Abstract
Hsp110s are divergent relatives of Hsp70 chaperones that hydrolyze ATP. Hsp110s serve as Hsp70 nucleotide exchange factors and act directly to maintain polypeptide solubility. To date, the impact of peptide binding on Hsp110 ATPase activity is unknown and an Hsp110/peptide affinity has not been measured. We now report on a peptide that binds to the yeast Hsp110, Sse1p, with a KD of ~2 nM. Surprisingly, the binding of this peptide fails to stimulate Sse1p ATP hydrolysis. Moreover, an Hsp70-binding peptide is unable to associate with Sse1p, suggesting that Hsp70s and Hsp110s possess partially distinct peptide recognition motifs.
Keywords: Hsp70, molecular chaperone, nucleotide exchange factor, fluorescence, ATPase
1. Introduction
Hsp70s are a ubiquitous class of molecular chaperones that are involved in protein folding, transport, degradation, and assembly and disassembly of protein complexes, both in times of cell stress and under normal growth conditions [1,2]. Hsp70s contain a highly conserved, N-terminal nucleotide-binding domain (NBD), which allosterically regulates the peptide binding state in the C-terminal substrate binding domain (SBD). In the ATP-bound state, peptide affinity is low, allowing substrate binding and release, whereas in the ADP-bound state the peptide affinity is higher. This cycle can be modulated by DnaJ/Hsp40 co-chaperones, which possess an Hsp70 interaction motif known as a J-domain. The stimulation of Hsp70 ATP hydrolysis, which leads to peptide capture, can be achieved by J-domain binding or by peptide binding. Subsequently, peptide release can be triggered by nucleotide exchange factors (NEFs) that help to liberate the bound ADP, facilitating ATP rebinding.
The Hsp110s are divergent relatives of Hsp70s found only in eukaryotes. Like Hsp70s, Hsp110s exhibit ATPase activity [3,4]. Unlike Hsp70s, however, Hsp110s do not actively fold proteins, but rather act as holdases to maintain the solubility of denatured model protein substrates [3,5–7]. Hsp110s also function as NEFs for Hsp70s [3,8,9]. Hsp110s most likely play an important role in protein homeostasis because deletion of the predominant cytoplasmic Hsp110 in yeast, SSE1, leads to a slow growth phenotype [10,11], and because Hsp110 over-expression in mammalian tissue culture cells confers thermotolerance [6]. Sse1p has also been proposed to mediate nascent polypeptide folding at the ribosome in conjunction with Hsp70s [12].
Sse1p shares ~30% overall sequence identity with an abundant, cytosolic yeast Hsp70, Ssa1p, and the two chaperones associate [12,13]. The SBD in Sse1p is significantly longer than the analogous region in Ssa1p, and like other Hsp110-Hsp70 pairs, the sequence similarity between the proteins is largely confined to the NBDs. Nevertheless, the SBD of Sse1p, which is conserved among Hsp110s, is predicted to bind peptides [14] and the ability of the Sse1p SBD to exhibit holdase activity is consistent with the presence of a peptide-binding site in this domain. Thus, it was interesting that a putative peptide-binding groove in the recent Sse1p crystal structure lacked bound peptide [15]. This might have arisen because the adjoining NBD was ATP-bound and that Sse1p—like Hsp70s—undergoes a nucleotide-dependent conformational change that may catalyze peptide release [15,16]. Nevertheless, the orientation of SBD subdomains is quite different between Sse1p and Hsp70s, and it was not completely clear if the binding of peptides to Sse1p would activate the ATPase activity. Moreover, it was found that Sse1p nucleotide binding but not hydrolysis was essential to support the viability of yeast containing site-directed sse1 mutants in an sse1Δsse2Δ background, suggesting that a tight coupling of ATP hydrolytic cycles to peptide binding/release may not be vital for Hsp110 activity [17]. To date, there have been no reports measuring the peptide affinity of Sse1p/Hsp110 or indicating directly whether peptide binding activates ATP hydrolysis.
We now describe the identification of a peptide that binds to Sse1p with a KD of ~2 nM, and discovered that ATP hydrolysis was unchanged upon peptide addition. In contrast, a peptide substrate for Ssa1p activated the Hsp70’s ATPase activity but was unable to bind Sse1p. These data suggest that Hsp110s and Hsp70 possess distinct peptide preferences, hint that Hsp110s might exhibit uncoupled peptide binding and ATP hydrolysis, and provide further support that the mechanisms of action of Hsp70s and Hsp110s are unique.
2. Materials and methods
2.1 Protein purifications
Hexahistidine-tagged forms of Sse1p and Ssa1p were isolated from yeast cell extracts to a purity level >95% as described [5,18], and as noted in the text, the presence of this amino acid extension on Sse1p did not inhibit its ATPase or holdase activities. Likewise, tagged Ssa1p function in vitro or in vivo was unimpaired [18]. The absence of contaminating ATPases in the Sse1p preparation was confirmed by mass spectrometry of the purified protein sample. A soluble form of the ER-associated Hlj1p chaperone was purified as described [19]. Purified Sis1p was a generous gift from Dr. P. Needham (NIH) and the cysteine string protein (Csp) was kindly provided by Dr. Hui Zhang (University of Pittsburgh School of Medicine).
To remove the hexahistine tag from Sse1p, the protein was treated with recombinant bovine enterokinase followed by removal of the enterokinase with anti-enterokinase-agarose as per the manufacturer’s instructions (Sigma-Aldrich). The release of the hexahistidine tag from Sse1p did not alter the ATPase activity of the Hsp110 or the inability of Sse1p to be stimulated by J-domain-containing proteins or peptide (data not shown).
2.2 Peptides
Peptide ala-p5 (ALLLMYRR) is derived from the precursor of chicken mitochondrial aspartate amino-transferase [20] and was synthesized by the GenScript Corporation, Scotch Plains, NJ. A fluorescein isothiocyanate (FITC) derivative was prepared as described [21]. The “LIC” peptide (LICGFRVVLMYRF; amino acids 256–268 in firefly luciferase) was synthesized by the University of Pittsburgh Peptide Synthesis Facility and labeled with 6-carboxylflourescein (6CF) using an epsilon-aminohexanoic linker at the N terminus. The purity of each peptide was >89%, and the molecular masses were confirmed by mass spectrometry. The peptides were dissolved in DMSO (FITC-ala-p5) or DMF (6CF-LIC) and stored at a final concentration of 6mM at −20°C.
2.3 Assays for chaperone-mediated ATP hydrolysis and peptide binding
The ATPase activities of Ssa1p and Sse1p were assayed under steady-state conditions for 30 min at 30ºC as described [22]. Where indicated, a J-domain containing co-chaperone or an equivalent volume of buffer was added at the start of the reaction such that the final concentrations and ratio of the proteins were 2 μM co-chaperone: 1 μM Hsp70/Hsp110. In other cases, the FITC-ala-p5 or 6CF-LIC peptide (or an equal volume of solvent) was pre-incubated with the chaperones at the indicated concentration at 4°C for 15 min.
The binding of FITC-ala-p5 and 6CF-LIC to Ssa1p and Sse1p was analyzed by incubating increasing concentrations of chaperone with 5 nM peptide in black 96-well microtiter plates (Corning), and fluorescence polarization was measured in an Analyst AD instrument (Molecular Devices). Polarization values are expressed in millipolarization units (mPs) and were calculated using the equation mP=1000*[(IS−ISB)−(IP−IPB)]/[(IS−ISB)+(IP−IPB)], where IS is the parallel emission intensity measurement and IP is the perpendicular emission intensity sample measured, and ISB and IPB are the corresponding measurements for background (buffer).
3. Results and discussion
To assess whether the regulation of ATP hydrolysis by Sse1p is similar to that of Ssa1p, we first purified hexahistidine-tagged forms of both proteins and compared their ATPase activities in the presence and absence of select Hsp40 co-chaperones. As shown in Fig. 1, we found that the activity of the Ssa1p but not Sse1p was significantly enhanced by two cytosolic yeast Hsp40s, Hlj1p and Sis1p, which are respectively ER- and ribosome-associated Hsp70 co-chaperones that are known to interact with Ssa1p [19,23]. A mammalian Hsp40, Csp, which activates ATP hydrolysis by mammalian Hsc70 [24] also selectively stimulated ATP hydrolysis by Ssa1p. Notably, the unstimulated rates of ATP hydrolysis by Ssa1p and Sse1p were very similar; the turnover numbers were 0.33 per minute for Ssa1p and 0.34 per minute for Sse1p. This is consistent with the range of published values for the unstimulated rates of ATP hydrolysis by Hsp70 chaperones [25–27].
Fig. 1. The ATPase activity of yeast Hsp70 but not Hsp110 is stimulated by select Hsp40 co-chaperones.
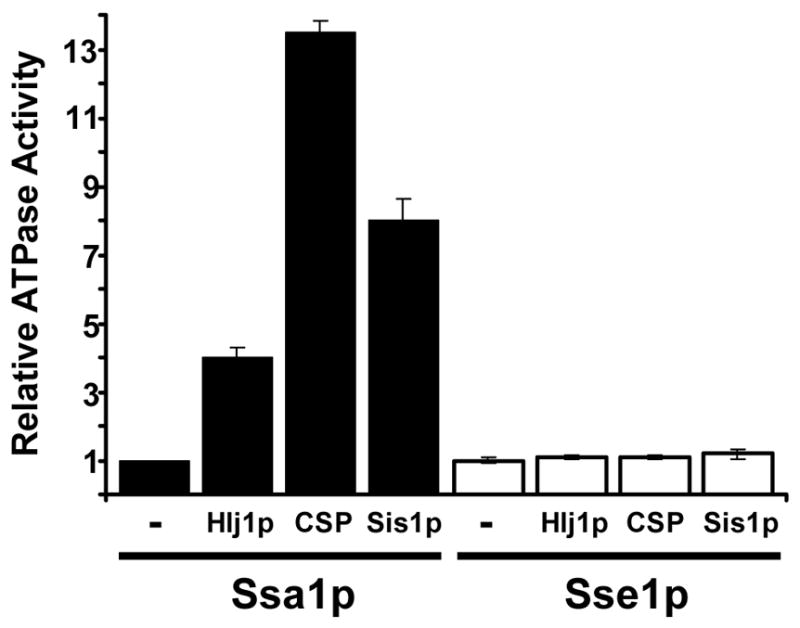
The relative steady-state ATPase activities of Ssa1p and Sse1p were examined in the presence or absence of the indicated Hsp40s, as described in the Materials and methods. The ATPase activity in each experiment was then standardized to the amount of ATP hydrolysis in the absence of added Hsp40. Data represent the means of 3–8 independent determinations, +/−SE; p<0.0001 for the Hlj1p-, Csp-, and Sis1p-mediated stimulation of Ssa1p; the Sis1p-mediated stimulation of Sse1p was not significant (p<0.16).
Next, we employed a method that was previously used to isolate peptides that bound to the E. coli Hsp70, DnaK [28] and Hsp40, DnaJ [29]. In brief, nitrocellulose blots containing partially overlapping 14 amino acid peptides derived from firefly luciferase, an Sse1p substrate [5,30], were probed with purified Sse1p. Peptides corresponding to select “hits” from this analysis were synthesized and one peptide, designated “LIC”, was conjugated to 6-carboxyfluorescein (6CF) so that binding could be assessed in solution. We found that 6CF-LIC associated with Sse1p with a KD of 2.3 nM (Fig. 2A, open circles). This is higher than the affinity of Hsp70 for other peptides, such as p5 and APPY, measured in the presence of ADP or in the absence of nucleotide (~60 nM;[31,32]), but the binding of 6CF-LIC to Ssa1p under these conditions (KD = 27 nM; Fig. 2A, closed circles) was quite similar to the published values for peptide binding. Furthermore, addition of unlabeled LIC peptide to a final concentration of 1 μM displaced ~25% of the bound 6CF-LIC under steady-state conditions; in contrast, incubation of Sse1p with 6CF-LIC in the presence of ATP or ADP had no statistically significant effect on peptide binding (data not shown), consistent with the conjecture that a tight coupling between Sse1p’s ATPase cycle and peptide binding may not be vital (see Introduction). Based on these data, we conclude that Sse1p has a significantly higher binding affinity for the LIC peptide than Ssa1p.
Fig. 2. Sse1p exhibits high-affinity peptide binding.
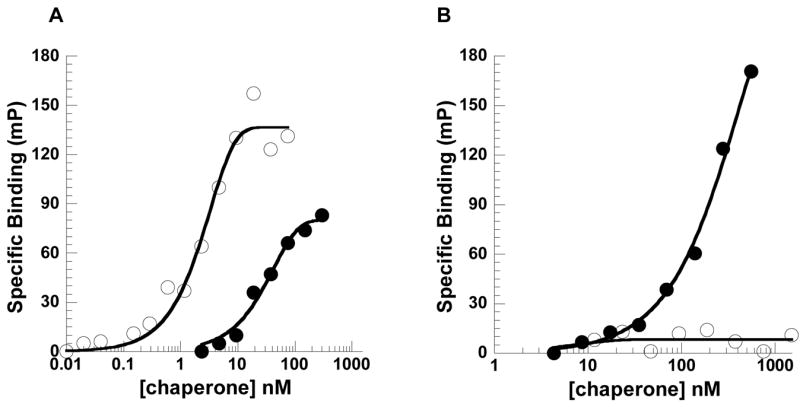
The binding of (A) 6CF-LIC and (B) FITC-ala-p5 to Ssa1p and Sse1p were assayed as described in the Materials and methods. Ssa1p data are represented by closed circles (●) and Sse1p data are indicated by open circles (○). Curve fits are indicated by solid black lines.
To determine whether Sse1p could also bind to an Hsp70 peptide substrate, we made use of a FITC-conjugate of ala-p5 (see Materials and methods). Although Ssa1p bound FITC-ala-p5 with a KD of ~100 nM, no interaction was observed between this peptide and Sse1p (Fig. 2B, compare closed to open circles). Of note, we recently reported that the binding of FITC-ala-p5 to Hsp70 is ATP-dependent [21], indicating the validity of using this modified substrate to measure the peptide binding affinity for the chaperone. Combined with the data presented in Fig. 2A, these results indicate that Sse1p exhibits peptide binding specificity and that the peptide preferences for Sse1p and Ssa1p are not entirely overlapping. A complete analysis of the peptide preferences of Sse1p will require continued analysis using peptide-display technologies, but based on our data we suggest that these efforts should prove informative.
Having identified 6CF-LIC as a high-affinity peptide substrate for Sse1p, we then examined whether the peptide stimulates the chaperone’s ATPase activity. Experiments using FITC-ala-p5 were performed in parallel and the combined data are summarized in Fig. 3. As anticipated, we found that a 30-fold molar excess of ala-p5 significantly enhanced the ATPase activity of Ssa1p. In contrast, neither FITC-ala-p5 nor the binding peptide 6CF-LIC affected the ATP hydrolytic rate of Sse1p (Fig. 3).
Fig. 3. The ATPase activity of Sse1p is not enhanced by the addition of a peptide substrate.
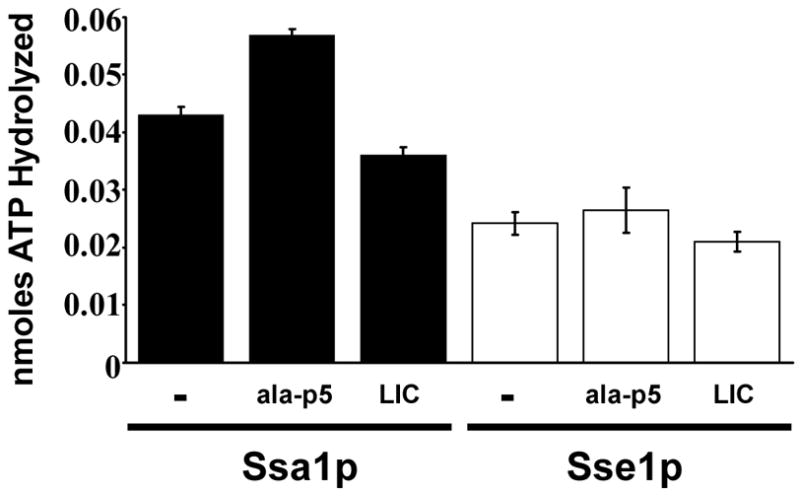
Steady-state ATPase assays were performed as described in the Materials and methods in the presence or absence of a 30-fold molar excess of FITC-ala-p5 or 6CF-LIC. Data represent the means of 3–6 independent experiments, +/−SE. Note that only the stimulation of Ssa1p by ala-p5 is significant (p<0.0004).
Based on the sensitivity of the fluorescence methods employed in this study, our data also establish a means to screen for small molecule compounds that might interfere with peptide binding to Hsp110s. Because defects in Sse1p function enhance the degradation of an atherosclerotic-inducing apolipoprotein (apoB) [33], we suggest that inhibitors that block Hsp110-substrate binding in hepatic cells might provide a means to lower circulating cholesterol levels. Furthermore, mammalian Hsp110 and an ER-localized Hsp110 homolog, Grp170, bind tumor-derived peptides and protein substrates, and these chaperones have been used to trigger innate and antigen-associated immunity [34]. Based on our data, one reason Hsp110/Grp170 may be highly effective in anti-cancer vaccines is that they have such a high affinity for peptide substrates. Further studies can now be directed to test this hypothesis.
Acknowledgments
This study was supported by grant CA119001 from the National Institutes of Health to G.C. and J.L.B. We thank Drs. P. Needham, R. Youker, and H. Zhang for reagents, and Dr. J. Schneider-Mergener (Jerini) for kindly providing the firefly luciferase peptide blots. We are ever grateful to Drs. J. Minden and S. Dowd of Carnegie Mellon University for mass spectrometry assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brodsky JL, Chiosis G. Hsp70 Molecular chaperones: emerging roles in human disease and identification of small molecule modulators. Curr Top Med Chem. 2006;6:1215–1225. doi: 10.2174/156802606777811997. [DOI] [PubMed] [Google Scholar]
- 2.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raviol SHH, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006;25:2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raviol H, Bukau B, Mayer MP. Human and yeast Hsp110 chaperones exhibit functional differences. FEBS Lett. 2006;580:168–174. doi: 10.1016/j.febslet.2005.11.069. [DOI] [PubMed] [Google Scholar]
- 5.Goeckeler JL, Stephens A, Lee P, Caplan AJ, Brodsky JL. Overexpression of yeast Hsp110 homolog Sse1p suppresses ydj1–151 thermosensitivity and restores Hsp90-dependent activity. Mol Biol Cell. 2002;13:2760–2770. doi: 10.1091/mbc.02-04-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh HJ, Chen X, Subjeck JR. Hsp110 protects heat-denatured proteins and confers cellular thermoresistance. J Biol Chem. 1997;272:31636–31640. doi: 10.1074/jbc.272.50.31636. [DOI] [PubMed] [Google Scholar]
- 7.Dragovic BSZ, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaner L, Sousa R, Morano KA. Characterization of Hsp70 binding and nucleotide exchange by the yeast Hsp110 chaperone Sse1. Biochemistry. 2006;45:15075–15084. doi: 10.1021/bi061279k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shomura Y, et al. Regulation of Hsp70 function by HspBP1: structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol Cell. 2005;17:367–79. doi: 10.1016/j.molcel.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 10.Mukai H, Kuno T, Tanaka H, Hirata D, Miyakawa T, Tanaka C. Isolation and characterization of SSE1 and SSE2, new members of the yeast HSP70 multigene family. Gene. 1993;132:57–66. doi: 10.1016/0378-1119(93)90514-4. [DOI] [PubMed] [Google Scholar]
- 11.Shirayama M, Kawakami K, Matsui Y, Tanaka K, Toh-e A. MSI3, a multicopy suppressor of mutants hyperactivated in the RAS-cAMP pathway, encodes a novel HSP70 protein of Saccharomyces cerevisiae. Mol Gen Genet. 1993;240:323–332. doi: 10.1007/BF00280382. [DOI] [PubMed] [Google Scholar]
- 12.Yam AYW, Albanese V, Lin HTJ, Frydman J. Hsp110 cooperates with different cytosolic HSP70 systems in a pathway for de novo folding. J Biol Chem. 2005;280:41252–41261. doi: 10.1074/jbc.M503615200. [DOI] [PubMed] [Google Scholar]
- 13.Shaner L, Wegele H, Buchner J, Morano KA. The yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb. J Biol Chem. 2005;280:41262–41269. doi: 10.1074/jbc.M503614200. [DOI] [PubMed] [Google Scholar]
- 14.Easton KYDP, Subjeck JR. The Hsp110 and Grp170 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones. 2000;5:276–290. doi: 10.1379/1466-1268(2000)005<0276:thagsp>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q, Hendrickson WA. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell. 2007;131:106–120. doi: 10.1016/j.cell.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kassenbrock CK, Kelly RB. Interaction of heavy chain binding protein (BiP/GRP78) with adenine nucleotides. EMBO J. 1989;8:1461–1467. doi: 10.1002/j.1460-2075.1989.tb03529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaner L, Trott A, Goeckeler JL, Brodsky JL, Morano KA. The function of the yeast molecular chaperone Sse1 is mechanistically distinct from the closely related Hsp70 family. J Biol Chem. 2004;279:21992–22001. doi: 10.1074/jbc.M313739200. [DOI] [PubMed] [Google Scholar]
- 18.McClellan AJ, Brodsky JL. Mutation of the ATP-binding pocket of SSA1 indicates that a functional interaction between Ssa1p and Ydj1p is required for post-translational translocation into the yeast endoplasmic reticulum. Genetics. 2000;156:501–512. doi: 10.1093/genetics/156.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Youker RT, Walsh P, Beilharz T, Lithgow T, Brodsky JL. Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol Biol Cell. 2004;15:4787–4797. doi: 10.1091/mbc.E04-07-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han W, Christen P. cis-Effect of DnaJ on DnaK in ternary complexes with chimeric DnaK/DnaJ-binding peptides. FEBS Lett. 2004;563:146–150. doi: 10.1016/S0014-5793(04)00290-X. [DOI] [PubMed] [Google Scholar]
- 21.Kang Y, Taldone T, Clement CC, Fewell SW, Aguirre J, Brodsky JL, Chiosis G. Design of a fluorescence polarization assay platform for the study of human Hsp70. Bioorg Med Chem Lett. 2008 doi: 10.1016/j.bmcl.2008.05.046. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McClellan AJ, Endres JB, Vogel JP, Palazzi D, Rose MD, Brodsky JL. Specific molecular chaperone interactions and an ATP-dependent conformational change are required during post-translational protein translocation into the yeast ER. Mol Biol Cell. 1998;9:3533–3545. doi: 10.1091/mbc.9.12.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu Z, Cyr DM. Protein folding activity of Hsp70 is modified differentially by the Hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem. 1998;273:27824–27830. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- 24.Chamberlain LH, Burgoyne RD. Activation of the ATPase activity of heat-shock proteins Hsc70/Hsp70 by cysteine-string protein. Biochem J. 1997;322:853–858. doi: 10.1042/bj3220853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ha JH, McKay DB. ATPase kinetics of recombinant bovine 70 kDa heat shock cognate protein and its amino-terminal ATPase domain. Biochemistry. 1994;33:14625–14635. doi: 10.1021/bi00252a031. [DOI] [PubMed] [Google Scholar]
- 26.Theyssen H, Schuster HP, Packschies L, Bukau B, Reinstein J. The second step of ATP binding to DnaK induces peptide release. J Mol Biol. 1996;263:657–670. doi: 10.1006/jmbi.1996.0606. [DOI] [PubMed] [Google Scholar]
- 27.McCarty JS, Buchberger A, Reinstein J, Bukau B. The role of ATP in the functional cycle of the DnaK chaperone system. J Mol Biol. 1995;249:126–137. doi: 10.1006/jmbi.1995.0284. [DOI] [PubMed] [Google Scholar]
- 28.Rudiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudiger S, Schneider-Mergener J, Bukau B. Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 2001;20:1042–1050. doi: 10.1093/emboj/20.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brodsky JL, Werner ED, Dubas ME, Goeckeler JL, Kruse KB, McCracken AA. The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J Biol Chem. 1999;274:3453–3460. doi: 10.1074/jbc.274.6.3453. [DOI] [PubMed] [Google Scholar]
- 31.Pierpaoli EV, Gisler SM, Christen P. Sequence-specific rates of interaction of target peptides with the molecular chaperones DnaK and DnaJ. Biochemistry. 1998;37:16741–16748. doi: 10.1021/bi981762y. [DOI] [PubMed] [Google Scholar]
- 32.Montgomery DL, Morimoto RI, Gierasch LM. Mutations in the substrate binding domain of the Escherichia coli 70 kDa molecular chaperone, DnaK, which alter substrate affinity or interdomain coupling. J Mol Biol. 1999;286:915–932. doi: 10.1006/jmbi.1998.2514. [DOI] [PubMed] [Google Scholar]
- 33.Hrizo SL, Gusarova V, Habiel DM, Goeckeler JL, Fisher EA, Brodsky JL. The Hsp110 molecular chaperone stabilizes apolipoproteinB from endoplasmic reticulum-associated degradation (ERAD) J Biol Chem. 2007;282:32665–32675. doi: 10.1074/jbc.M705216200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segal BH, Wang XY, Dennis CG, Youn R, Repasky EA, Manjili MH, Subjeck JR. Heat shock proteins as vaccine adjuvants in infections and cancer. Drug Discov Today. 2006;11:534–540. doi: 10.1016/j.drudis.2006.04.016. [DOI] [PubMed] [Google Scholar]


