Abstract
The final step in glycosylphosphatidylinositol (GPI) anchoring of cell surface proteins consists of a transamidation reaction in which preassembled GPI donors are substituted for C-terminal signal sequences in nascent polypeptides. In previous studies we described a human K562 cell mutant, termed class K, that accumulates fully assembled GPI units but is unable to transfer them to N-terminally processed proproteins. In further work we showed that, unlike wild-type microsomes, microsomes from these cells are unable to support C-terminal interaction of proproteins with the small nucleophiles hydrazine or hydroxylamine, and that the cells thus are defective in transamidation. In this study, using a modified recombinant vaccinia transient transfection system in conjunction with a composite cDNA prepared by 5′ extension of an existing GenBank sequence, we found that the genetic element affected in these cells corresponds to the human homolog of yGPI8, a gene affected in a yeast mutant strain exhibiting similar accumulation of GPI donors without transfer. hGPI8 gives rise to mRNAs of 1.6 and 1.9 kb, both encoding a protein of 395 amino acids that varies in cells with their ability to couple GPIs to proteins. The gene spans ≈25 kb of DNA on chromosome 1. Reconstitution of class K cells with hGPI8 abolishes their accumulation of GPI precursors and restores C-terminal processing of GPI-anchored proteins. Also, hGPI8 restores the ability of microsomes from the mutant cells to yield an active carbonyl in the presence of a proprotein which is considered to be an intermediate in catalysis by a transamidase.
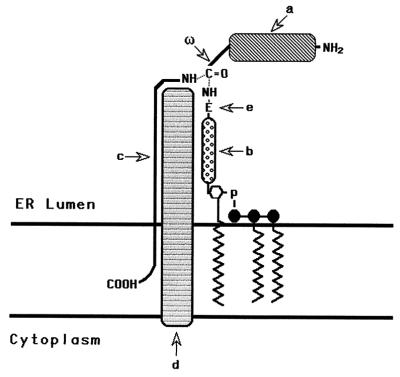
Glycosylphosphatidylinositol (GPI) anchors are posttranslationally added units that serve as general structures for attaching cell surface molecules to the plasma membrane (reviewed in refs. 1 and 2). They link proteins with a wide range of functions and are ubiquitously utilized by all eukaryotic cell types that have been studied to date. They are synthesized in the endoplasmic reticulum (ER) by sequential addition of glucosamine, mannose, and ethanolamine phosphate to phosphatidylinositol (PI). Once preassembled and appropriately positioned for protein transfer, they enter with N-terminally processed polypeptides into a concerted reaction (Fig. 1) in which they replace C-terminal peptide sequences at specific upstream (ω) sites (3, 4). Several lines of evidence (5, 6) have established that this final transfer step consists of a transamidation.
Figure 1.
Schematic representation of the GPI transfer reaction. The GPI transamidase catalyzes substitution of the GPI donor for the C-terminal peptide at the ω site in the N-terminally processed proprotein. a, Proprotein; ω, site of cleavage and GPI addition to the proprotein; b, carbohydrate portion of the GPI donor; c, C-terminal signal peptide; d, transamidase; e, terminal ethanolamine group of the GPI.
In previous reports (7, 8), we described a human K562 cell mutant initially termed IVEE and subsequently assigned to GPI complementation class K. This mutant synthesizes all of the known intermediates of the GPI fabrication process including the fully assembled putative mammalian GPI donors H7 and H8 (9, 10) but fails to incorporate GPI anchors into nascent polypeptides. Using microsomes from these cells in conjunction with an in vitro translation system employing cDNA encoding miniPLAP, an engineered form of GPI-anchored placental alkaline phosphatase (PLAP), we showed that these cells normally produce N-terminally processed prominiPLAP (8). However, unlike all other mutants affected at upstream GPI assembly steps, they show no detectable C-terminally processed mature miniPLAP, irrespective of reaction conditions (11). We further showed (8) that in distinction to other described GPI anchoring mutants, they fail to support C-terminal signal peptide replacement by hydrazine (HDZ) or hydroxylamine, alternative nucleophiles that can substitute for GPI donors (6). This latter finding excluded a compartmentalization defect affecting accessibility of the GPI donor to nascent polypeptides and thereby provided evidence that the lesion in mutant K cells resides in the transamidase itself or an associated factor essential for its enzymatic activity.
In the present study, we report on the identification of the genetic element affected in mutant K cells.¶ For this work, we employed a modified transient expression system based on the use of recombinant T7 RNA polymerase-containing vaccinia virus (12) in conjunction with T7-driven cDNA encoding a GPI-anchored reporter. This reporter, CD8⋅DAF (DAF, decay-accelerating factor), had been prepared previously (13) by appending DAF’s 3′ end-sequence to the extracellular domains of human lymphocyte CD8. The identification and characterization of the gene affected in class K cells is not only important regarding ER biology, but could have clinical relevance, in that comparative studies of C-terminal peptide processing signals in mammalian cells and trypanosomes (14) have indicated that the transamidase recognition sites in these organisms differ, making the transamidase a potential enzyme for therapeutic targeting.
METHODS
Cloning of Upstream and Downstream Human GPI8 (hGPI8) Sequence.
The upstream hGPI8 sequence was PCR-cloned using a HeLa cell library in pCDNA3 vector (Invitrogen). The vector-based T7 forward primer and an hGPI8-specific reverse primer P1 (cgatatgactagcggccacgctgcc) (0.5 μM each) were used in conjunction with 0.2 μg of library cDNA and 2.5 units of Taq polymerase in 10 mM Tris⋅HCl, pH 9.0/50 mM KCl/0.1% Triton X-100/2 mM MgCl2/200 μM dNTPs. After denaturation at 95°C for 3 min, the reaction mixture was amplified by 35 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C, followed by a final cycle of 72°C for 7 min. PCR-amplified products were subcloned into pGEM-T vector (Promega). The downstream GPI8 sequence was cloned by screening a JY-1 cell cDNA library in Charon BS(−) vector (American Type Culture Collection) with the full-length coding region as a probe. Duplicate plaque lifts on colony/plaque screen membranes were hybridized overnight at 43°C in 50% formamide/2× Pipes (0.4 M NaCl/10 mM Pipes, pH 6.5)/1% SDS containing 0.1 mg/ml denatured sonicated salmon sperm DNA. Hybridized transfers were rinsed twice at 55°C in 2× standard saline citrate/0.1% SDS. Positive clones in Bluescript were released with NotI and, after ligation, were transformed with DH5α. Inserts in pGEM-T or Bluescript were sequenced by the dideoxy termination method using a Sequenase version 2.0 kit (Amersham).
Vaccinia Transient Transfection System.
After overnight incubation in a 175-ml flask at 5–8 × 105/ml, cells were readjusted to 5 × 106/ml, mixed at a multiplicity of infection of 1 virus VTF7.3 per cell in RPMI 1640 medium containing 2% newborn calf serum, and infected for 1 h with shaking at 15-min intervals. The cells then were washed in serum-free RPMI 1640 medium, adjusted to 1 × 106 cells in 0.8 ml of Hepes-buffered saline (HeBS) transfection buffer, and 30 μg of CD8⋅DAF/pcDNA1, 30 μg of salmon sperm DNA, and 10 μg of hGPI8/pcDNA3 were added. The DNA was introduced by electroporation at 250 V and 960 μF in a Gene-Pulser (Bio-Rad). Electroporated cells were cultured in complete RPMI 1640 medium, and after 48 h, cells were stained with anti-CD8 mAb/fluorescein isothiocyanate-anti-mouse Ig and with propidium iodide and were analyzed on a FACStarPlus (Becton Dickinson) flow cytometer. In some experiments, cells were treated for 30 min at 37°C with PI-specific phospholipase C (PI-PLC) in PBS or PBS alone, before staining.
Stable Transfectants.
Linearized (XhoI) hGPI8/pcDNA3 (50 μg) was electroporated into 107 mutant K cells in 0.8 ml of HeBS transfection buffer under conditions identical to those described above. Transfected cells were cultured for 24 h in complete RPMI 1640 medium and G418 (0.8 mg/ml) was then added. After 3 wk of selection, refreshing half of the medium every fourth day, surviving neomycin-resistant (NeoR) cells were stained with murine IF5 (15) anti-CD59 mAb/fluorescein isothiocyanate-anti-mouse Ig and analyzed by flow cytometry.
Northern Blot Analyses.
Poly(A)+ mRNA was purified directly from cells with the Oligotex MiniKit (Qiagen, Chatsworth, CA) with two cycles of column purification. Purified products (2 μg) were electrophoresed on 1% agarose/2.2 M formaldehyde gels and transferred to Nitrobind nitrocellulose membranes (Micron Separations, Westboro, MA). After prehybridization at 42°C for 18 h in 5× SSPE/50% formamide/0.1% SDS/100 μg/ml denatured salmon sperm DNA/5× Denhardt’s solution, membranes were hybridized at 42°C with a 32P-labeled 1.2-kb full-length or 0.6-kb central coding region fragment of hGPI8 in 5× SSPE/50% formamide/0.1% SDS/10% dextran sulfate (Mr > 300,000)/100 μg/ml denatured sonicated salmon sperm DNA/5× Denhardt’s solution. Hybridized membranes were washed at 20°C for 2 h with 1× standard saline citrate containing 0.1% SDS.
Southern Blot Analyses.
Genomic DNA restricted with BamHI, HindIII, EcoRI, PstI, and KpnI and electrophoresed on 0.9% agarose gels was transferred to Zetaprobe nylon membranes (Bio-Rad). After prehybridization for 1 h in 1 mM EDTA/0.25 M Na2HPO4, pH 7.2/7% SDS, transfers were hybridized at 65°C for 18 h with a 32P-labeled 1.2-kb coding region fragment of hGPI8, and the membranes were washed twice at 55°C for 1 h in 2× SSC/1% SDS.
Chromosomal Localization.
Genomic DNA (1 μg) from human/rodent somatic cell hybrids [mutant panel #2, version III mini (Coriell Cell Repository, Camden, NJ)] was amplified using hGPI8 forward primer G1 (tgtgtttctcttgtatgaat, positions 1,282–1,301), and reverse primer G2 (tgataatttaatgtcttcac, positions 1,538–1,519) (0.5 μM each). PCR was performed with conditions identical to those for recovery of upstream hGPI8 sequence.
Biosynthetic Labeling.
Cells (107) were pretreated with tunicamycin and labeled as previously described (7, 8). 3H-labeled mannolipids were resolved on TLC plates developed with chloroform/methanol/water (10:10:3) and were analyzed in a Berthold (Nashua, NH) LB285 linear analyzer.
Preparation of Microsomes and Cotranslational Processing Studies of MiniPLAP.
Rough microsomal membranes were prepared as described (16). Briefly pelleted and washed cells, resuspended in 10 mM triethanolamine (TEA), pH 7.5/250 mM sucrose, were disrupted for 90 min in a N2 cavitation bomb charged to 1,200–1,500 psi. The supernatant of the low speed pellet of the resulting cell lysate in 10 mM TEA was recentrifuged for 90 min at 78,000 × g in a Ti70 rotor, and the rough microsomal membranes-containing pellet was resuspended in 250 mM sucrose/50 mM TEA, pH 7.5 and stored at −70°C.
MiniPLAP mRNA was prepared with HindIII-linearized pGEM4Z priming with SP6 RNA polymerase. In vitro translation of miniPLAP mRNA was performed with rabbit reticulocyte lysate. Cotranslational processing assays were carried out as described (16). Typically, reactions were conducted in a 25-μl translation mixture containing 1 μl of 100 ng/μl miniPLAP mRNA, 1.75 μl of [35S]methionine (15 mCi/ml, 1,100 Ci/mmol, Amersham; 1 Ci = 37 GBq), 12.5 μl of (nuclease-treated) rabbit reticulocyte lysate, 0.75 μl of a 1 mM amino acid mix minus methionine, and 0.5 μl of RNase inhibitor at 40 units/μl (all supplied by Promega). To this was added 2.5 μl of buffer A (100 mM KOAc/4 mM MgOAc) containing 20 μg/ml each of the protease inhibitors aprotinin, antipain, bestatin, chymostatin, leupeptin, and pepstatin, and 4 μl of rough microsomal membranes from a stock containing 50 A280 units per ml. The mixture then was incubated at 30°C for the times indicated. Immunoprecipitations were performed with rabbit anti-PLAP antibody (Accurate Chemicals, Westbury, NY) and protein A-Sepharose 4B (Pharmacia), and products were separated on 15% SDS/PAGE gels and analyzed by autoradiography and phosphoimaging.
RESULTS
Modified Vaccinia Transfection System.
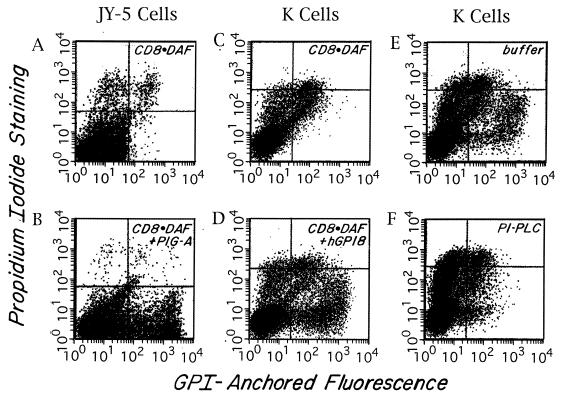
As indicated above, to identify the affected gene in mutant K cells, we utilized a transient expression system employing recombinant Vaccinia virus producing T7 RNA polymerase in conjunction with T7-driven CD8⋅DAF cDNA encoding GPI-anchored CD8 (13) as a reporter. With this system, a directionally oriented cDNA library or other test cDNA inserted downstream of the T7 promoter is cotransfected with the CD8⋅DAF reporter cDNA into virally infected cells (see Methods) and, after 48 h, transfectants are assayed for rescued GPI-anchored CD8 expression. For initial validation of the methodology and optimization of experimental conditions, we first performed studies with GPI-defective JY-5 lymphoblastoid cells (17) mutated in PIG-A (a gene required in the first step of GPI assembly) and PIG-A cDNA (18) in the T7-containing pCDNA1 vector. As shown in Fig. 2 A and B, these control studies verified that introduction of the appropriate rescuing element (i.e., PIG-A cDNA) induced high-level CD8 expression. Further control experiments showed that a 3:1 ratio of CD8⋅DAF cDNA to PIG-A cDNA gave optimal results, and that rescue of up to 20% of the JY-5 cell population was produced reproducibly. The staining was abolished by preincubation of the cells with PI-PLC (data not shown), confirming that GPI anchor assembly in the PIG-A-deficient JY-5 cells had been reconstituted.
Figure 2.
Rescue of GPI anchoring as assessed in the Vaccinia transient transfection system. Two-color FACS (Becton Dickinson) analyses of anti-CD8 and propidium iodide-stained cells. FL1 (x axis) = CD8 fluorescence intensity; FL2 (y axis) = propidium iodide fluorescence. Cells in the upper quadrants are propidium iodide-positive dead cells. Cells in the lower right quadrant are CD8+ and propidium iodide-negative rescued cells. The small number of cells in the lower right quadrant seen with CD8⋅DAF alone also stained with nonrelevant antibody. (A and B) JY-5 cells transfected with CD8⋅DAF alone or CD8⋅DAF and PIG-A cDNA. (C and D) K cells transfected with CD8⋅DAF alone or CD8⋅DAF and hGPI8. (E and F) CD8⋅DAF and hGPI8 transfected transients treated with buffer or PI-PLC. About 70% of the CD8+ cells were cleaved.
Rescue of Mutant K Cells.
Based on the above results, we applied the Vaccinia/CD8⋅DAF system for use with the mutant K line [first establishing optimal virus to cell ratio and electroporation conditions for K562 cells (see Methods)]. In the course of work with library cDNA using the system, we became aware (A. Conzelmann, personal communication) that a partial human homolog of the yeast GPI8 gene that rescued a yeast mutant (gpi-8) exhibiting a phenotype similar to mutant K cells was present in the GenBank database. It initially had been isolated as an unknown sequence as part of the Washington University Merck Expressed Sequence Tag project (accession no. R18975) and subsequently further sequenced by B. Mohamed (University of Fribourg, Fribourg, Switzerland) (accession no. Y07596). Using our pcDNA3 library cDNA, we PCR-cloned this 1.55-kb element, which showed 43% translated homology to yGPI8 but lacked an upstream ATG. By means of further PCR with specific reverse primers and a vector-based forward primer in conjunction with library cDNA (Fig. 3), we recovered a 150-bp segment containing 27 bp of sequence upstream of this GenBank sequence, including a change resulting in an ATG. We prepared a composite sequence using an internal restriction site and inserted the combined sequence into pcDNA3. We then cotransfected this plasmid together with CD8⋅DAF/pcDNA1 into mutant K cells and assayed for CD8 expression. As shown in Fig. 2 C and D, rescue of CD8 expression in mutant K cells was observed with efficiency similar to that obtained with PIG-A cDNA in JY-5 cells (Fig. 2 A and B). Additionally, as shown in Fig. 2 E and F, the expressed CD8 epitope was PI-PLC-sensitive, indicative of restored GPI-anchor assembly.
Figure 3.
Cloning of hGPI8 sequence. The accession uppercase sequence is the newly cloned and corrected sequence and the lowercase sequence was PCR cloned from a GenBank sequence (accession no. Y07596). The bold, underlined, capitalized ATG gives the start codon and the bold, underlined, lowercase tag gives the stop codon. The underlines show the alternative polyadenylation signals.
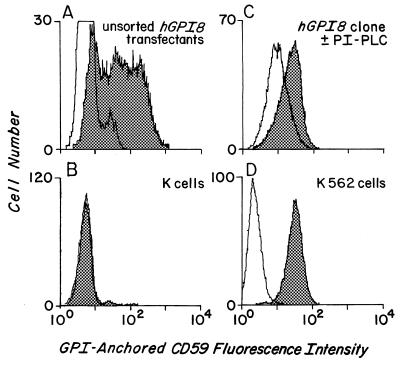
In view of these results, hGPI8/pcNA3 was introduced into mutant K cells in the absence of virus, and stable transfectants were selected with G418. As seen in Fig. 4, flow cytometric assays (i) of the resulting (unsorted) cells (Fig. 4 A and B) showed reconstitution of endogenous CD59 expression and (ii) of a cloned line, surface CD59 levels approaching those in parental K562 cells. PI-PLC digestion analyses (Fig. 4C) showed ≈70% enzyme sensitivity of the expressed CD59 protein, the extent of cleavage that is characteristic of the K562 cell line (8, 9, 13).
Figure 4.
Stable hGPI8/K cell transfectants. The x axis shows CD59 fluorescence, and the y axis shows cell number. (A) Unsorted Neo-resistant stable transfectants. (B) Nontransfected mutant K cells. (C) Cloned transfectant treated with buffer (shaded) or PI-PLC (unshaded) prior to staining. (D) Wild-type K562 cells. Shaded areas represent staining with anti-CD59 mAb and, except for C, unshaded areas represent staining with nonrelevant RPC5 control mAb.
To verify that hGPI8 was affected in mutant K cells, mRNA from K cells was reverse-transcribed, and after PCR using hGPI8-specific primers, the resulting cDNA was sequenced. Deletion of A105T106 resulting in a stop codon (TAG) at position 131 was seen. In addition, a second coding region mutation (deletion of one of six tandem T residues starting at T407) further downstream was also uncovered. Analysis of four independent clones gave identical results.
Biochemical Properties of hGPI8/K Cell Transfectant.
[3H]Man biosynthetic labeling of parental K562, untransfected mutant K cells, and hGPI8/K cell transfectants were performed. Reconstitution with hGPI8 decreased accumulation of the terminally ethanolamine-phosphate-substituted GPI putative donors H7 and H8 and shifted the GPI mannolipid pattern toward that of parental K562 cells (7–9) (data not shown).
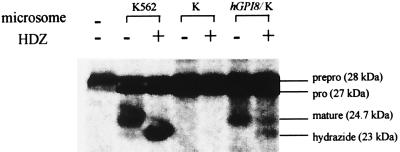
To determine if introduction of hGPI8 enabled mutant K cells to interact with HDZ and thus restored their transamidation capability, we conducted in vitro translation and processing studies with microsomes from the hGPI8/K cell transfectants using the miniPLAP system that permits cell-free assessment of C-terminal transamidation (3, 5, 8, 11, 16). As shown in Fig. 5, microsomes from untransfected mutant K cells supported formation only of the 28-kDa translation product and of 27-kDa N-terminally processed prominiPLAP but failed to further process this product irrespective of the presence of HDZ. In contrast, microsomes of the hGPI8/K cell transfectant showed C-terminal processing (transamidation) of prominiPLAP to 24.7-kDa mature GPI-anchored miniPLAP and, in the presence of HDZ, supported formation of the 23.0-kDa miniPLAP hydrazide. The products were the same as those formed by microsomes from parental K562 cells (8). Although the processing efficiencies were not equal, these experiments were not intended to be quantitative, as the amounts of hGPI8 mRNA and of microsomes were not standardized.
Figure 5.
Effect of hGPI8 on C-terminal processing of miniPLAP. Cotranslational processing of miniPLAP mRNA by microsomes of parental K562, untransfected K, and hGPI8/K transfectants in the absence and presence of HDZ. The microsome preparations were made at different times and fewer microsomes (35 vs. 50 OD) were used in the case of hGPI8/K transfectants. The positions of 28-kDa preprominiPLAP, 27-kDa prominiPLAP, 24.7-kDa GPI-anchored miniPLAP, and the 23.0-kDa miniPLAP hydrazide are shown.
The Human hGPI8 Gene.
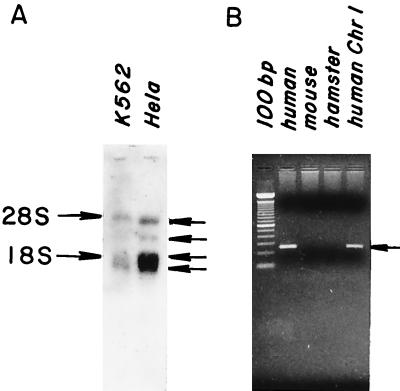
As a first step in further characterizing hGPI8, Northern blot analyses were performed with poly(A)+ mRNA from K562 and HeLa cells. Major (polyadenylated) bands at 1.6 and 1.9 kb, and minor bands at 3.0 and 4.5 kb (Fig. 6A) were seen in both cell types. Screening of a λ cDNA library (American Type Culture Collection) showed that the 1.6- and 1.9-kb transcripts derived from the use of alternative polyadenylation signals (see Fig. 3). All bands were more prominent in HeLa cells that are more active in GPI transfer (8). The 1.9-kb band overlapping 18S rRNA was similar in intensity when total RNA was used instead of poly(A)+ RNA, and the same results were seen with a shorter internal coding region probe (data not shown). In view of the detection of the 3.0- and 4.5-kb RNA bands, several additional PCR amplifications of the pcDNA3 library cDNA and rapid amplification of cDNA ends analyses with reverse-transcribed HeLa mRNA were performed to determine whether more 5′ sequence was present in these RNAs. With either method, no further upstream sequence was isolated. Amplification of the coding region yielded one band and amplifications with a 3′ primer in conjunction with the pcDNA3 library did not yield longer products, leaving the nature of these two minor species unclarified.
Figure 6.
Northern blot analyses and chromosomal localization of hGPI8. (A) Poly(A)+ mRNA probed with 1.2-kb coding segment of hGPI8. (B) DNA from chromosome-specific mouse– or hamster–human hybrids probed with a 257-bp 3′ untranslated region hGPI8 segment.
Southern blot hybridization of human genomic DNA with the 1.2-kb hGPI8 composite cDNA showed bands consistent with a single gene 20–25 kb in length. PCR amplifications with genomic DNA from human/rodent chromosome-specific hybrids (see Methods) localized the gene to chromosome 1 (Fig. 6B).
DISCUSSION
In this study, we cloned missing upstream sequence of a human GenBank element showing 43% translated homology to yGPI8 cDNA. We found that (i) the composite hGPI8 sequence complements GPI anchoring in class K cells, and (ii) endogenous hGPI8 sequence in K cells is mutated, indicating that hGPI8 corresponds to the affected genetic element in these cells. We demonstrated that hGPI8 restores the ability of class K cells to support C-terminal processing by GPI donors and by HDZ and thus corresponds to the GPI transamidase or to an essential component required for activity of its enzymatic site. Finally, we showed that GPI8 mRNA derives from a ≈25-kb gene that resides on chromosome 1.
For cloning of the affected element in the K cell line, we employed a recombinant vaccinia-based transient transfection system producing T7 RNA polymerase. Based on initial work with PIG-A– JY-5 cells and PIG-A cDNA, showing that endogenous CD59 expression was restored but expression levels were low, we utilized cotransfected CD8⋅DAF cDNA encoding an exogenous GPI-anchored reporter. This strategy was adopted to place expression of both the gene of interest and the GPI-anchored reporter under control of the T7 promoter. We placed CD8⋅DAF cDNA in T7-driven pcDNA1 containing the SupF dominant marker and used library cDNA or candidate cDNA in T7-driven pcDNA3 containing AmpR and NeoR elements. This allowed for (i) isolation of rescuing DNA from sorted CD8+ cells without the complication of contaminating CD8⋅DAF in work with library cDNA, and (ii) the use of the rescuing cDNA for the production of stable transfectants.
Analysis of endogenous hGPI8 mRNA in class K cells showed two mutations: (i) deletion of an upstream AT resulting in a stop codon 25 bp downstream of this point, and (ii) deletion of one of 6 tandem T residues ≈300 bp further downstream. The presence of two mutations is consistent with the fact that K cells were isolated after N-methyl-N′-nitro-N-nitrosoguanidine mutagenesis (7). Neither mutation created or deleted a restriction site. Although four clones showed the same changes, the nature of mutation(s) on other chromosomes, if present, was not further investigated.
Studies with yeast have shown that at least two gene products are involved in the final C-terminal GPI transamidation step. The first of these genes to be identified, termed GAA1 (20), was isolated from a Saccharomyces cervisiae mutant (gaa1) that initially was noted to be defective in endocytosis. It subsequently was found to synthesize the fully assembled yeast GPI precursor CP2 but, unlike wild-type cells, failed to transfer it to the proform of GPI-anchored gas1p (21). It is a 614-amino acid-long multimembrane-spanning ER protein with a large lumenal domain near its cytoplasmically oriented N terminus. A human homolog of the protein has been identified that has 28% overall but greater amino acid homology in the lumenal domain [N. Inoue and T. Kinoshita, presented at FASEB Summer Research Conference (August 5–9, 1995), Copper Mountain, CO; Y. Hiroi and I. Komuro, personal communication]. The second of these proteins, yGPI8, the human homolog that is the subject of this report, was recently cloned from a mutagenized S. cervisiae clone gpi-8 that exhibited a similar phenotype (22). The yGPI8 gene encodes a 47-kDa 411-amino acid-long type I transmembrane ER protein which has a large N-terminal luminal domain and a short cytosolic C-terminal domain of ≈14 amino acids. The functions of yeast gaa1p and of gpi8p are unknown. The observation (23) that yeast gpi8p has 27.5% identity to a jackbean asparaginyl endopeptidase, which has been found to exhibit transamidase activity in vitro, has suggested that this protein is the transamidase itself. Previously characterized transamidases do not have cofactors nor are they composed of more than one subunit (19, 24).
Our finding that hGPI8 restores the ability of class K microsomes to support nascent protein uptake of GPI and interaction with the potent nucleophile HDZ concomitant with cleavage of the protein at its ω site provides fairly convincing evidence that hGPI8 codes for the GPI transamidase. The development of a reconstituted system with purified proteins (transamidase complex + proprotein + GPI) would establish this firmly.
Acknowledgments
We thank Sara Cechner for manuscript preparation and Dr. Robert Peterson for help with sequence analysis. This investigation was supported by National Institutes of Health Grants P01 DK38181 and R01 AI23598.
ABBREVIATIONS
- GPI
glycosylphosphatidylinositol
- ER
endoplasmic reticulum
- HDZ
hydrazine
- DAF
decay-accelerating factor
- PI
phosphatidylinositol
- PLAP
placental alkaline phosphatase
- PI-PLC
PI-specific phospholipase C
- TEA
triethanolamine
- Neo
neomycin
Note Added in Proof
Following submission of this paper, evidence was presented in a talk by T. Kinoshita at the International Meeting on Interactions of GPI Anchors with Biological Membranes, Sept. 14-17, 1997, that ω site recognition is dependent on gaa1p structure.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF022913).
This work was presented in abstract form at the 17th International Congress of Biochemistry and Molecular Biology (FASEB), Aug. 24–29, 1997, San Francisco.
References
- 1.Englund P T. Annu Rev Biochem. 1993;62:121–138. doi: 10.1146/annurev.bi.62.070193.001005. [DOI] [PubMed] [Google Scholar]
- 2.Stevens V L. Biochem J. 1995;310:361–370. doi: 10.1042/bj3100361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerber L D, Kodukula K, Udenfriend S. J Biol Chem. 1992;267:12168–12173. [PubMed] [Google Scholar]
- 4.Moran P, Raab H, Kohr W J, Caras I W. J Biol Chem. 1991;266:1250–1257. [PubMed] [Google Scholar]
- 5.Maxwell S E, Ramalingam S, Gerber L D, Udenfriend S. Proc Natl Acad Sci USA. 1995;92:1550–1554. doi: 10.1073/pnas.92.5.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maxwell S E, Ramalingam S, Gerber L D, Brink L, Udenfriend S. J Biol Chem. 1995;270:19576–19582. doi: 10.1074/jbc.270.33.19576. [DOI] [PubMed] [Google Scholar]
- 7.Mohney R P, Knez J J, Ravi L, Sevlever D, Rosenberry T L, Hirose S, Medof M E. J Biol Chem. 1994;269:6536–6542. [PubMed] [Google Scholar]
- 8.Chen R, Udenfriend S, Prince G M, Maxwell S E, Ramalingam S, Gerber L D, Knez J, Medof M E. Proc Natl Acad Sci USA. 1996;93:2280–2284. doi: 10.1073/pnas.93.6.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirose S, Prince G M, Sevlever D, Ravi L, Rosenberry T L, Ueda E, Medof M E. J Biol Chem. 1992;267:16968–16974. [PubMed] [Google Scholar]
- 10.Ueda E, Sevlever D, Prince G M, Rosenberry T L, Hirose S, Medof M E. J Biol Chem. 1993;268:9998–10002. [PubMed] [Google Scholar]
- 11.Amthauer R, Kodukula K, Brink L, Udenfriend S. Proc Natl Acad Sci USA. 1992;89:6124–6128. doi: 10.1073/pnas.89.13.6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elroy-Stein O, Moss B. In: Current Protocols in Molecular Biology. Janssen K, editor. Vol. 2. New York: Wiley; 1992. pp. 16.19.1–16.19.9. [Google Scholar]
- 13.Tykocinski M L, Shu H-K, Ayers D J, Walter E I, Getty R R, Groger R K, Hauer C A, Medof M E. Proc Natl Acad Sci USA. 1988;85:3555–3559. doi: 10.1073/pnas.85.10.3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caras I W, Moran P. J Med Biol Res. 1994;27:185–188. [PubMed] [Google Scholar]
- 15.Okada N, Harada R, Fujita T, Okada H. J Immunol. 1989;143:2262–2266. [PubMed] [Google Scholar]
- 16.Kodukula K, Cines D, Amthauer R, Gerber L, Udenfriend S. Proc Natl Acad Sci USA. 1992;89:1350–1353. doi: 10.1073/pnas.89.4.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollander N, Selvaraj P, Springer T P. J Immunol. 1988;141:4283–4290. [PubMed] [Google Scholar]
- 18.Miyata T, Takeda J, Iida Y, Yamada N, Inuoe N, Takahashi M, Maeda K, Kitani T, Kinoshita T. Science. 1993;259:1318–1320. doi: 10.1126/science.7680492. [DOI] [PubMed] [Google Scholar]
- 19.Buchanan J M. Adv Enzymol. 1973;39:91–183. doi: 10.1002/9780470122846.ch2. [DOI] [PubMed] [Google Scholar]
- 20.Hamburger D, Egerton M, Riezman H. J Cell Biol. 1995;129:629–639. doi: 10.1083/jcb.129.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nuoffer C, Jenö P, Conzelmann A, Riezman H. Mol Cell Biol. 1991;11:27–37. doi: 10.1128/mcb.11.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benghezal M, Benachour A, Rusconi S, Aebi M, Conzelmann A. EMBO J. 1996;15:6575–6583. [PMC free article] [PubMed] [Google Scholar]
- 23.Abe Y, Shirane K, Yokosawa H, Matsushita H, Mitta M, Kato J, Ishii S. J Biol Chem. 1993;268:3525–3529. [PubMed] [Google Scholar]
- 24.Tate S S, Meister A. Proc Natl Acad Sci USA. 1974;71:3329–3333. doi: 10.1073/pnas.71.9.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]