molecular elucidation of anthocyanin biosynthesis and regulation in plants has been facilitated by the easily discernible phenotypes such as the altered pigmentation in flowers, seeds, and leaves caused by changes in anthocyanin production (Shirley et al., 1995). Consequently, in the last two decades most structural genes and many regulatory proteins involved in anthocyanin synthesis have been identified and functionally characterized. Anthocyanins are produced by the phenylpropanoid pathway, as illustrated in Fig. 1. This pathway starts with phenylalanine being converted to cinnamic acid by phenylalanine ammonia lyase (PAL), and diverges into several branches at ρ-coumaroyl CoA. One branch is the flavonoid pathway, in which chalcone synthase (CHS) catalyses the formation of the flavonoid skeleton from ρ-coumaroyl CoA, and subsequently leads to flavonol, cyanidin, and anthocyanin synthesis. Two branches initiated from ρ-coumaroyl CoA are responsible for the production of three monolignols, the essential precursors for lignin synthesis. Lignins are aromatic heteropolymers and embedded in plant cell walls to provide mechanical support and water transport capacity. Besseau et al. (2007) reported that repression of lignin synthesis in Arabidopsis significantly enhanced flavonoid production, indicating that the metabolic flux in the phenylpropanoid pathway can be switched among different branches for the production of anthocyanins or lignins.
Fig. 1.
The phenylpropanoid pathway for the biosynthesis of anthocyanin and three monoligols. ρ-Coumaroyl CoA is at the junction of the metabolic routes leading to anthocyanin synthesis or to lignin production. Enzymes in each step: PAL, phenylalanine ammonia lyase; C4H, cinnamic acid 4-hydroxylase; 4CL, ρ-coumaroyl-CoA synthase; CHS, chalcone synthase; CHI, chalcone-flavanone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavanone 3′-hydroxylase; DRF, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase; AGT, anthocyanin glycosyltransferase; HCT, hydroxycinnamoyl-CoA:skimimate/quinate hydroxycinnamoyltransferase; C3H, ρ-coumarate 3-hydroxylase; CCoAOMT, caffeoyl-CoA 3-O-methyltransferase; CCR, cinnamoyl-CoA reductase; F5H, ferulate 5-hydroxylase; COMT, caffeic acid O-methyltransferase; CAD, cinnamyl alcohol dehydrogenase.
It has been shown that plant anthocyanin production is induced by a wide range of biotic and abiotic stressors such as pathogen attack, wounding, UV light, low temperature, heavy metal contamination, and nutrient stress such as phosphorus (Pi) limitation (Steyn et al., 2002; Gould, 2004). Therefore, anthocyanin accumulation has been commonly used to indicate that plants are subjected to some biotic or abiotic stressors, and plants are most probably equipped with specific pathways to activate anthocyanin synthesis to cope with different stressors. This possibility was confirmed by the finding that Arabidopsis has a pathway controlling sucrose-specific induction of anthocyanin synthesis (Ohto et al., 2002; Teng et al., 2005; Solfanelli et al., 2006). When grown with a high sucrose supply in vitro, Arabidopsis seedlings accumulated anthocyanins (Ohto et al., 2002), and at least one structural gene in each step of the anthocyanin synthesis pathway was up-regulated by sucrose (Solfanelli et al., 2006). In contrast, the Arabidopsis seedlings with a mutated AtMYB75/PAP1 (Production of Anthocyanin Pigment 1) gene did not accumulate anthocyanins although abundant sucrose was supplied (Teng et al., 2005). Although it has not been determined whether other stressors can induce the pap1 mutant seedlings to synthesize anthocyanins, the study by Teng et al. (2005) indicates that Arabidopsis is equipped with a sucrose-induced anthocyanin synthesis pathway, which is controlled by PAP1.
N limitation has been found to induce anthocyanin accumulation and up-regulate anthocyanin synthesis genes in plants (Bongue-Bartelsman and Phillips, 1995; Chalker-Scott, 1999; Stewart et al., 2001; Peng et al., 2007a). However, it is not known whether plants are equipped with a specific pathway to control N limitation-induced anthocyanin production, and the physiological significance of anthocyanin accumulation for adaptation to N limitation remains undetermined. In this study, the two issues were addressed by further characterizing the nla mutant for anthocyanin synthesis. Although the nla mutant could not accumulate anthocyanins under N limitation, Pi limitation induced the synthesis of abundant anthocyanins in this mutant even when grown in a limiting N condition. Further, the Pi limitation-induced anthocyanin accumulation allowed the nla mutant to adapt to N limitation and not to produce the N limitation-induced early senescence phenotype. When the nla mutant was grown under N limitation, the anthocyanin synthesis genes were significantly down-regulated compared with wild-type plants, while the expression of many genes in the lignin synthesis pathway was markedly enhanced. While grown under a combined N and Pi limitation, the nla mutant and wild-type plants were similar in anthocyanin and lignin synthesis. All the results demonstrate that Arabidopsis plants are equipped with a specific pathway to control N limitation-induced anthocyanin synthesis, in which NLA is an essential molecular component, and anthocyanin accumulation is important for Arabidopsis adaptation to N limitation.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia (Col) wild type and the nla mutant were grown in the nutrient-free soil LB2 (SunGro Horticulture Canada Ltd, BC, Canada) supplied with limiting N and/or Pi nutrient within a controlled environmental condition as described (Peng et al., 2007a).
For N limitation:
The plants were supplied with 3 mM KNO3 in the nutrient solution (10 mM KH2PO4, pH 5.6, 2 mM MgSO4, 1 mM CaCl2, 0.1 mM Fe-EDTA, 50 μM H3BO4, 12 μM MnSO4, 1 μM ZnCl2, 1 μM CuSO4, 0.2 μM Na2MoO4), and 10 mM KNO3 was used as the sufficient N supply control.
Pi limitation:
The plants were grown with 0.5 mM KH2PO4 in the nutrient solution (10 mM KNO3, 2 mM MgSO4, 1 mM CaCl2, 0.1 mM Fe-EDTA, 50 μM H3BO4, 12 μM MnSO4, 1 μM ZnCl2, 1 μM CuSO4, 0.2 μM Na2MoO4, pH 5.6), and 10 mM KH2PO4 was used as the sufficient Pi supply control.
Combined Pi and N limitation:
The plants were supplied with 3 mM KNO3 + 0.5 mM KH2PO4 in the nutrient solution (10 mM KCl, 2 mM MgSO4, 1 mM CaCl2, 0.1 mM Fe-EDTA, 50 μM H3BO4, 12 μM MnSO4, 1 μM ZnCl2, 1 μM CuSO4, 0.2 μM Na2MoO4, pH 5.6), and 10 mM KNO3 + 10 mM KH2PO4 was used as the sufficient N and Pi control. The nutrient solutions were provided once a week for 4 weeks. At 24 and 28 days after germination (DAG), the fifth to the eighth rosette leaves were harvested, frozen in liquid N, and stored at – 80 °C for the following biochemical assays and RNA extraction.
Anthocyanin measurement and extraction of soluble phenolic compounds
Anthocyanins were extracted from the rosette leaf samples (100 mg per sample) of the nla mutant and wild-type plants, and assayed as described in Noh and Spalding (1998). The soluble phenolic compounds were extracted from the rosette leaf samples (100 mg per sample) according to Besseua et al. (2007), and used for the following high-pressure liquid chromatography (HPLC) and liquid chromatography–mass spectrometry (LC-MS) analysis.
Analysis of phenolic compounds by HPLC and LC-MS
An Agilent Technology 1100 Series HPLC system with a diode array detector (DAD) was used for quantification and identification of polyphenolics in the samples. Separation of various phenolics was carried out in a Phenomenex® Luna C18(2) column (250×4.6 mm i.d.; particle size, 5 μm) with a C18 guard column. The binary mobile phase consisted of 6% acetic acid and acetonitrile (solvent B). The flow rate was 1.0 ml min−1 for 45 min. The system was run in the following program: 8% B isocratically for 20 min, 8% B to 15% B in 5 min, and then 15% B isocratically for 20 min. There was a 5 min post-run for re-conditioning. The detector was set at 360 nm and 520 nm for simultaneous monitoring of flavonols and cyanidins. The polyphonolics were expressed as quercetin 3-glucoside/kaempferol 3-glucoside and cyanidin 3-glucoside equivalent, respectively.
LC-MS-MS was performed using HPLC coupled to a photodiode array UV detector (Finnigan MAT Spectra System UV6000LP, San Jose, CA, USA) equipped with a Finnigan LCQ Deca electrospray ionization-mass spectrometer (HPLC-ESI-MS) operated in a negative ion mode. The same separation conditions as described in the HPLC analysis were used in the LC-MS experiment. The instrument parameters were optimized against kaempferol 3-glucoside prior to sample analysis.
Lignin staining and measurement
The nla mutant and wild-ype plants grown under various nutrient conditions and of the same size (described above) were used for lignin staining at 21 and 24 DAG. Sections were taken from the primary inflorescences at the same positions, stained with 1% phloroglucinol-HCl for 10 min, and mounted on slides with 50% glycerol+6 N HCl. The slides were viewed immediately on a Leica DM LS2 microscope (Leica, Wetzler, Germany).
Lignin was measured quantitatively according to Lange et al. (1995). The stems (200–300 mg) from the nla mutant and wild-type plants were harvested at 21 DAG (2–3 cm) and 24 DAG (6–8 cm), homogenized in liquid N, suspended in 1.5 ml of methanol, and vigorously stirred for 1 h. Following 5 min centrifugation, the pellet was washed with 1.5 ml of the following solvents by vortexing for 15 min: (i) methanol twice; (ii) 1 M NaCl; (iii) 1% SDS; (iv) water twice; (v) CHCl3/CH3OH (1:1); and (vi) tert-butyl methyl ether. The final pellet was freeze-dried overnight, and 10 mg was taken for the following assay. The powder was treated with 1 ml of 2 M HCl and 0.2 ml of thioglycolic acid for 4 h at 95 °C. The mixture was cooled down and centrifuged for 10 min. The pellet was washed with water three times, and suspended in 1 ml of NaOH (0.5 M). The suspension was shaken vigorously overnight at 4 °C to extract the lignin thioglycolic acid derivatives (LTGAs), and centrifuged for 10 min. The supernatant was collected, and the pellet was mixed with 0.5 ml of NaOH (0.5 M) for 1 h. Following centrifugation, the supernatant was combined with the previous one, and acidified with 0.3 M concentrated HCl to precipitate LTGAs at 4 °C for 4 h. The mixture was centrifuged, and the brown pellet was vacuum dried. The LTGA pellet was dissolved in 1.5 ml of NaOH (0.5 M). The absorption at 280 nm was measured and used as the indicator of lignin content. To compare the lignin content in the wild type and in the nla mutant directly, the lignin content of the wild type was set as 100%, and the relative lignin content in the nla mutant = (the nla mutant lignin content/wild type lignin content)×100%.
RNA extraction and real-time quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from the rosette leaf samples (100 mg per sample) using TriZol reagent (Invitrogen, Carlsbad, CA, USA). The first-strand cDNA was synthesized from total RNA with a kit (Fermantas) and used for qRT-PCR analysis. The PCR primers were designed using the Applied Biosystems (Foster City, CA, USA) software Primer Express 2.0, and their sequences are available upon request. The qRT-PCR was carried out and analysed with a 7300 real-time PCR system (Applied Biosystems) according to the manufacturer's instruction. Actin-8 was used as endogenous control. For comparing the gene expression levels under varying experimental conditions including different DAGs, nitrate application and genotype were expressed as the fold changes relative to the transcript abundance of the tested genes in wild-type plants grown with 10 mM nitrate for 24 DAG, and visualized as a row of coloured boxes using BAR Heatmapper Plus software (http://bbc.botany.utoronto.ca/ntools/cgi-bin/ntools_heatmapper_plus.cgi). Black to brighter black colour indicates no or a small change in gene expression levels. Yellow to brighter yellow colour means increasingly significant up-regulation of gene expression.
Results
Pi limitation induced anthocyanin accumulation and prevented the N limitation-induced early senescence phenotype in the nla mutant
Nitrate at 10 mM provides Arabidopsis plants with sufficient N nutrient, while 3 mM nitrate limits the generative growth of Arabidopsis plants significantly, and causes them to develop a set of adaptive responses to N limitation, such as reducing growth and photosynthesis, recycling N from old, mature organs to actively growing ones, and producing substantial quantities of anthocyanins (Bi et al., 2005; Peng et al., 2007a). The nla mutant cannot adapt to the N limitation growth condition. As shown in Fig. 2A and B, when supplied with 3 mM nitrate, the nla mutant produced the N limitation-induced early senescence phenotype and failed to accumulate anthocyanins as compared with the wild type. This observation is consistent with a previous report (Peng et al., 2007a). Based on the physiological function of anthocyanins in plants (Bongue-Bartelsman and Phillips, 1995; Chalker-Scott, 1999; Smillie and Hetherington, 1999; Hoch et al., 2003), it can be speculated that the major defect caused by the nla mutation may be the failure to accumulate anthocaynins under N limitation. To test this hypothesis, it is necessary to determine whether increasing anthocyanin accumulation in the nla mutant could prevent its N limitation-induced early senescence phenotype, and allow this mutant to adapt to N limitation. For this purpose, CHS, PAP1, and PAP2, which play important roles in anthocyanin synthesis (Shirley et al., 1995; Borevitz et al., 2000), were transformed into the nla mutant. However, the transgenic nla mutant plants which overexpressed CHS, PAP1, and PAP2, respectively, did not accumulate anthocyanins under N limitation, and still produced the N limitation-induced early senescence phenotype (data not shown).
Fig. 2.
The effect of N limitation and/or Pi limitation on the growth and anthocyanin accumulation in the nla and the wild type (Col). The plants of the two genotypes were grown with sufficient Pi (10 mM)+limiting N (3 mM) (A), limiting Pi (0.5 mM)+sufficient N (10 mM) (C), or limiting Pi+limiting N (E). The anthocyanin contents in the rosette leaves were determined at different days after germination (B, D, and F), and values were the means of five replicates with standard deviations.
Pi limitation induces anthocyanin accumulation (Steyn et al., 2002; Gould, 2004), and in a previous study (Peng et al., 2007a) it was found that 0.5 mM Pi limited Arabidopsis generative growth significantly while 10 mM Pi was sufficient. Indeed, grown with limiting Pi (0.5 mM) and sufficient N (10 mM nitrate) for 36 DAG, both wild-type plants and the nla mutant produced >24 U g−1 fresh weight (FW) anthocyanins, and their rosette leaf colour was dark green (Fig. 2C, D). The detailed developing process of such dark green rosette leaves was delineated in previous studies (Peng et al., 2007a, b). This result indicates that although the nla mutant is defective in activating anthocyanin production under N limitation, it still has the functional Pi limitation-induced anthocyanin synthesis pathway. To determine whether the Pi limitation-induced anthocyanin accumulation can prevent the N limitation-induced early senescence phenotype in the nla mutant, this mutant and wild-type plants were subjected to a combined Pi and N limitation growth condition by supplying 0.5 mM Pi and 3 mM nitrate. As shown in Fig. 2E and F, grown with limiting Pi and N, the nla mutant and wild-type plants accumulated abundant anthocyanins. At 24 DAG, the two genotypes produced 8.2–8.6 U g−1 FW anthocyanins, and contained >30 U g−1 FW anthocyanins at 36 DAG. Most importantly, similar to the wild type, the nla mutant plants had dark green rosette leaves and did not show the N limitation-induced early senescence phenotype (Fig. 2E). Further, the nla mutant and wild-type plants grown with 0.5 mM Pi and 3 mM nitrate had much smaller rosette leaves than those supplied with 0.5 mM Pi but 10 mM nitrate (comparing Fig. 2C and E). Therefore, 3 mM nitrate still subjected the two genotype plants to N limitation under a Pi limitation grown condition. Because sufficient Pi (10 mM) did not induce the nla mutant plants grown with limiting nitrate (3 mM) to accumulate anthocyanins from 20 DAG to 28 DAG (Fig. 2B), these mutant plants had pale green rather than dark-green rosette leaves and displayed the N limitation-induced early senescence phenotype (Fig. 2A). In contrast, under the same nutrient condition, the wild-type plants increased anthocyanin content by >11-fold from 20 DAG to 28 DAG (Fig. 2B), and did not show senescence symptoms until 32 DAG (data not shown). Taken together, all the data indicate that anthocyanin accumulation is essential for Arabidopsis plants to adapt to N limitation, and the major defect caused by the nla mutation is the failure to accumulate anthocyanins under N limitation. However, the nla mutation has no effect on the Pi limitation-induced anthocyanin synthesis pathway, which induces the nla mutant to accumulate anthocyanins under N and Pi limitation, and prevents N limitation-induced early senescence phenotype.
The nla mutation down-regulated the N limitation-induced anthocyanin synthesis pathway
The finding that the major defect caused by the nla mutation is the failure to accumulate anthocyanins under N limitation makes it necessary to determine whether the genes involved in anthocyanin synthesis were not up-regulated by N limitation in the nla mutant. The nla mutant and wild-type plants grown with 3 mM or 10 mM nitrate were harvested at 24 DAG (before N limitation-induced early senescence occurs in the nla mutant) and 28 DAG (after N limitation-induced early senescence occurs in the nla mutant) to prepare RNA. Subsequently, real-time qRT-PCR was used to analyse the expression of seven major structural genes functioning in each step of the anthocyanin synthesis pathway (Solfanelli et al., 2006). The transcription of PAP1 and PAP2, the two positive regulators for anthocyanin synthesis (Borevitz et al., 2000), was also determined. The seven major structural anthocyanin synthesis genes are CHS (At5g13930), CHI (At5g05270), F3H (At3g51240), F3′H (At3g07990), DFR (At5g42800), ANS (At4g22880), and AGT (At5g17050) (Fig. 1). As shown in Fig. 3, comparing the expression of the nine anthocyanin synthesis genes in the wild-type plants grown with 3 mM nitrate with that in the wild-type plants supplied with 10 mM nitrate revealed that N limitation up-regulated the nine genes by 4- to 124-fold at 24 and 28 DAG. This result was consistent with the fact that N limitation markedly increased the anthocyanin content in wild-type plants (Fig. 2B). In the nla mutant, although N limitation failed to induce anthocyanin accumulation (Fig. 2B), this stress still enhanced the expression of the nine anthocyanin synthesis genes (Fig. 3). However, the expression levels of the nine genes up-regulated by N limitation were markedly lower in the nla mutant than that in wild-type plants. In the nla mutant N limitation increased the expression of CHS, CHI, F3H, F3′H, DFR, ANS, AGT, PAP1, and PAP2 by 9.8-, 13.0-, 7.2-, 9.5-, 7.1-, 8.4-, 5.2,-, 5.1-, and 79.5-fold, respectively, at 28 DAG. In contrast, in wild-type plants N limitation up-regulated the nine genes by 21.2-, 10.1-, 22.8-, 43.4-, 36.7-, 71.3-, 10.8-, 6.8-, and 127-fold, respectively, at 28 DAG (Fig. 3).
Fig. 3.
Effect of the nla mutation on N limitation-induced up-regulation of anthocyanin synthesis genes. The nla mutant and wild-type (Col) plants were grown with 10 mM or 3 mM nitrate for 24 and 28 DAG. Gene expression levels were determined by qRT-PCR, and expressed as fold change relative to the transcript abundance in wild-type plants grown with 100 mM nitrate for 24 DAG, which was set as 1. Values are the means of three replicates, with standard deviations in parentheses, and were visualized as a row of coloured boxes using BAR Heatmapper Plus software (http://bbc.botany.utoronto.ca/ntools/cgi-bin/ntools_heatmapper_plus.cgi). Black to brighter black colour indicates no or a small change in gene expression, and yellow to brighter yellow colour means increasingly significant up-regulation of gene expression. PAP1, Production of Anthocyanin Pigment 1 gene; PAP2, Production of Anthocyanin Pigment 2 gene.
The production of flavonoids was investigated in both the nla mutant and wild-type plants. For this purpose, flavonoids were extracted from the rosette leaves of the nla mutant and wild-type plants grown with 3 mM or 10 mM nitrate, and detected by HPLC analysis. Comparing the flavonoid HPLC profile from the nla mutant with that from wild-type plants showed that the two genotypes had the same flavonoid composition: two quercetin-type flavonols, three kaempferol-type flanonols, and one cyanindin derivative (Fig. 4A–D). However, under the N limitation growth condition, the nla mutant had much lower flavonoid contents than did wild-type plants. As shown in Fig. 4E–G, the nla mutant grown with 3 mM nitrate for 24 and 28 DAG contained 49.8–91.4 ppm kaemfperol-type flavonols, 2–3.1 ppm quercetin-type flavonols, and 0–5.4 ppm cyanidin derivatives. In comparison, the wild-type counterpart contained 80–108 ppm kaemfperol-type flavonols, 3.7–8.7 ppm quercetin-type flavonols, and 11–35.7 ppm cyanidin derivatives. The nla mutant and wild-type plants grown with 10 mM nitrate had similar expression levels of the nine anthocyanin synthesis genes (Fig. 3); the two genotypes produced similar and low amounts of flavonols, and their cyanidin derivative contents were not detectable (Fig. 4E–G). All these results demonstrate that the nla mutation down-regulated the N limitation-induced anthocyanin synthesis pathway by reducing the expression of anthocyanin synthesis genes and flavonoid production. In particular, the production of the cyanidin derivatives is most markedly reduced in the nla mutant under N limitation.
Fig. 4.
Effect of the nla mutation on N limitation-induced flavonoid production. The nla mutant and wild-type (Col) plants were grown with 10 mM or 3 mM nitrate for 24 and 28 DAG. Flavonoids were extracted and analysed by HPLC. The flavonoid components were identified by LC-MS. (A) and (B) HPLC profiles at 360 nm of the extracts from Col and the nla mutant grown with 3 mM nitrate for 28 DAG, respectively. (C) and (D) HPLC profiles at 520 nm of the extracts from Col and the nla mutant grown with 3 mM nitrate for 28 DAG, respectively. Q1, quercetin 3,7-dirhamnoglucoside; Q2, quercetin 3-glucoside 7-rhamnoside; K1, kaempferol 3-rhanmoglucoside 7-rhamnoside; K2, kaempferol 3-glucoside 7-rhamnoside; K3, kaempferol 3,7-dirhamnoside; C, cyanidin 3-[2-(sinapoyl)-xylosyl-6-ρ-coumaryol-glucoside] 5-malonyl-glucoside. MAU, milliabsorbance unit. The contents of quercetin-type flavonol (E), kaempferol-type flavonol (F), and cyanidin (G) were measured by HPLC. Statistical analysis by t-test indicates that Col contains significantly higher amounts of flavonoids than does the nla mutant at *P=0.05 or **P=0.01.
N limitation up-regulated lignin synthesis genes and increased lignin production in the nla mutant
The phenylpropanoid pathway has two main branches. One is for anthocyanin synthesis and the other for lignin production. The common substrate for the two branches is ρ-coumaroyl CoA (Fig. 1). Three enzymes, PAL, C4H, and 4CL, catalyse the reactions to convert phenylalanine to ρ-coumaroyl CoA. The previous microarray analysis of the nla mutant found that N limitation significantly increased the expression of PAL1 (At2g37040), C4H (At2g30490), and 4CL3 (At1g65060) in nla mutant and wild-type plants (Peng et al., 2007b). Consistent with this, the qRT-PCR analysis in this study showed that N limitation increased the expression of PAL1 and C4H by ∼5- to 6-fold and 4- to 8-fold, respectively, in the two genotypes (Fig. 5). The N limitation-induced up-regulation level of 4CL3 in the nla mutant was ∼2-fold lower than that in wild-type plants, while the expression of 4CL4 was increased >4-fold by N limitation in the nla mutant, but not in wild-type plants (Fig. 5). This result implies that the nla mutant and wild-type plants have no difference in the expression of the genes whose products catalyse the synthesis of ρ-coumaroyl CoA. However, the nla mutant has a drastic reduction in anthocyanin accumulation under N limitation, and the simplest explanation is that the ρ-coumaroyl CoA used for anthocyanin synthesis is decreased, and the ρ-coumaroyl CoA flux entering the lignin synthesis branch is increased. This would indicate that the nla mutation may up-regulate the lignin synthesis pathway under N limitation.
Fig. 5.
Effect of the nla mutation on the expression of the genes involved in lignin synthesis under N limitation. The nla mutant and wild-type (Col) plants were grown with 10 mM or 3 mM nitrate for 24 and 28 DAG. Gene expression levels were determined by qRT-PCR, and expressed as fold change relative to the transcript abundance in wild-type plants grown with 10 mM nitrate for 24 DAG, which was set as 1. Values are the means of three replicates, with standard deviations in parentheses, and are visualized as a row of coloured boxes using BAR Heatmapper Plus software (http://bbc.botany.utoronto.ca/ntools/cgibin/ntools_ heatmapper_plus.cgi). Black to brighter black colour indicates no or a small change in gene expression, and yellow to brighter yellow colour means increasingly significant up-regulation of gene expression.
In the microarray analysis of the nla mutant response to N limitation (Peng et al., 2007b), six structural genes in the lignin synthesis pathway were found to be up-regulated 5- to 10-fold by N limitation in the nla mutant, but not in wild-type plants. The six genes are At1g67980, Ag1g33030, At1g76470, At1g80820, At14g34230, and At1g72680, which encode CCoAOMT6, COMT-like 5, CCR2, CCR-like1, CAD6, and CAD9 (Fig. 1), respectively. This finding, which indicates that the nla mutation indeed activates the lignin synthesis pathway under N limitation, was confirmed by qRT-PCR analysis. As shown in Fig. 5, the expression of the six lignin synthesis genes in the nla mutant grown with 3 mM and 10 mM nitrate was compared. This revealed that N limitation up-regulated CCoAOMT6, COMT-like 5, CCR2, CCR-like1, CAD6, and CAD9 by 72.30-, 15-, 42-, 11.7-, 6.5-, and 11.2-fold, respectively, at 28 DAG. In contrast, in wild-type plants, the expression of the six lignin synthesis genes was only increased 1.6- to 6-fold by N limitation at 28 DAG (Fig. 5).
The significant up-regulation of lignin synthesis genes by N limitation in the nla mutant suggests that the nla mutation may enhance lignin production under N limitation. Therefore, the lignin content in the nla mutant and wild-type plants under sufficient and limiting N conditions was assessed by staining the stem sections from the two genotypes with phloroglucinol/HCl. Grown with 3 mM nitrate, the nla mutant had strong lignin staining at 21 DAG when the primary inflorescence was 2–3 cm high and the lignin staining intensity was increased markedly at 24 DAG (Fig. 6A). In contrast, in the wild-type plants grown with 3 mM nitrate, lignin staining was very weak at 21 DAG and the lignin staining was still less than that in the nla mutant at 24 DAG (Fig. 6A). On the other hand, supplied with sufficient nitrate (10 mM), the nla mutant and wild-type plants had similar lignin staining intensity at 21 and 24 DAG (Fig. 6A). To confirm the lignin staining results, the lignin contents in the nla mutant and wild-type plants were quantitatively analysed according to Lange et al. (1995). As shown in Fig. 6B, the lignin content in the nla mutant grown with limiting N was >2-fold greater than that in the wild-type plants, while the two genotypes produced similar amounts of lignin when supplied with sufficient nitrate (10 mM). These results indicate that the lignin synthesis pathway was up-regulated by N limitation in the nla mutant, and implies that the nla mutation switches the phenylpropanoid metabolic flux from anthocyanin synthesis to lignin production under N limitation.
Fig. 6.
Effect of the nla mutation on lignin production under N limitation. The nla mutant and wild-type (Col) plants were grown with 3 mM or 10 mM nitrate. The primary inflorescences were harvested at 21 and 24 DAG for assaying lignin production. (A) Visualization of lignin in the stem sections by phloroglucinol-HCl staining. (B) The relative lignin content in the stems of Col and the nla mutant. Lignin contents were determined by using the thioglycollate method, and the calculation of the relative lignin contents in the two genotypes is described in Materials and methods. The values were the means of five replicates with standard deviations. An asterisk indicates that the nla mutant has significantly higher lignin contents than does Col (t-test, P=0.01).
The Pi limitation switched the phenylpropanoid metabolic flux back to anthocyanin synthesis from lignin production in the nla mutant under N limitation
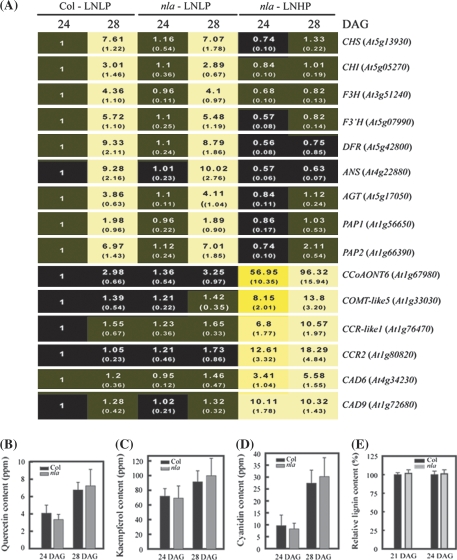
As described earlier, Pi limitation induced the nla mutant to accumulate anthocyanin under N limitation and prevented the N limitation-induced early senescence phenotype (Fig. 2). Correspondingly, the nla mutant was similar to the wild type for the expression of phenylpropanoid synthesis genes and the production of flavonoids and lignin under combined Pi and N limitation (Fig. 7). The qRT-PCR analysis demonstrated that the nla mutant and wild-type plants grown with both limiting Pi and N had similar expression levels of the anthocyanin synthesis genes (Fig. 7A). In comparison, the nla mutant supplied with limiting N but sufficient Pi had much lower expression of these genes (Fig. 3). Further, the nla mutant and wild-type plants supplied with limiting Pi and N had a similar amount of flavonols (quercetin-type and kaemferol-type) and anthocyanin (Figs 7B–D and 2B). At the same time, the six lignin synthesis genes, which were up-regulated markedly in the nla mutant supplied with limiting N (Fig. 5), had similar and low expression levels in both the nla mutant and wild-type plants under combined Pi and N limitation (Fig. 7A), and they also had similar lignin content (Fig. 7E). This was in striking contrast to the nla mutant grown with limiting N but sufficient Pi which contained much more lignin than the wild-type counterpart (Fig. 6B). These results suggest that while the phenylpropanoid metabolic flux is channelled to lignin synthesis in the nla mutant grown with limiting N, Pi limitation switches the phenylpropanoid metabolic flux back to anthocyanin synthesis. Thus, under the condition of combined N and Pi limitation, the nla mutant was induced to accumulate anthocyanins by Pi limitation, which allows this mutant to adapt to N limitation (Fig. 2E).
Fig. 7.
The effect of combined Pi and N limitation on the phenylpropanoid pathway. The nla mutant and wild-type (Col) plants were grown with 0.5 mM Pi and 3 mM nitrate, and subjected to gene expression, and flavonoid and lignin analysis. (A) qRT-PCR analysis of the expression of the genes involved in anthocyanin and lignin synthesis. The expression levels are presented as fold change relative to the transcript abundance in Col grown with 10 mM nitrate for 24 DAG, which was set as 1. Values are the means of three replicates, with standard deviations in parentheses, and were visualized as a row of coloured boxes using BAR Heatmapper Plus software (http://bbc.botany.utoronto.ca/ntools/cgi-bin/ntools_heatmapper_plus.cgi). Black to brighter black colour indicates no or a samll change in gene expression, and yellow to brighter yellow colour means increasingly significant up-regulation of gene expression. LNLP, 3 mM nitrate+0.5 mM Pi; LNHP, 3 mM nitrate+10 mM Pi. The content of quercetin-type flavonol (B), kaempferol-type flavonol (C), cyanidin (D), and the relative lignin contents were determined in the nla mutant and Col grown with combined N and Pi for 24 and 28 DAG. The values in (B–E) were the means of four replicates with standard deviations. Statistical analysis by the t-test reveals that Col and the nla mutant are not significantly different in flavonoid and lignin contents (P=0.05).
Discussion
Anthocyanins are the most widely produced secondary metabolites in plants, and their synthesis is induced by various biotic and abiotic stressors (Steyn et al., 2002; Gould, 2004). Among them, N limitation has been well known to enhance anthocyanin production and up-regulate anthocyanin synthesis genes in plants (Bongue-Bartelsman and Phillips, 1995; Chalker-Scott, 1999; Stewart et al., 2001; Diaz et al., 2006; Lea et al., 2007; Peng et al., 2007a). However, it has not been demonstrated that plants have a specific pathway to control N limitation-induced anthocyanin synthesis. To address this issue, this study further characterized the previously identified nla mutant (Peng et al., 2007a) for anthocyanin synthesis, since this mutant is defective in accumulating anthocyanin under N limitation (Fig. 2B; Peng et al., 2007a). Pi limitation can also induce Arabidopsis plants to accumulate anthocyanins (Chalker-Scott, 1999; Steyn et al., 2002), and this study found that the nla mutant produced abundant anthocyanins under Pi limitation no matter whether sufficient or limiting N was supplied (Fig. 2C–F). This suggests that Arabidopsis plants have an N limitation-induced anthocyanin synthesis pathway, which can be specifically disrupted by the nla mutation. However, this mutation has no effect on the Pi limitation-induced anthocyanin synthesis pathway, which induces the nla mutant to accumulate anthocyanins even under N limitation.
Anthocyanins have diverse physiological functions in plants, ranging from attracting pollinators in flowers to assisting plants to acquire tolerance or resistance to various biotic and abiotic stressors (Gould, 2004). This study demonstrated that anthocyanin accumulation was important for Arabidopsis plants to adapt to N limitation. With the disrupted N limitation-induced anthocyanin synthesis pathway, the nla mutant did not accumulate anthocyanins under N limitation, and did not adapt to N limitation, but instead produced an early senescence phenotype (Fig. 2A; Peng et al., 2007a). In contrast, when the nla mutant was grown under combined Pi and N limitation, the functional Pi limitation-mediated anthocyanin synthesis pathway induced the nla mutant to produce abundant anthocyanins (Fig. 2C–F). This anthocyanin accumulation allowed the nla mutant to adapt to N limitation, with the nla mutant not producing the N limitation-induced early senescence phenotype (Fig. 2E). Therefore, these results indicate that anthocyanin accumulation is indeed important for Arabidopsis adaptability to N limitation, and the major defect caused by the nla mutation is the lack of N limitation-induced anthocyanin accumulation.
The structural and regulatory genes for anthocyanin synthesis are highly up-regulated by N limitation in Arabidopsis (Lea et al., 2007; Peng et al., 2007b). The finding that the nla mutant is mainly defective in anthocyanin accumulation under N limitation implies that the nla mutation may cause the failure of N limitation to up-regulate anthocyanin synthesis genes. However, this study found that N limitation still up-regulated anthocyanin synthesis genes in this mutant (Fig. 3). Therefore, it is not surprising to find that overexpressing CHS, PAP1, and PAP2 in the nla mutant could not enhance anthocyanin synthesis and rescue the N limitation-induced early senescence phenotype. Nevertheless, consistent with a previous microarray analysis of the nla mutant response to N limitation (Peng et al., 2007b), the nla mutation significantly decreased the N limitation-induced up-regulation levels of the anthocyanin synthesis genes as compared with that in wild-type plants (Fig. 3). Correspondingly, the nla mutant contained much lower flavonol and cyanidin contents than did the wild-type plants under N limitation (Fig. 4). Further, the expression was most severely affected in the genes whose products function in the late steps in anthocyanin synthesis such as DFR and ANS (Fig. 3), and very little cyanidin was detected in the nla mutant grown with limiting N (Fig. 4G). Therefore, it is most likely that the nla mutation mainly leads to the disruption in the conversion of flavonols to cyanidin in the N limitation-induced anthocyanin synthesis pathway.
Anthocyanin synthesis is one major branch of the phenylpropanoid pathway, and the other important branch is for lignin synthesis. The two branches diverge at and use the common substrate ρ-coumaroyl CoA (Fig. 1). This makes it possible to regulate the ρ-coumaroyl CoA flux into the two branches based on plant phenylpropanoid metabolic status. Besseau et al. (2007) reported that repressing lignin synthesis in Arabidopsis significantly enhanced flavonoid and anthocyanin production, and the metabolic flux in the phenylpropanoid pathway was mainly channelled into the anthocyanin synthesis branch. In this study, the genes PAL1, C4H, and 4CL, which are involved in converting phenylalanine to ρ-coumaroyl CoA in the phenylpropanoid pathway (Fig. 1), had similar expression levels in the nla mutant and wild-type plants under N limitation (Fig. 5). This suggests that the two genotypes have a similar capacity to produce ρ-coumaroyl CoA under N limitation. Accordingly, the large reduction of anthocyanin synthesis in the nla mutant under N limitation implies that the ρ-coumaroyl CoA used for anthocyanin synthesis is reduced, while more ρ-coumaroyl CoA is channelled to the lignin synthesis branch. Indeed, six lignin synthesis genes were significantly up-regulated by N limitation in the nla mutant but not in wild-type plants (Fig. 5), and the lignin content in the nla mutant was >2-fold that in wild type plants under N limitation (Fig. 6). These results reveal that, in wild-type plants, the phenylpropanoid metabolic flux goes to anthocyanin synthesis under N limitation, while the nla mutation switches this flux to lignin production. Pi limitation can reroute the nla mutation-mediated phenylpropanoid metabolic flux back to anthocyanin synthesis under N limitation (Fig. 7). Under combined Pi and N limitation, the nla mutant and wild-type plants were similar in the expression of the anthocyanin and lignin synthesis genes and in the production of anthocyanins and lignin (Fig. 7).
In conclusion, all the data presented in this study demonstrate that Arabidopsis plants are equipped with an N limitation specifically induced anthocyanin synthesis pathway, in which NLA is an essential molecular component. NLA is involved in channelling the phenylpropanoid metabolic flux into anthocyanin synthesis, which allows wild-type plants to accumulate abundant anthocyanins and adapt to N limitation. However, the nla mutation specifically disrupts the N limitation-induced anthocyanin synthesis pathway and switches the phenylpropanoid metabolic flux to lignin synthesis. Accordingly, the nla mutant fails to accumulate anthocyanins and adapt to N limitation, but enhances lignin production, and this in turn leads to an early senescence phenotype. Taken together, this study suggests that NLA controls the N limitation-induced anthocyanin synthesis pathway, and anthocyanin accumulation is important for Arabidopsis adaptability to N limitation.
Crop cultivars with strengthened N limitation adaptability are important given that in current agriculture practice, increasing amounts of nitrogen fertilizers are applied to crops worldwide and have caused severe N pollution to the environment and increased the cost of crop production (Good et al., 2004). Delineating the molecular mechanism governing plant adaptability to N limitation is essential for developing crop cultivars with enhanced yields under lower N fertilization regimes. In a previous study, the cloning and functional characterization of NLA revealed that plants have a molecular mechanism to adapt to N limitation (Peng et al., 2007a). In this work it was found that anthocyanin accumulation is essential for Arabidopsis adaptability to N limitation, and NLA controls the N limitation-induced anthocyanin synthesis pathway. These results will be used to explore the molecular mechanism controlling plant adaptation to N limitation further.
Acknowledgments
This work was supported by the Natural Sciences and Engineering Research Council (NSERC) and industrial and government partners through the Green Crop Networks (GCN) Research Network.
References
- Besseau S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M. Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. The Plant Cell. 2007;19:148–162. doi: 10.1105/tpc.106.044495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y-M, Zhang Y, Signorelli T, Zhao R, Zhu T, Rothstein SJ. Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. The Plant Journal. 2005;44:680–692. doi: 10.1111/j.1365-313X.2005.02568.x. [DOI] [PubMed] [Google Scholar]
- Bongue-Bartelsman M, Phillips DA. Nitrogen stress regulates gene expression of enzymes in the flavonoid biosynthetic pathway of tomato. Plant Physiology and Biochemistry. 1995;33:539–546. [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. The Plant Cell. 2000;12:2383–2393. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker-Scott L. Environmental significance of anthocyanins in plant stress responses. Photochemistry and Photobiology. 1999;70:1–9. [Google Scholar]
- Diaz C, Saliba-Colombani V, Loudet O, Belluomo P, Moreau L, Daniel-Vedele F, Morot-Gaudry JF, Masclaux-Daubresse C. Leaf yellowing and anthocyanin accumulation are two genetically independent strategies in response to nitrogen limitation in Arabidopsis thaliana. Plant and Cell Physiology. 2006;47:74–83. doi: 10.1093/pcp/pci225. [DOI] [PubMed] [Google Scholar]
- Ding L, Wang KJ, Jiang GM, Biswas DK, Xu H, Li LF, Li YH. Effects of nitrogen deficiency on photosynthetic traits of maize hybrids released in different years. Annals of Botany. 2005;96:925–930. doi: 10.1093/aob/mci244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick DN. Review of the symposium on developing drought and low-N tolerant maize. In: Edmeades GO, Banziger M, Mickelson HR, Pena-Valdicia CB, editors. Developing drought and low N-tolerant maize. El Batan. Mexico: CIMMYT; 1997. pp. 554–556. [Google Scholar]
- Good AG, Shrawat AK, Muench DG. Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends in Plant Science. 2004;9:597–605. doi: 10.1016/j.tplants.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Gould KS. Nature's Swiss Army knife: the diverse protective roles of anthocyanins in leaves. Journal of Biomedical Biotechnology. 2004;2004:314–320. doi: 10.1155/S1110724304406147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch WA, Singsaas EL, McCown BH. Resorption protection. Anthocyanins facilitate nutrient recovery in autumn by shielding leaves from potentially damaging light levels. Plant Physiology. 2003;133:1296–1305. doi: 10.1104/pp.103.027631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamis S, Lamaze T, Lemoine Y, Foyer C. Adaptation of the photosynthetic apparatus in maize leaves as a result of nitrogen limitation. Plant Physiology. 1990;9:1436–1443. doi: 10.1104/pp.94.3.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Lapierre C, Sandermann H. Elicitor-induced spruce stress lignin, structural similarity to early developmental lignins. Plant Physiology. 1995;108:1277–1287. doi: 10.1104/pp.108.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea US, Slimestad R, Smedvig P, Lillo C. Nitrogen deficiency enhances expression of specific MYB and bHLH transcription factors and accumulation of end products in the flavonoid pathway. Planta. 2007;225:1245–1253. doi: 10.1007/s00425-006-0414-x. [DOI] [PubMed] [Google Scholar]
- Marschner H. Mineral nutrition of higher plants. London: Academic Press; 1995. [Google Scholar]
- Noh B, Spalding EP. Anion channels and the stimulation of anthocyanin accumulation by blue light in Arabidopsis seedlings. Plant Physiology. 1998;116:503–509. doi: 10.1104/pp.116.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto M-A, Onai K, Furukawa Y, Aoki E, Araki T, Nakamura K. Effects of sugar on vegetative development and floral transition in Arabidopsis. Plant Physiology. 2001;127:252–261. doi: 10.1104/pp.127.1.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Bi Y-M, Zhu T, Rothstein SJ. Genome-wide analysis of Arabidopsis responsive transcriptome to nitrogen limitation and its regulation by the ubiquitin ligase gene NLA. Plant Molecular Biology. 2007b;65:775–797. doi: 10.1007/s11103-007-9241-0. [DOI] [PubMed] [Google Scholar]
- Peng M, Hannam C, Gu H, Bi Y-M, Rothstein SJ. A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts Arabidopsis adaptability to nitrogen limitation. The Plant Journal. 2007a;50:320–337. doi: 10.1111/j.1365-313X.2007.03050.x. [DOI] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. The Plant Journal. 1995;8:659–671. doi: 10.1046/j.1365-313x.1995.08050659.x. [DOI] [PubMed] [Google Scholar]
- Smillie RM, Hetherington SE. Photoabatement by anthocaynin shields photosynthetic systems from light stress. Photosynthetica. 1999;36:451–463. [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P. Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiology. 2006;140:637–646. doi: 10.1104/pp.105.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart AJ, Chapman W, Jenkins GI, Graham I, Martin T, Crozier A. The effect of nitrogen and phosphorus deficiency on flavonol accumulation in plant tissues. Plant, Cell and Environment. 2001;24:1189–1197. [Google Scholar]
- Steyn WJ, Wand SJE, Holcroft DM, Jacobs G. Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytologist. 2002;155:349–361. doi: 10.1046/j.1469-8137.2002.00482.x. [DOI] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiology. 2005;139:1840–1852. doi: 10.1104/pp.105.066688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollenaar M, Wu J. Yield improvement in temperate maize is attributable to greater stress tolerance. Crop Science. 1999;39:1597–1604. [Google Scholar]
- Winkel-Shirley B. Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]