Abstract
Clinically isolated syndrome (CIS) refers to the earliest clinical manifestation of multiple sclerosis (MS). Currently there are no prognostic biological markers that accurately predict conversion of CIS to clinically definite MS (CDMS). Furthermore, the earliest molecular events in MS are still unknown. We used microarrays to study gene expression in naïve CD4+ T cells from 37 CIS patients at time of diagnosis and after 1 year. Supervised machine-learning methods were used to build predictive models of disease conversion. We identified 975 genes whose expression segregated CIS patients into four distinct subgroups. A subset of 108 genes further discriminated patients in one of these (group 1) from other CIS patients. Remarkably, 92% of patients in group 1 converted to CDMS within 9 months. Consistent down-regulation of TOB1, a critical regulator of cell proliferation, was characteristic of group 1 patients. Decreased TOB1 expression at the RNA and protein levels also was confirmed in experimental autoimmune encephalomyelitis. Finally, a genetic association was observed between TOB1 variation and MS progression in an independent cohort. These results indicate that CIS patients at high risk of conversion have impaired regulation of T cell quiescence, possibly resulting in earlier activation of pathogenic CD4+ cells.
Keywords: clinically isolated syndrome, gene expression
Multiple sclerosis (MS) is a common disabling neurologic disease of young adults (1). Most patients with MS initially present with a clinically isolated syndrome (CIS) caused by an inflammatory demyelinating insult in the central nervous system (CNS). Approximately one-third of CIS patients progress to clinically definite MS (CDMS) within 1 year after diagnosis, and approximately half do so after 2 years (2, 3). It is estimated that ≈10% of CIS patients remain free of further demyelinating attacks and neurological complications even in the presence of radiological evidence of white matter lesions (4). Although structural neuroimaging studies are invaluable in the diagnosis and clinical surveillance of MS (3, 5), there currently is no biological marker that accurately predicts MS conversion in CIS patients. Individualized early prognosis and prediction of CDMS would be of substantial value because patients at high risk for rapid progression could be offered disease-modifying therapy, an approach shown to be beneficial in early MS (2). In addition, CIS patients represent a unique population for the study of early molecular events that lead to demyelination and axonal degeneration.
In this article we describe the analysis of gene expression in naïve (unstimulated) CD4+ T cells isolated from well characterized CIS patients at the time of diagnosis and after 1 year and show a distinct molecular signature that differentiates CIS patients from healthy controls. The expression of a subset of those genes successfully identifies a group of patients at high risk of MS conversion. Molecular changes observed in individuals who rapidly converted to MS are consistent with the inability of CD4+ T cells to regulate the transition from quiescence to activation.
Results
Gene expression microarray analysis was performed in negatively isolated naïve CD4+ T cells obtained from 37 CIS patients after initial clinical presentation (mean 4.5 ± 2.6 months) and from 29 controls matched for age and gender. Four arrays failed to pass our quality control protocol and thus were excluded from further analysis. Demographic characteristics of the remaining patients (n = 34) and controls (n = 28) were similar (Table 1). Analysis was focused on the 1,718 probe sets that showed at least a 2-fold change from each gene's median value in >20% of the samples.
Table 1.
Characteristics of subjects at baseline
| Characteristic | CIS (n = 34) | Control (n = 28) | P |
|---|---|---|---|
| Age, y (SD) | 37 (10) | 35 (11) | 0.36 |
| Female, no. (%) | 25 (74%) | 18 (64%) | 0.43 |
| Whites, no. (%) | 31 (91%) | 26 (93%) | 0.81 |
| HLA-DRB1*1501 positive, n (%) | 13 (39%) | 6 (21%) | 0.15 |
Principal component analysis (PCA), using expression values from these 1718 probe sets showed a clear segregation between controls and CIS samples (Fig. 1A). Furthermore, a hierarchical clustering of all samples based on the expression of the same probe sets discriminated CIS from controls with high accuracy (only 3 samples were misclassified: 2 controls and 1 patient) (Fig. 1B). The robustness index for this classification was 99.8% (see Material and Methods). PCA and hierarchical clustering were performed with the 1,718 probe sets with the highest variance across all samples but without any prior statistical testing for differences between cases and controls. When subjected to a t test, 975 probe sets were found to be differentially expressed in CIS and controls after correction for multiple comparisons (false discovery rate (FDR) < 0.1] [supporting information (SI) Table S1]. Interestingly, most of the discriminating transcripts (70%) were underexpressed in CIS, whereas the remaining 30% were overexpressed. This finding is in agreement with previous observations that down-regulated genes greatly outnumber up-regulated genes in T lymphocytes from MS patients when studied by gene expression microarrays (6, 7) or by FACS (8).
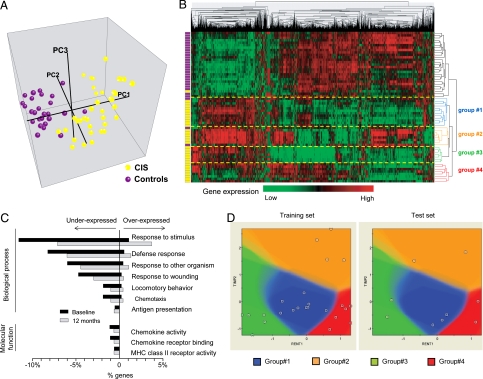
Fig. 1.
Molecular signature in CD4+ cells segregates CIS patients from controls. (A) Three-dimensional plot of the first 3 principal components (PC1, PC2, and PC3) computed from expression values of the 1718 genes with the highest variance (see text). Each dot represents a sample: purple = control; yellow = CIS. (B) Hierarchical clustering of expression values from the same genes and samples as in A. The colored bars on the left indicate the class of each sample: purple = control; yellow = CIS. Distances between genes (or samples) were computed by using Pearson correlation and average linkage. The length of each dendrogram arm is proportional to the computed distance. The four groups of CIS corresponding to the first four splits of the dendrogram (groups 1, 2, 3, and 4) are indicated by dashed yellow lines. (C) Gene ontology categories significantly enriched in CIS patients at baseline. The percentage of genes in each gene oncology category is plotted for baseline (black bars) and 12 months (gray bars). (D) A representative model for the prediction of the four CIS groups, using the Integrated Bayesian Inference System (IBIS). Among the 1718 genes with high variance, gene pairs predictive of CIS groups were selected (see Materials and Methods). The model shown based on the expression of TIMP2 and RENT1 was created with the training test (left panel) and was applied to the test set (right panel).
Gene ontology (GO) enrichment of these 975 differentially expressed genes revealed alteration of major molecular functions and biological processes (Fig. 1C). Unexpectedly for an autoimmune disease, most genes involved in inflammatory responses, including proinflammatory cytokines, chemokines, integrins, and HLA class II molecules, were down-regulated in CIS individuals (Table S1). One possible explanation for overall reduced transcript abundance is cell death. However, we did not find evidence that this generalized down-regulation was caused by increased apoptosis. Quite the opposite: transcripts coding for the anti-apoptotic molecule BAX were increased by 2-fold, whereas those coding for the proapoptotic molecules BCL-2 and cytochrome C (CYCS) were decreased by 2-fold. Overexpressed genes were involved mostly in protein metabolism (27% of overexpressed genes) and/or nucleotide binding (23% of overexpressed genes). Considered together, these results suggest a decreased inflammatory activity in CD4+ T cells from CIS patients.
On the basis of transcriptional activity, samples from CIS patients segregated into four groups (groups 1, 2, 3, 4) corresponding to the first four splits of the dendrogram (robustness index 99.4%) (Fig. 1B). Furthermore, we investigated the likelihood that this segregation occurred by chance, using the Integrated Bayesian Inference System, a supervised machine-learning approach (9). In short, we measured the accuracy of 2-gene Bayesian models created by using only a “training” set (70% samples) to classify a left-out “test” set (30% samples) into the four defined CIS groups (Fig. 1D). The mean accuracy after 10 randomized splits of the samples for the top 7 gene pairs was >80% (Table S2).
Patients from each of the four CIS transcriptional groups did not differ significantly according to age, gender, ethnic background, time from initial clinical event, or HLA-DRB1*1501 status (Fig. 2A). In contrast, time to conversion into CDMS was significantly shorter in patients from group 1 (Fig. 2A). The proportional Cox-regression hazard ratio for patients in group 1 was 3.5 (95% confidence interval 1.4–8.8, P = 0.008) indicating a much higher risk of MS conversion for these individuals. Gadolinium enhancement on brain MRI shortly after clinical presentation was significantly higher in patients from group 1 than in those from other groups, also indicating a higher disease activity (Fig. 2A). However, only 58% of the patients in group 1 showed gadolinium enhancement within 3 months of diagnosis.
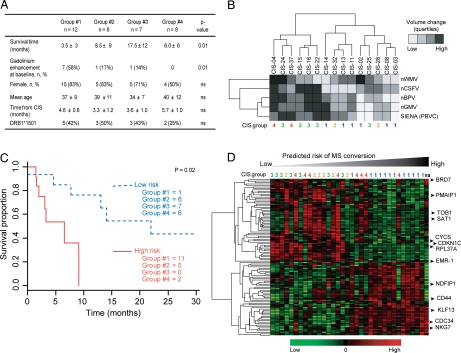
Fig. 2.
Clinical and radiological characteristics of the four CIS groups. (A) Clinical and MRI characteristics of the four CIS groups defined by gene expression: survival time (MS conversion), brain MRI gadolinium enhancement within 3 months of clinical onset, demographic characteristics, time from initial demyelinating event, and HLA-DRB1*1501 status of CIS patients from the four groups. P values indicate significance between groups. (B) Hierarchical clustering of differences (Δ) between measurements at baseline and 12 months later of brain parenchyma (nBPV), gray matter (nGMV), white matter (nWMV), and CSF (nCSFV) volumes, and Structural Image Evaluation, using Normalization, of Atrophy (SIENA) (percentage brain volume change, PBVC) expressed as quartiles. To avoid treatment bias, only patients who had not received disease-modifying therapy after 1 year were used for this analysis. MRI quartiles and patients were clustered by using Euclidian distance and average linkage. The last row indicates the group of samples to which each patient belongs. (C) Kaplan-Meyer curve of high-risk (solid red line) and low-risk (dashed blue line) groups of MS conversion as predicted by the expression of 28 genes. The number of patients from the predefined four groups of CIS is given for each survival group. One patient was not allocated to any group (na). (D) Hierarchical clustering of the 108 genes that characterize patients in group 1. Samples are sorted according to the MS conversion risk as defined in C. Expression-defined CIS groups are shown atop the heatmap. Symbols for genes discussed in the text are indicated by arrowheads.
To evaluate the concordance of gene expression with neurodegeneration, we investigated to what extent changes in brain volume differed across the four CIS groups. To avoid biases related to therapy, only the 15 patients who did not receive disease-modifying therapy during the first year after diagnosis were included in this analysis. Normalized brain parenchyma, white matter, gray matter, and cerebrospinal fluid (CSF) volumes were not significantly different among the four groups at baseline. However, hierarchical clustering of changes after 1 year in these quantitative MRI parameters segregated patients into 2 major groups (Fig. 2B). One group displayed higher volume change (i.e., an increase in CSF and decrease in brain volume), indicating a larger degree of neurodegeneration. The other group was characterized by relatively low change in CSF or brain volume. Interestingly, all group 1 patients were clustered in the latter group (χ2 test, P = 0.01).
To explore further what information about conversion to MS is contained in gene expression at the CIS stage, we built a predictive model of survival (i.e., conversion to MS) based on supervised principal components (10). The resulting model contained 28 genes and allowed a segregation of CIS into high- and low-risk groups (Fig. 2C). The separation remained significant even after adjustment for age, gender, and HLA-DRB1*1501 status (data not shown). Patients segregated into the high-risk group based on their gene expression (red line in Fig. 2C) converted to MS by 9 months of follow-up. Remarkably, 11 of the 12 patients from group 1 were clustered into the high-risk group, thus resulting in a sensitivity of 92% and a specificity of 86%. This study confirms our previous observation that the gene expression–based segregation of group 1 patients is associated with high risk of MS conversion.
Differential gene expression observed shortly after CIS diagnosis may reflect either an acute and transient biological response to the disease and/or a predisposing causative signature. To investigate the significance of this differential gene expression further and to confirm our observations, we collected and processed samples from the same individuals 1 year (mean, 11 ± 3 months) after diagnosis of CIS. For this follow-up, 31 persons who had CIS and 9 controls were available. Hierarchical clustering of these samples performed with the same 1718 genes identified at baseline still discriminated those affected with CIS from controls (Fig. S1A). Among them, differential expression in 461 transcripts was statistically significant, including 270 (59%) of the 975 genes originally found to be differentially expressed at baseline (Fig. S1B). Remarkably, the first split of the samples' dendrogram segregated 100% of the previously identified group 1 patients from the rest of CIS patients and controls (yellow box in Fig. S1A). In contrast, previously identified CIS groups 2, 3, and 4 were no longer detected. To measure the robustness of the group 1 signature over time, we built a support vector machine classifier (with 10-fold leave-one-out cross-validation) with the 108 expression values obtained at baseline (training set) and used those values to predict the status of samples obtained at 12 months (test set). This model classified group 1 samples at baseline with 100% accuracy, positive predictive value, and negative predictive value (Fig. S1C). Although less powerful after 12 months, the same model was able to predict group 1 patients with an accuracy of 86%, a positive predictive value of 78%, and a negative predictive value of 90% (Fig. S1C). Together, these results suggest that the molecular signature found in naïve CD4+ T cells of group 1 CIS patients is stable for at least 1 year.
Because the group 1 signature persists over time, we focused on this group, which also seems to constitute a relatively consistent biological entity. Among the 975 genes whose expression differentiates CIS from controls at baseline, 108 also were differentially expressed between group 1 and the other CIS groups combined (Fig. 2D, Table S3). With the exception of a ribosomal protein (RPL37A), the most underexpressed gene in group 1 patients was transducer of ERBB-2, 1 (TOB1) a finding confirmed by RT-PCR (Fig. 3A). TOB1 is a member of the anti-proliferative APRO family and has been shown to repress T cell proliferation (11). We observed a strong down-regulation of TOB1 on in vitro activation of peripheral-blood CD4+ T cells from control individuals (n = 3, Fig. 3B), in accordance with previous studies (12). To examine whether down-regulation of TOB1 is associated with T cell proliferation in vivo, we immunized C57/Bl6 mice with either MOG35–55 or complete Freund's adjuvant (CFA) and investigated TOB1 protein levels in the lymph nodes by immunofluorescence. In agreement with our molecular and in vitro data, TOB1 immunostaining was decreased in both groups 3 days after immunization, whereas higher levels of the protein were detected in the lymph nodes of naïve mice (Fig. 3C). These results suggest that TOB1 down-regulation can be detected as T cells proliferate in response to either a specific (MOG peptide) or unspecific (CFA) antigenic stimulus.
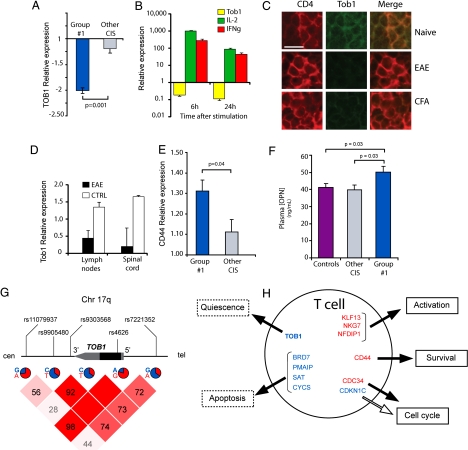
Fig. 3.
TOB1 abrogates T cell quiescence. (A) Relative expression (fold change compared with controls) of TOB1 in CIS patients from group 1 and CIS patients from other groups assessed by RT-PCR. (B) Relative expression of TOB1 (yellow bars), IL-2 (green bars), and IFN-γ (red bars) assessed by RT-PCR in CD4+ T cells cultured for 6 or 24 h in plates coated with 1 μg/μl anti-CD3 and 1 μg/μl anti-CD28 antibodies (n = 3). (C) Immunostaining for TOB1 and CD4 in lymph nodes of mice injected with MOG35–55 (EAE), CFA alone, or vehicle (Naïve). Lymph nodes were dissected and stained 3 days after immunization. (D) Microarray-based TOB1 expression in lymph nodes and spinal cords from mice immunized with MOG35–55 plus adjuvant (EAE) or adjuvant only (CFA). Data were mined from refs. 13 and 14. (E) Relative expression (fold change compared with controls) of CD44 in CIS patients from group 1 and CIS patients from other groups assessed by RT-PCR. (F) Plasma OPN concentration in group 1 patients, other CIS patients, and controls measured by ELISA. (G) Genomic map of TOB1 showing the relative position of the 5 SNPs used for association analysis. The frequency of the major and minor alleles is indicated as a pie chart below each marker. The Haploview matrix indicating pairwise D′ among all markers is shown below. (H) Schematic representation of gene expression signature in T cells from group 1 patients. Genes involved in quiescence, apoptosis, cell cycle, or related to T-cell activation are shown. Red = overexpressed; blue = underexpressed. Solid arrows represent induction; open arrows represent repression. Error bars represent SD.
C57/Bl6 mice injected with MOG35–55 reproducibly develop experimental autoimmune encephalomyelitis (EAE), a widely used laboratory model for MS. In contrast to the sustained inflammation endured by animals with EAE, the response to CFA is expected to be transient, and we hypothesized that patterns of Tob1 expression should reflect this difference. To test this hypothesis, we searched our database from two previous experiments in which we measured high-throughput gene expression in lymph nodes and spinal cords at the peak of EAE (13, 14). As expected, Tob1 mRNA expression was decreased in both lymph nodes and spinal cords of EAE animals compared with CFA controls (Fig. 3D).
In addition to intramolecular mechanisms, engagement of transmembrane receptors may contribute in regulating T-cell homeostasis. Among the 3 differentially expressed genes coding for transmembrane receptors (SIGLEC10, EMR1, and CD44), only CD44 was overexpressed in CIS patients (Table S3). This up-regulation was confirmed by quantitative RT-PCR (Fig. 3E). Of interest, CD44 serves as a receptor for osteopontin (OPN or SPP1), a pleiotropic molecule that has been shown to be highly expressed in both MS plaques and EAE lesions (15). OPN acts as a key promoter of disease severity in EAE by directing the differentiation and survival of T-helper type 1 cells (16). Plasma OPN levels were significantly higher in CIS group 1 patients than in other CIS groups or in controls (Fig. 3F).
Because CIS patients classified as group 1 converted to CDMS earlier than other CIS patients, we hypothesized that TOB1 also is implicated in the progression of disease once established. A genetic effect then would be expected in CDMS patients showing extreme phenotypes (mild or severe). To test this hypothesis, we genotyped 5 single-nucleotide polymorphisms (SNPs) located within or near the gene (Fig. 4G) in individuals selected from a cohort of >1200 patients who had relapsing/remitting MS classified clinically as either “mild” [Expanded Disability Status Scale (EDSS) score < 3 15 years after onset, n = 62] or “severe” (EDSS score > 6 10 years after onset, n = 74). Allelic frequencies were analyzed by logistic regression and case-control association. Differences in allelic frequencies for marker rs4626 (coding, synonymous) between mild and severe cases were statistically significant by logistic regression for both genotype and trend tests (marker rs7221352 also showed both effects, although without reaching statistical significance). The same 2 markers showed statistical significance in the allele case-control (mild versus severe) analysis (Table S4). Haplotype analysis with these two markers also showed statistical significance for the associated markers but not for the neutral alleles (G-A: exact P value = 0.0115, A-G: exact P value = 0.0353). Together, these data suggest that, although TOB1 down-regulation identifies CIS patients at higher risk of conversion to CDMS, there also is a genetic association between markers in this gene and the clinical progression of CDMS patients.
Discussion
Here, we show that early molecular changes in circulating CD4+ T cells differentiate CIS patients from healthy controls and correlate with disease progression. The molecular signature detected at presentation identifies a group of CIS patients at high risk of MS conversion with high accuracy. Although MRI assessment is used routinely to monitor and forecast conversion into MS, its specificity remains moderate (3). The utility and predictive value of oligoclonal bands in the CSF (17) and anti-myelin antibodies (18) remain controversial (19). Thus, in the absence of reliable biological markers associated with MS conversion in CIS patients, our results support the development of potentially valuable tools for personalized disease management. Although the persistence of this molecular signature over time suggests that it is a robust marker, we acknowledge that independent confirmation is needed.
Overall, the CIS group was characterized by a profound down-regulation of the inflammatory transcriptome. This counterintuitive observation may reflect a physiological response by the adaptive immune response to restore immunological homeostasis and to control incipient pathogenic autoimmunity. A similar mechanism may operate in type 1 diabetes (20). Regulation of T cell quiescence may play a critical role in maximizing the ability of adaptive immune responses to defend the host while maintaining immunologic tolerance to prevent autoimmunity (21). We hypothesize that in CIS patients the down-regulation of genes involved in proinflammatory responses may reflect the activity of physiological pathways to keep naïve T cells quiescent.
Several lines of evidence suggest that quiescence of naive T cells is tightly regulated by the activity of transcription factors (21). In particular, TOB1, a transcriptional repressor, has been shown to play a crucial role in keeping naïve T cells from proliferating (11). TOB1 interacts with homologs of the mothers against decapentaplegic, drosophila (SMAD) proteins, a family of transcription factors downstream of the TGF-β family receptor. A critical function of TOB1–SMAD complexes is to suppress IL-2 gene transcription directly and ultimately inhibit T-cell proliferation (12). In vitro, TOB1 transcriptional inhibition has been shown to be sufficient to decrease the threshold of T cell receptor (TCR) activation (11). Decreased expression of TOB1 in group 1 CIS patients suggests that one of the early events facilitating conversion to MS is a release of the brake posed by TOB1 on the activation and proliferation of naïve T cells. Supporting this hypothesis, patients from CIS group 1 showed decreased expression of the cell-cycle inhibitor CDKCN1 (also called “p57” and “kip2”) (22) and increased expression of CDC34, which promotes cell-cycle progression (23). In addition, expression of several key proapoptotic genes (CYCS, SAT, PMAIP1, BRD7) also was decreased in group 1 CIS patients, consistent with a pro-cell survival transcriptional profile. In an independent study that highlights the central importance of some ubiquitous molecules, the anti-apoptotic gene alpha B-crystallin (CRYAB) recently has been found to be highly expressed in MS brain lesions and has been shown to act as a brake on several inflammatory pathways in both the immune system and the central nervous system (24).
Decreased expression of TOB1 in group 1 CIS patients may reflect either a genetic modifier for the disease or a defect in other mechanisms involved in maintenance of T-cell quiescence. The genetic association of two markers with the dicothomous trait “benign” or “severe” suggests that, at least in part, TOB1 is responsible for disease progression. Although a direct relationship between genotypic markers and TOB1 expression was not demonstrated in this study, our results warrant further studies in this direction.
A recent study in EAE shows that Opn promotes progression of EAE through increased proliferation and survival of T lymphocytes (16). The authors show that this effect is associated with the phosphorylation of a quiescence factor, Foxoa3, induced by Opn (21). The role of OPN in MS has been supported by its increased expression in demyelinating lesions and plasma levels in patients who have active MS (25). In CIS patients at high risk of conversion to MS, increased plasma OPN concentration and the increased expression of its receptor (CD44) in T cells may be functionally associated with TOB1 down-regulation.
In conclusion, we identified a gene-expression signature implicated in the regulation of the quiescence of CD4+ T cells that characterizes patients at high risk of MS conversion after a first neurological event. Our results highlight TOB1 deregulation as one of the earliest events involved in individuals who convert rapidly to MS. Targeting pathways that regulate T cell quiescence may represent an interesting therapeutic strategy for preventing MS progression.
Materials and Methods
Patients and Samples.
The study cohort consisted of 37 untreated CIS patients and 29 healthy control subjects matched for age and sex, evaluated at the University of California, San Francisco Multiple Sclerosis Center. CIS patients were identified as subjects presenting with a first, well defined neurological event persisting for >48 h involving the optic nerve, brain parenchyma, brainstem, cerebellum, or spinal cord. All CIS patients demonstrated at least two abnormalities measuring >3 mm2 on brain MRI. Patients were followed for an average of 20 (± 8) months. Time to conversion was defined as the delay between recruitment and next clinical event or the date of identified MRI changes fulfilling the McDonald criteria (5). Written informed consent was obtained from all study participants.
MRI.
MRI scans for all subjects were acquired on a 1.5-T MRI scanner (GE) with a standard head coil. All CIS subjects were scanned every 3 months during the first year of follow-up and then every 6 months during the second year. T2 hyperintense lesions were identified on simultaneously viewed T2- and proton density-weighted dual echo images (1 mm × 1 mm × 3 mm pixels, interleaved slices, 20 ms and 80 ms echo times) with regions of interest drawn based on a semiautomated threshold with manual editing as described elsewhere (26). Annual percentage brain volume change was calculated from high-resolution 3D T1-weighted spoiled gradient recalled echo volumes (pixel size 1 mm × 1 mm × 1.5 mm, 124 slices, flip angle 40°), using Structural Image Evaluation, using Normalization, of Atrophy (SIENA) (27).
RNA Preparation and Hybridization.
Blood samples were collected at the time of recruitment into the study (baseline) and after 12 months. Peripheral blood mononuclear cells were separated in a ficoll gradient and frozen in liquid nitrogen until needed. Naïve CD4+ T cells were isolated by negative selection, using Dynabeads (Invitrogen). CD4+ T cells purity was assessed by FACS (> 95%, data not shown). RNA then was extracted by using RNeasy Mini kit (Quiagen), amplified with MessageAmp II aRNA kit (Ambion), and labeled with Bio-11-UTP for subsequent hybridization onto Affymetrix Human Genome U133 Plus2.0 arrays (TGEN).
Statistical Analysis.
Quality control analysis of the arrays was performed by using the Bioconductor package. To pass quality control, arrays had to have at least 40% of their probe sets called present and had to have similar RNA degradation slopes, GAPDH and beta-actin ratios, scaling factors, histograms, and box plot of intensities. Arrays were normalized by using robust multi-array normalization (RMA) (28). Statistical analyses were carried out by using BRB-array Tools (Biometrics Research Branch). For multiple comparison correction, genes were considered differentially expressed if the univariate P value was <0.001 and the FDR was < 0.1 (29). Genes predicting MS conversion were determined by using the Survival Analysis Prediction Tool of BRB-array Tools. The two survival risk groups were built by using PCA with a P value set at 0.001 for univariably correlated genes with survival and leave-one-out cross-validation (10). For cases with risk greater or lower than the average risk (50th percentile), Kaplan–Meier survival curves were used. Hierarchical clustering was performed by using Genes@work software (IBM Research). To gauge robustness in the classification, we perturbed the dataset by adding random (white) Gaussian noise, using the median variance of the dataset, and reclustered the samples 100 times. The index of robustness is the mean percentage of the times a pair of samples remained in the same cluster. To investigate the likelihood that segregation into four groups occurred by chance, we used the Integrated Bayesian Inference System, a supervised machine-learning approach (9).
Quantitative RT-PCR.
A master mix was prepared, essentially as described (9), with the addition of 200 μM ROX (Sigma) and overlaid on top of each well of a freshly thawed 384-well plate containing 5 ng of RNA in each well. Reactions were performed in triplicates, using an ABI 7900 Sequence Detection System (Applied Biosystems).
Immunofluorescence and ELISA.
Draining lymph nodes from either naive or injected (MOG35–55 or CFA alone) C57/Bl6 mice were removed, washed in PBS, and then embedded in optimal cutting temperature (OCT) compound and frozen. Sections were cut at 6 μm on a cryostat and stained for immunofluorescence examination, using either a rabbit anti-TOB1 polyclonal antibody (H-70, Santa Cruz Biotechnology) or a purified rat anti CD4 antibody (BD PharMingen). Secondary antibodies were anti-rabbit Alexa Fluor 488 (Molecular Probes) and anti-rat Alexa Fluor 594 (Molecular Probes). ELISAs for OPN were carried out by using the Quantikine kit (R&D Systems) according to manufacturer's instructions.
Genotyping of TOB1 SNPs.
Five SNPs located within or near TOB1 were selected for genotyping in 62 patients who had mild MS and in 74 patients who had severe MS. Mild disease was defined as an EDSS score < 3 after 15 years of onset. Severe disease was defined as an EDSS score > 6 after 10 years of onset. Genotyping assays were carried out in 384-well plates, using TaqMan Universal PCR Master Mix on an ABI GeneAmp PCR System 7900 (Applied Biosystems). Statistical tests were carried out in SAS and Jmp Genomics suite (SAS). For haplotype analysis, exact P values were calculated by using the expectation-maximization algorithm in a Monte Carlo approach with 10,000 permutations.
Supplementary Material
Acknowledgments.
We are grateful to the patients and controls for participating in the study. This work was supported by the National Multiple Sclerosis Society (NMSS) Collaborative Research Center (CA) Awards 1035-A-7 (to J.R.O., D.P., and S.E.B.), RG 3240-A-1 (to R.G.H.), and JF 2122-A-1 (to D.P.). J.-C.C. is the recipient of fellowship awards from Leem Recherche, Lilly Institute, French Ministry of Foreign Affairs (Lavoisier Program), and the Foundation Bettencourt Schueller. D.P. is a Harry Weaver Neuroscience Scholar of the NMSS.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805065105/DCSupplemental.
References
- 1.Hauser SL, Goodin DS. In: Harrison's Principles in Internal Medicine. Braunwald E, et al., editors. New York: McGraw-Hill; 2005. pp. 2461–2471. [Google Scholar]
- 2.Kappos L, et al. Treatment with interferon beta-1b delays conversion to clinically definite and McDonald MS in patients with clinically isolated syndromes. Neurology. 2006;67:1242–1249. doi: 10.1212/01.wnl.0000237641.33768.8d. [DOI] [PubMed] [Google Scholar]
- 3.Korteweg T, et al. MRI criteria for dissemination in space in patients with clinically isolated syndromes: A multicentre follow-up study. Lancet Neurol. 2006;5:221–227. doi: 10.1016/S1474-4422(06)70353-2. [DOI] [PubMed] [Google Scholar]
- 4.Brex PA, et al. A longitudinal study of abnormalities on MRI and disability from multiple sclerosis. N Engl J Med. 2002;346:158–164. doi: 10.1056/NEJMoa011341. [DOI] [PubMed] [Google Scholar]
- 5.McDonald WI, et al. Recommended diagnostic criteria for multiple sclerosis: Guidelines from the International Panel on the Diagnosis of Multiple Sclerosis. Ann Neurol. 2001;50:121–127. doi: 10.1002/ana.1032. [DOI] [PubMed] [Google Scholar]
- 6.Satoh J, et al. Microarray analysis identifies an aberrant expression of apoptosis and DNA damage-regulatory genes in multiple sclerosis. Neurobiol Dis. 2005;18:537–550. doi: 10.1016/j.nbd.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Satoh J, et al. T cell gene expression profiling identifies distinct subgroups of Japanese multiple sclerosis patients. J Neuroimmunol. 2006;174:108–118. doi: 10.1016/j.jneuroim.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Kantor AB, et al. Identification of short-term pharmacodynamic effects of interferon-beta-1a in multiple sclerosis subjects with broad-based phenotypic profiling. J Neuroimmunol. 2007;188:103–116. doi: 10.1016/j.jneuroim.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Baranzini SE, et al. Transcription-based prediction of response to IFNbeta using supervised computational methods. PLoS Biol. 2005;3:e2. doi: 10.1371/journal.pbio.0030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004;2:E108. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tzachanis D, et al. Tob is a negative regulator of activation that is expressed in anergic and quiescent T cells. Nat Immunol. 2001;2:1174–1182. doi: 10.1038/ni730. [DOI] [PubMed] [Google Scholar]
- 12.Yusuf I, Fruman DA. Regulation of quiescence in lymphocytes. Trends Immunol. 2003;24:380–386. doi: 10.1016/s1471-4906(03)00141-8. [DOI] [PubMed] [Google Scholar]
- 13.Baranzini SE, Bernard CC, Oksenberg JR. Modular transcriptional activity characterizes the initiation and progression of autoimmune encephalomyelitis. J Immunol. 2005;174:7412–7422. doi: 10.4049/jimmunol.174.11.7412. [DOI] [PubMed] [Google Scholar]
- 14.Otaegui D, et al. Increased transcriptional activity of milk-related genes following the active phase of experimental autoimmune encephalomyelitis and multiple sclerosis. J Immunol. 2007;179:4074–4082. doi: 10.4049/jimmunol.179.6.4074. [DOI] [PubMed] [Google Scholar]
- 15.Chabas D, et al. The influence of the proinflammatory cytokine, osteopontin, on autoimmune demyelinating disease. Science. 2001;294:1731–1735. doi: 10.1126/science.1062960. [DOI] [PubMed] [Google Scholar]
- 16.Hur EM, et al. Osteopontin-induced relapse and progression of autoimmune brain disease through enhanced survival of activated T cells. Nat Immunol. 2007;8:74–83. doi: 10.1038/ni1415. [DOI] [PubMed] [Google Scholar]
- 17.Tintoré M, et al. Isolated demyelinating syndromes: Comparison of CSF oligoclonal bands and different MR imaging criteria to predict conversion to CDMS. Mult Scler. 2001;7:359–363. doi: 10.1177/135245850100700603. [DOI] [PubMed] [Google Scholar]
- 18.Berger T, et al. Antimyelin antibodies as a predictor of clinically definite multiple sclerosis after a first demyelinating event. N Engl J Med. 2003;349:139–145. doi: 10.1056/NEJMoa022328. [DOI] [PubMed] [Google Scholar]
- 19.Kuhle J, et al. Lack of association between antimyelin antibodies and progression to multiple sclerosis. N Engl J Med. 2007;356:371–378. doi: 10.1056/NEJMoa063602. [DOI] [PubMed] [Google Scholar]
- 20.Orban T, et al. Reduced CD4+ T-cell-specific gene expression in human type 1 diabetes mellitus. J Autoimmun. 2007;28:177–187. doi: 10.1016/j.jaut.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 21.Tzachanis D, Lafuente EM, Li L, Boussiotis VA. Intrinsic and extrinsic regulation of T lymphocyte quiescence. Leuk Lymphoma. 2004;45:1959–1967. doi: 10.1080/1042819042000219494. [DOI] [PubMed] [Google Scholar]
- 22.Matsuoka S, et al. p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev. 1995;9:650–662. doi: 10.1101/gad.9.6.650. [DOI] [PubMed] [Google Scholar]
- 23.Plon SE, Leppig KA, Do HN, Groudine M. Cloning of the human homolog of the CDC34 cell cycle gene by complementation in yeast. Proc Natl Acad Sci USA. 1993;90:10484–10488. doi: 10.1073/pnas.90.22.10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ousman SS, et al. Protective and therapeutic role for alphaB-crystallin in autoimmune demyelination. Nature. 2007;448:474–479. doi: 10.1038/nature05935. [DOI] [PubMed] [Google Scholar]
- 25.Vogt MH, Lopatinskaya L, Smits M, Polman CH, Nagelkerken L. Elevated osteopontin levels in active relapsing-remitting multiple sclerosis. Ann Neurol. 2003;53:819–822. doi: 10.1002/ana.10606. [DOI] [PubMed] [Google Scholar]
- 26.Blum D, et al. Dissociating perceptual and conceptual implicit memory in multiple sclerosis patients. Brain Cogn. 2002;50:51–61. doi: 10.1016/s0278-2626(02)00009-x. [DOI] [PubMed] [Google Scholar]
- 27.Smith SM, De Stefano N, Jenkinson M, Matthews PM. Normalized accurate measurement of longitudinal brain change. J Comput Assist Tomogr. 2001;25:466–475. doi: 10.1097/00004728-200105000-00022. [DOI] [PubMed] [Google Scholar]
- 28.Irizarry RA, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Roy Stat Soc. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.