Abstract
The penta-subunit retromer complex of yeast mediates selective retrieval of membrane proteins from the prevacuolar endosome to the trans Golgi network. In this study, we set out to generate a panel of vps35 dominant-negative mutants that disrupt retromer-mediated cargo sorting. Mapping of the mutations revealed two types of alterations leading to dominant-negative behavior of the 944-amino acid protein: (i) mutations at or near the R98 residue or (ii) C-terminal truncations exemplified by a nonsense mutation at codon 733. Both could be suppressed by overexpression of wild-type Vps35p, suggesting that these dominant-negative mutants compete for interactions with other retromer subunits. Interestingly, Vps35-R98W expression destabilized Vps26p while having no effect on Vps29p stability, while Vps35-Q733* expression affected Vps29p stability but had no effect on Vps26p. Measurement of Vps35/Vps26 and Vps35/Vps29 pairwise associations by coimmunoprecipitation in the presence or absence of other retromer subunits indicated that the R98 residue, which is part of a conserved PRLYL motif, is critical for Vps35p binding to Vps26p, while both R98 and residues 733–944 are needed for efficient binding to Vps29p.
Keywords: endosome, protein sorting, retromer, Saccharomyces cerevisiae, trans Golgi network, vesicle coat
The carboxypeptidase Y (CPY) receptor Vps10p and Ste13p (dipeptidyl aminopeptidase A) are membrane proteins that traffic between the trans Golgi network (TGN) and endosomal system of yeast. Vps10p is transported to the prevacuolar endosome (PVC) in a manner that is at least partially dependent on clathrin and associated Gga adaptors (1). While TGN-to-PVC trafficking of Vps10p appears to be direct, Ste13p instead is transported from the TGN to the early endosome in a Gga-independent manner (2). Once at the early endosome, Ste13p can be retrieved back to the TGN in a clathrin- and adaptor protein-1-dependent process (2) or can traffic to the PVC. Upon reaching the PVC, both Vps10p and Ste13p, as well as the TGN membrane protein Kex2p, are selectively retrieved back to the TGN in a manner dependent on aromatic sorting signals in their cytosolic domains (3–7).
Retrieval from the PVC of Vps10p and A-ALP (a model protein based on Ste13p) is mediated by an endosomal complex of five subunits termed the retromer: Vps35p, Vps29p, Vps26, Vps5p and Vps17p (8–12). Another sub-unit, sorting nexin-3 (Snx3p/Grd19p), is required for retrieval of A-ALP but not Vps10p (13), and recent data suggest that it acts as cargo-specific adaptor for the retromer (14). Mammalian orthologs of retromer subunits with the exception of Vps17p have been identified and characterized (15,16). Much like its yeast counterpart, the mammalian retromer is required for retrieval of the functional homolog of Vps10p, the cation-independent man-nose 6-phosphate receptor (MPR), from endosomes to the TGN (17,18). In addition, retromer function has been implicated in transcytosis of the polymeric immunoglobulin receptor in polarized epithelial cells (19).
Recent advances have shed light on retromer structure/function relationships. Salt-mediated dissociation of the retromer complex from the cytosolic face of yeast endosomal membranes causes a subcomplex comprised of Vps35p/Vps29p/Vps26p to separate from another sub-complex containing Vps5p and Vps17p (9). Likewise association of the mammalian Vps35p/Vps29p/Vps26p subcomplex with Snx1/Snx2 appears to be relatively weak as it can be detected by two-hybrid assay but not by native immunoprecipitation (20). This suggests that these two subcomplexes have distinct functional roles. The Vps35p/Vps29p/Vps26p subcomplex has a role in cargo selectivity as the Vps35p subunit has been shown by both genetic and biochemical approaches to associate with the retrieval signals of A-ALP and Vps10p (21) and with the cytosolic domain of the MPR in mammalian cells (17). In contrast, the Vps5/Vps17 subcomplex appears to be involved in membrane vesicle or tubule formation. Vps5p, at least in vitro, has the ability to self-assemble into very large complexes (9). Vps5p and its mammalian homolog Snx1 associate with endosomal membranes through phox homology domains that bind to phosphatidylinositol (3) phosphate and contain Bin/Amphiphysin/Rvs domains that have been shown to sense and induce membrane curvature (22–24). The retromer localizes to tubular–vesicular regions of endosomes in both yeast and mammalian cells (9,17,18). Taken together, these data suggest that the retromer functions as a cargo-selective membrane coat that facilitates transport through vesicles or tubules.
Vps35p has a central role in retromer assembly and function. Two-hybrid assays and in vitro pull-down experiments suggest that an N-terminal domain of human Vps35p associates with Vps26p, while a C-terminal region associates with Vps29p (15,25). Both N- and C-terminal regions of human Vps35p associated with Snx1 through the two-hybrid system (15). The structural basis for cargo binding by yeast Vps35p appears to be complex as distinct regions appear to be involved in recognition of different cargo molecules (12). In this study, we describe dominant-negative alleles of Vps35p that interfere with retromer-mediated cargo sorting and destabilize other retromer subunits. Study of the interaction between these mutant Vps35p proteins and other retromer subunits provides new insights into the functional domains of Vps35p.
Results
Identification of mutations that cause dominant-negative phenotypesin Vps35p
In order to identify structural domains within yeast Vps35p, we randomly mutagenized the VPS35 gene and screened for dominant-negative alleles. This was achieved by transforming multicopy, 2μ plasmids containing the randomly mutagenized alleles into a wild-type strain and screening transformants for CPY sorting defects. Comparison of the phenotypes of the 14 clones identified in the screen indicated that they all exhibited marked phenotypes on a 2μ plasmid (Figure 1, right panel), although the dominant-negative phenotypes were less extreme than the sorting defect of a vps35Δ strain carrying an empty vector. Most of the alleles also exhibited discernable phenotypes when carried on low-copy centromeric plasmids (Figure 1, middle panel). No significant effects on yeast cell growth by the dominant-negative alleles were observed (data not shown).
Figure 1. CPY sorting defects in a wild-type strain expressing dominant-negative alleles from both CEN and 2μ plasmids.
A vps35Δ strain (bottom row, middle position) was analyzed as a control. All others were Vps+ strain SNY36-9A expressing wild-type or the indicated dominant-negative versions of VPS35 from either centromeric (middle panel) or 2μ, (right-hand panel) plasmids. Control strains carrying empty parental vectors are indicated (Vector). Each strain was patched in duplicate onto selective media, and CPY colony immunoblots were performed to assess the extent of CPY secretion as described in Materials and Methods.
DNA sequencing of the dominant-negative alleles (Table 1) indicated that two general types of alterations resulted in a dominant-negative phenotype. The first were missense mutations at or nearby the R98 residue as exemplified by the hp5, hp8, hp10, hp12 and hp14 alleles. Interestingly, R98 resides within a PRLYL motif that is 100% conserved between yeast, plant, nematode and mammalian Vps35p (Figure 2). The second type is a truncation of the C-terminal conserved region as exemplified by the hp1, hp3, hp6, hp7, hp9, hp11, hp14and hp16 alleles. The only alteration in the hp11 allele was the Q733* truncation mutation, therefore deletion of a C-terminal region clearly converted Vps35p into a dominant-negative protein.
Table 1.
Amino acid substitutions identified in each dominant-negative mutant allelea
| Allele name | Amino acid substitution(s) |
|---|---|
| hp1 | S108R, R632C, Q733* |
| hp3 | M40I, I227F, V322I, N352I, E463D, T812(f. s.) |
| hp5 | Q35K, R98S, L174W, I898M, H921Q |
| hp6 | A431V, Y582F, K831* |
| hp7 | I758F, V769M, K831(f. s.) |
| hp8 | R98W, L330I, H414Q, Q422H, L702I, A746S, G893* |
| hp9 | I530T, N612S, L792I, K831(f. s.) |
| hp10 | K53I, I61M, R98S, L243V, Q294R, L830F |
| hp11 | Q733* |
| hp12 | L20I, R98S, H180N, I212F, L387M, Y696F, I758V, L825V |
| hp13 | Q391E, K413R, C720I, C720I, G821D, L911M, Q913* |
| hp14 | C24Y, V96L, Y708* |
| hp15 | K533R, A657V, E756D, I819F, L911M |
| hp16 | A252S, E771* |
The missense, nonsense (*) and frame-shift (f. s.) mutations found in each vps35 dominant-negative mutant allele are indicated. Silent mutations are not shown. Mutations at or near position 98 are indicated in bold.
Figure 2. The R98 residue of Saccharomyces cerevisiae Vps35p is contained with a conserved PRLYL motif.
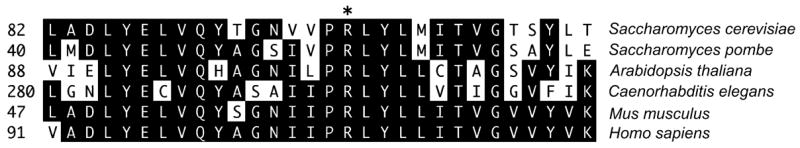
Thirty residue regions from the indicated Vps35p orthologs were aligned. The R98 residue of S. cerevisiae Vps35p is indicated by an asterisk. Residues that match the majority consensus are shaded.
To test whether a single missense mutation at codon 98 would cause a dominant-negative phenotype, the R98W mutation was moved into an otherwise wild-type Vps35p context. Strains expressing wild-type or mutant Vps35p were pulsed for 10 min with 35S-met/cys followed by a chase for various times and immunoprecipitation using an anti-Vps35p antibody. Vps35p is an extremely stable protein with very little degradation apparent after 90 min (Figure 3A). The Vps35-R98W and Vps35-Q733* proteins exhibited similar stability to wild type, while a R98W, Q733* double mutant protein was only slightly destabilized. The Vps35-Q733* and Vps35-R98W, Q733* proteins exhibited accelerated migration on SDS–PAGE compared with wild type, consistent with the deletion of amino acids 733–944.
Figure 3. Assessment of dominant-negative and functional properties of the vps35-R98Wand vps35-Q733* alleles.

A) A vps35Δ strain, SNY80, expressing the indicated forms of Vps35p from centromeric plasmids was analyzed. Stability of wild-type and mutant forms of Vps35p was assessed by pulsing with [35S]methionine/cysteine for 10 min and chasing for the indicated times. Immunoprecipitation with anti-Vps35-N antibody (see Materials and Methods) was then carried out and proteins analyzed by SDS–PAGE. B) Strains SNY36-9A (VPS35) and SNY80 (vps35Δ) expressing the indicated forms of Vps35p from centromeric plasmids were pulsed as in A and chased for 45 min. CPY was then immunoprecipitated from intracellular (I) and extracellular (E) fractions, separated by SDS–PAGE and the percentage of CPY secreted quantified by phosphorimager analysis. C) Strain SNY36-9A expressing the indicated forms of Vps35p from 2μ plasmids in the absence (−) or presence (+) of wild-type Vps35p overexpression through the TDH3 promoter was analyzed as in B. D) Strain SHY35 carrying 2μ plasmids harboring the indicated VPS35 alleles or no insert (−) was pulsed as in A and chased for the indicated times. Cps1p was then immunoprecipitated, treated with Endoglycosidase H and separated by SDS–PAGE. The precursor (pCps1, p2) and mature, processed (mCps1, m) forms of Cps1p and CPY are indicated.
To analyze these alleles further, a more quantitative CPY sorting assay was used in which the amount of newly synthesized CPY in the intracellular and extracellular (media) fractions was compared. Expression of the vps35-R98W and vps35-Q733* alleles from centromeric plasmids in a wild-type strain caused 4 and 10% CPY secretion, respectively (Figure 3B), while expression of these alleles from multicopy plasmids caused much higher CPY secretion (54% each; Figure 3C). Similar results were obtained using an assay in which the sorting of a model TGN protein, A-ALP, that undergoes retromer-mediated retrieval from the PVC (12,21), was assessed (Figure 4). While expression of the vps35-R98W and vps35-Q733* alleles on centromeric plasmids resulted in only a slight acceleration in vacuolar processing in an otherwise wild-type strain (Figure 4A), these alleles on 2μ plasmids exhibited much stronger sorting defects (Figure 4B). There was little or no effect on the rate of vacuolar processing of a protein (Cps1p) that is sorted from the TGN to the late endosome before being delivered to the vacuole (Figure 3D). Thus, expression of the VPS35 mutants appeared to specifically affect PVC-to-TGN retrieval. In summary, these results confirmed that the mutation of the R98 residue converts Vps35p into a dominant-negative protein. Thus for the remainder of the study, we focused on the R98W and Q733* mutations.
Figure 4. Effect of Vps35p dominant-negative alleles expressed on A-ALP sorting.

Strains SNY36-9A (VPS35) and SNY80 (vps35Δ) carrying the indicated VPS35 alleles on centromeric plasmids, as well as the A-ALP expression plasmid pSN55 (3), were pulsed with [35S]methionine/cysteine for 10 min and chased for the indicated times. A-ALP was immunoprecipitated, and the precursor (p) and mature (m) forms were separated by SDS–PAGE. Chr., chromosomal allele.
The VPS35 dominant-negative mutant proteins are limited in function but can compete with wild-type Vps35p
The vps35-R98W and vps35-R98W, Q733* alleles were completely non-functional based on analysis of their ability to complement a vps35Δ allele (Figure 3B). However, the vps35-Q733* allele was partially functional as a strain containing vps35-Q733* as the sole VPS35 allele exhibited 26% CPY secretion as compared with 78% secretion for a strain not expressing any form Vps35p whatsoever. Together, these data underscore the importance of the R98 residue for function.
It is expected that Vps35-R98W and Vps35-Q733* proteins are dominant negative because they compete with wild-type Vps35p for interactions with other retromer subunits. This would drive formation of defective retromer complexes that cannot fully engage, impairing coat assembly, vesicle formation or trafficking. If this were the case, then the dominant-negative effect should be suppressed by increasing expression of wild-type Vps35p. Consistent with this idea, we found that the introduction of a construct in which VPS35 was placed under control of the strong TDH3 promoter did indeed lessen the dominant-negative phenotype caused by the Vps35-R98W and Vps35-Q733* proteins (Figure 3C).
Expression of VPS35 dominant-negative alleles causes specific effects on Vps26p and Vps29p membrane association
To determine if the membrane association of retromer subunits is affected by expression of dominant-negative Vps35p proteins, we used previously generated hemag-glutinin (HA)-tagged alleles of Vps5p, Vps26p and SnxSp known to be fully functional (10,13,26). In addition, Vps35p, Vps29p and Vps17p were similarly tagged with HA, resulting in fully functional proteins as shown by CPY sorting assays (data not shown). Cells expressing tagged retromer alleles in the presence or absence of wild-type or mutant Vps35p exogenously expressed from multicopy plasmids were pulsed for 30 min and chased for 30 min before being lysed and the membranes sedimented at 150 000 × g for 60 min. Retromer subunits were then immunoprecipitated from pellet and supernatant fractions using an anti-HA epitope antibody. In a wild-type strain in which there was no exogenous expression of either wild-type or mutant Vps35p, the majority of the retromer subunits were found in the membrane fraction (Figure 5A).
Figure 5. Membrane association of retromer subunits in the presence of dominant-negative Vps35p.
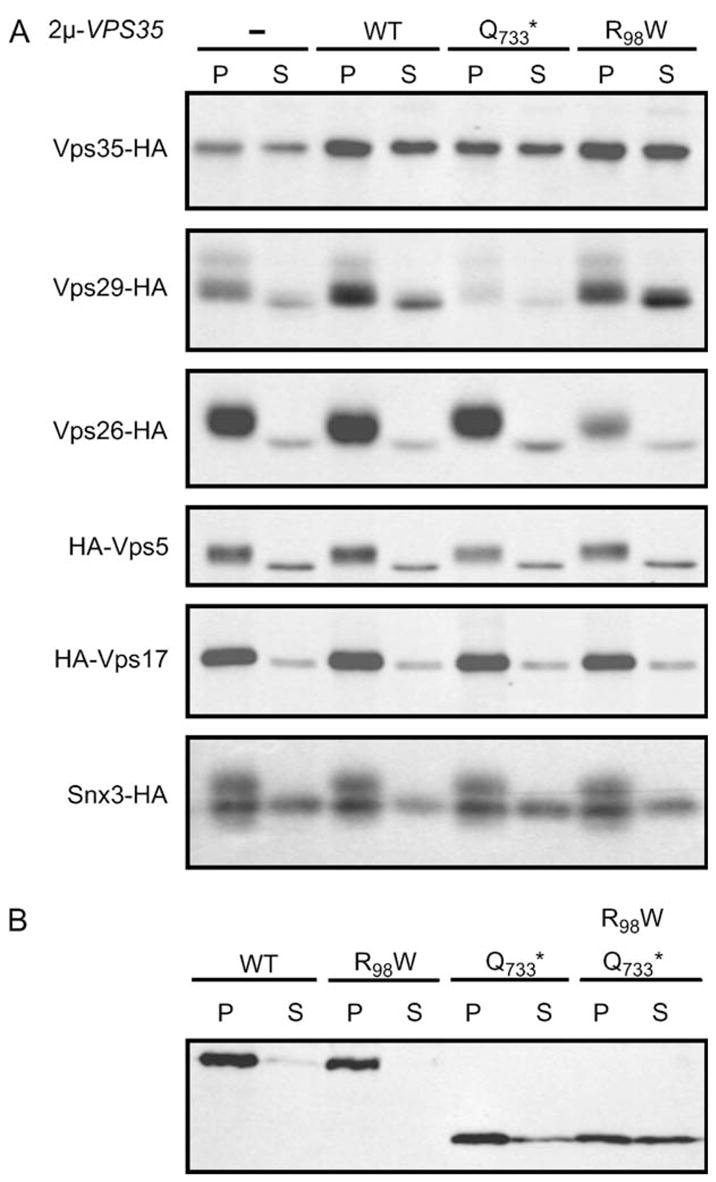
A) Strains PBY15/pAH58, SNY183, SNY205/pVps26-HA, PBY35/pAH31 and ZHY1 (from top to bottom panels) carrying the indicated 2μ plasmid-borne VPS35 alleles were spheroplasted, pulsed with [35S]methionine/cysteine for 30 min and chased for 30 min. Equivalent percentages of the membrane pellet (P) and supernatant (S) fractions generated from lysed spheroplasts were then subjected to immunoprecipitation with rabbit anti-HA antibody followed by separation by SDS–PAGE. B) Strain PBY15 carrying the indicated centromeric plasmid-borne VPS35 alleles was analyzed as described in A except the immunoprecipitation that was carried out with anti-Vps35-N antibody (see Materials and Methods).
Interestingly, the membrane-associated pools of Vps29-HA and Snx3-HA clearly exhibited electrophoretic forms with decreased SDS-PAGE mobility relative to the cytosolic pools, suggesting that a posttranslational modification might regulate membrane association of these subunits. While expression of VPS35 dominant-negative alleles generally did not have a dramatic effect on membrane association, the Vps35-R98W protein did shift a portion of Vps29-HA from the membrane-associated to the cytosolic pool. Similarly, expression of the Vps35-Q733* protein may have caused a slight redistribution of Vps26-HA from the membrane to the cytosolic fraction. Expression of the dominant-negative Vps35p proteins from multicopy plasmids had no discernable effect on membrane association of Vps35-HA expressed at normal levels.
To assess the membrane association of the dominant-negative Vps35p proteins themselves, wild-type or mutant Vps35p proteins were expressed from centromeric plasmids in a vps35Δ strain. The untagged Vps35p analyzed in this experiment (Figure 5B) exhibited improved membrane association compared with the HA-tagged allele in Figure 5A. Thus, while the tag does not interfere with function, it may reduce the efficiency of membrane association. The R98W single mutation did not disrupt membrane association, the Q733* single mutation reduced membrane association somewhat and the double mutant was affected even more severely (Figure 5B). These results suggest that both the 733–944 region and the R98 residue play a role in membrane association of Vps35p with progressively decreased overall membrane binding as the mutations cause the protein to become increasingly disabled in binding to other retromer subunits.
The VPS35 dominant-negative alleles specifically affect stability of some retromer subunits
Another observation made from the membrane association data was that there appeared to be a reduction in the level of Vps29-HA in a strain expressing Vps35-Q733* as well as a reduction in Vps26-HA in a strain expressing Vps35-R98W (Figure 5A). This instability is most likely because of proteasome-mediated turnover of the retromer as it is a cytosolic complex. In addition, the instability is apparently independent of vacuolar degradation as it occurred despite the fact that the strains used in experiment in Figure 5 were vacuolar-protease-deficient pep4Δ strains.
To more carefully investigate this effect, we subjected strains with HA-tagged alleles of VPS35, VPS29or VPS26, with or without expression of wild-type or mutant Vps35p from 2μ plasmids, to a pulse-chase regimen followed by immunoprecipitation with an anti-HA antibody. As was observed for untagged Vps35p (Figure 3A), Vps35-HA expressed on a low-copy centromeric plasmid in the absence of any other Vps35p construct was very stable over the 90-min chase period (Figure 6). Introduction of a multicopy plasmid expressing wild-type untagged Vps35p reduced the stability of Vps35-HA, suggesting that when Vps35p overexpression produces stoichiometric excess of functional Vps35p with respect to other retromer subunits, the cells limit that excess through degradation without altering the levels of other retromer subunits. Multicopy expression of dominant-negative vps35 alleles also decreased the stability of a coexpressed Vps35-HA allele (Figure 6); however, the effect was actually less than that seen for overexpression of wild-type Vps35p. On the other hand (and consistent with results from Figure 5), overexpression of dominant-negative Vps35 mutant alleles Q733* and R98W uniquely perturbed protein stability in trans for Vps29-HA and Vps26-HA, respectively (Figure 6). Both Vps29-HA and Vps26-HA were destabilized by the Vps35-R98W, Q733* double mutant. Expression of the dominant-negative Vps35p mutants had little or no effect on the other retromer subunits (Vps5p, Vps17p and Snx3p; data not shown), consistent with the idea that Vps35p forms a subcomplex with Vps29p and Vps26p and is less directly associated with the other retromer subunits (9). The differential effect on Vps29p and Vps26p degradation and membrane association because of expression of each of the dominant-negative Vps35p alleles suggests that each mutation may inactivate a distinct functional domain involved in interaction with each subunit.
Figure 6. Specific dominant-negative forms of Vps35p destabilize Vps26p and Vps29p.
A) Strains SNY80/pAH58, SNY183 carrying pTS18 [CEN-PEP4; (38)] and SNY204/pVps26-HA (from top) transformed with the indicated 2μ plasmid-borne VPS35 alleles were analyzed. These strains were assessed for stability of the retromer subunits using the pulse–chase regime described in Figure 3 followed by immunoprecipitation using a rabbit anti-HA antibody. B) Strain SNY205 carrying pVps26-HA, and either the 2μ plasmid YEp351 (Vector) or the vps35-R98W allele in YEp351 (vps35-R98W), was pulsed and chased for the indicated times before being analyzed for membrane association of Vps26-HA as described in the legend to Figure 5. Phosphorimager analysis was used to determine the half-times of degradation, shown below each panel in A and in the table B, and to determine the percentage of Vps26-HA that was membrane associated as indicated below the panel in B. The table in B indicates the rate of degradation for Vps26-HA in each strain background calculated for the pellets (P), supernatants (S) or as a total value (S + P).
To determine whether the membrane-associated or cytosolic form of Vps26-HA was destabilized upon expression of Vps35-R98W, we combined the pulse–chase analysis in Figure 6A with the fractionation regime used for Figure 5. There was a modest decrease in Vps26-HA membrane association throughout the time–course when Vps35-R98W was expressed than in the control (Figure 6B). In addition, the difference in Vps26-HA stability between the control and Vps35-R98W strain was most dramatically revealed when the membrane-associated form of Vps26-HA was compared. This result is consistent with the idea that Vps35-R98W exerts its destabilizing effect on Vps26-HA on the endosome membrane.
The R98W and Q733* mutations cause distinct defects in Vps35p interaction with other retromer subunits
To assess whether the R98W and Q733* mutations affect the association of Vps35p with other retromer subunits, a total of 20 vps35Δ strains were generated containing plasmid-based untagged wild-type or mutant VPS35 alleles (or empty vector) combined with HA-tagged VPS29, VPS26, VPS17or VPS5 alleles. Each HA-tagged retromer protein was either expressed from a tagged chromosomal allele or from a centromeric plasmid-borne allele driven by its own promoter. A native immunoprecipitation using anti-Vps35p antibody was carried out on each strain, and both the immunoprecipitated proteins and non-immunoprecipitated supernatant proteins were run on gels and blotted with anti-HA or anti-Vps35p antibodies. As expected, a large percentage of each of the four HA-tagged retromer proteins was coimmunoprecipitated with wild-type Vps35p (Figure 7). However, the Vps35p dominant-negative mutants were generally less effective in coimmunoprecipitating the other subunits. For example, all of the Vps35p mutants exhibited dramatic reductions of HA-Vps17 and HA-Vps5 coimmunoprecipitation compared with wild-type Vps35p. The amount of coimmunoprecipitated Vps29-HA was decreased somewhat in the presence of all of the Vps35p mutants expressed, but even in the presence of the Vps35-Q733* mutant that causes Vps29-HA to be destabilized (Figure 5), the remaining Vps29-HA could clearly associate with the Vps35p mutants. Most interestingly, little or no Vps26-HA was coimmunoprecipitated by Vps35-R98W and Vps35-R98W, Q733*, while only a modest reduction in coimmunoprecipitation was observed for Vps35-Q733*. This result underscores the importance of the R98 residue of Vps35p for association with Vps26p.
Figure 7. Specific effects of R98W and Q733* mutations on Vps35 association with retromer subunits.

A) Extracts from strains SNY195 (Vps29-HA), SNY207-1C/pSN420 (Vps26-HA), SNY193 (HA-Vps17) and SNY202b/pAH31 (HA-Vps5) carrying centromeric plasmids containing the indicated wild-type or mutant VPS35 alleles were immunoprecipitated with a mixture of rabbit anti-Vps35-N and rabbit anti-Vps35-C anti-bodies of roughly equal titre (see Materials and Methods). The immunoprecipitates (I) and supernatants (S, equivalent to 6.4% of the I samples) were separated by SDS-PAGE and blotted with a mouse anti-HA antibody (bottom four panels). In the top panel, the SNY195 (Vps29-HA) blot was stripped and reprobed with the anti-Vps35-N/anti-Vps35-C antibody mixture to show the relative level of the full-length (FL) and truncated (T) forms of Vps35p that were immunoprecipitated. Anti-Vps35-N/anti-Vps35-C blots of the Vps26-HA, HA-Vps17 and HA-Vps5 samples were indistinguishable from that of the Vps29-HA samples (data not shown). B) Strains SNY215 and SNY216 contained chromosomal deletions of all retromer alleles but expressed either Vps29-HA (SNY215) or Vps26-HA (SNY216) on multicopy plasmids as well as the indicated VPS35 alleles from centromeric plasmids. Cell extracts were subjected to the same coimmunoprecipitation procedure as described in A. The percentage of Vps29-HA and Vps26-HA from each strain that were coimmunoprecipitated was calculated by quantification of the Western blot (see Materials and Methods) and is indicated below each panel. C and D) The strains shown in the upper panel of B were analyzed in C. Likewise, the strains in the lower panel of B were analyzed in D. Whole cell extracts were separated by SDS–PAGE and blotted using an anti-Vps35p (upper panels) and anti-glucose 6-phosphate dehydrogenase (G6PDH, lower panels) antibody.
It is likely that each retromer subunit contains binding interfaces with multiple other retromer subunits. Thus, the binding between individual subunits such as Vps35p and Vps29p might very well be influenced by the presence of associations with additional subunits. To examine Vps35p/Vps29p and Vps35p/Vps26p pairwise interactions in the absence of other retromer subunits, we generated a penta-knockout vps35Δ vps29Δ vps26Δ vps17Δ vps5Δ strain containing a multicopy Vps29-HA construct, and another penta-knockout strain containing a multicopy Vps26-HA construct, each driven by the endogenous promoters, respectively. These strains were viable but grew much more slowly than wild type (data not shown). Each strain was then transformed with multicopy untagged wild-type or mutant VPS35 alleles (or empty vector), also using the endogenous VPS35 promoter. The same native coimmunoprecipitation regimen as that described above was then performed using these strains, where both binding partners were overexpressed and the remaining retromer sub-units absent. In all cases, the extent of pairwise coimmunoprecipitation in the absence of other subunits was relatively low, especially for Vps26-HA. Consistent with Figure 7 A, the extent of coimmunoprecipitation of Vps29-HA with the Vps35-Q733* mutant was further decreased. Moreover, in the case of the double Vps35 mutant, the coimmunoprecipitation of Vps29-HA was even less, and the Vps29-HA band of slower mobility was no longer apparent (Figure 7B). In the case of Vps26-HA, although the extent of coimmunoprecipitation was low in all cases, the most notable decrease in coimmunoprecipitation was seen with the Vps35-R98W mutant. Consistent with Figure 7A, the data in Figure 7B suggest that the dominant-negative mutant subunits are likely to exhibit impaired association with Vps29p or Vps26p, while the presence of other subunits are likely to help produce more complete (yet still defective) retromer protein complexes.
To address whether differences in coimmunoprecipitation in Figure 7B could be explained by instability of mutant forms of Vps35p, we analyzed whole cell extracts of these strains by immunoblotting using anti-Vps35p and anti-glucose 6-phosphate dehydrogenase as a loading control. The efficiency of coimmunoprecipitation of Vps29p by the various mutants does not clearly correlate with the levels of expressed Vps35p (Figure 7C), suggesting that in these strains, Vps35p expression levels were sufficient to reflect intrinsic Vps29p-binding capability rather than being limited by Vps35p stability. Likewise, reduction in coimmunoprecipitation of Vps26-HA by Vps26-R98W (Figure 7B) was also not limited by Vps35-R98W protein expression level (Figure 7D).
Assessment of the effect of overexpressing the dominant-negative Vps35p mutants on interaction between wild-type Vps35p and retromer subunits
We wished to determine whether dominant-negative Vps35 proteins blocked endogenous retromer assembly or allowed assembly while disrupting retromer function. To examine this, we assessed the ability of untagged Vps35 dominant-negative mutants to disrupt interactions between Vps35-13xMyc and HA-tagged retromer subunits in a series of strains carrying a chromosomal copy of VPS35 tagged with the 13xMyc epitope and one of the HA-tagged retromer alleles (plus all of the other retromer subunits in untagged form). Cells were lysed under native conditions and subjected to immunoprecipitation using an anti-HA antibody followed by immunoblotting with either anti-myc or anti-Vps35 antibodies.
In the absence of any overexpressed Vps35p, immunoprecipitation of each retromer subunit coimmunoprecipitated significant amounts of Vps35-13xMyc expressed at endogenous levels. Vps29-HA exhibited the most coimmunoprecipitation and SnxS-HA exhibited the least. The modest but significant Vps35-13xMyc/Snx3p coimmunoprecipitation is consistent with a recent study proposing that Snx3p is a cargo-specific adaptor of the retromer (14). Curiously, the results in Figure 8 suggest that Snx3p may bind even more of Vps35-13xMyc in the presence of the Vps35-R98W mutant. Overexpression of neither dominant-negative mutant of Vps35p exerted major impact on the extent of association measured between ‘endogenous’ retromer subunits (Figure 8), even though Vps35-Q733* interacted with Vps26-HA, while Vps35-R98W did not. Taken together, the data in Figures 7 and 8 highlight the significance of residue R98 of Vps35p in binding to Vps26p yet at the same time indicate that dominant-negative Vps35p mutants actually allow assembly of endogenous (yet defective) retromer subunit protein complexes.
Figure 8. Influence of dominant-negative VPS35 alleles on the coimmunoprecipitation of wild-type Vps35p and other retromer subunits.

Extracts from strains SNY201-5, SNY208-9D/pSN420, SNY206, SNY209/pAH31, SNY200 and SNY197 (shown in A–F, respectively) carrying the indicated 2μ plasmid-borne VPS35 alleles were immunoprecipitated with a rabbit anti-HA antibody. The immunoprecipitated proteins (I) and non-immunoprecipitated supernatant proteins (S, equivalent to 6.4% of the I samples) were separated by SDS–PAGE and immunoblotted with a mouse anti-myc antibody. For the samples in which Vps26-HA was immunoprecipitated, the blot was stripped and reprobed with a mixture of anti-Vps35-N and anti-Vps35-C antibodies (see Materials and Methods). The full-length (FL) and truncated (T) forms of Vps35p are indicated for this blot. The diffuse band migrating more slowly than that of full-length untagged Vps35p is likely to contain the Vps35-13xMyc protein.
Discussion
Vps35p clearly plays a central role in retromer function, but compared with other retromer subunits, relatively little is known about its structure–function relationships. In carrying out this study, our goal was to identify features on Vps35p that mediate its interaction with other retromer subunits or cargo using yeast genetics to screen for dominant-negative Vps35p alleles. The rate of mutagenesis was high as most of the mutants we obtained contained several amino acid changes within the 944-amino acid ORF. Of the 14 mutants obtained, five contained mutations at or near R98, and of the remaining nine alleles, all but one had a C-terminal truncation. This taken together with our phenotypic analysis suggests that the motif surrounding the R98 residue and the C-terminal region represent major structural features engaged in Vps35p function. Vps35p sequence comparisons from various organisms (Figure 2) indicate that R98 is embedded within a PRLYL motif that is absolutely conserved across species. As an extension of these studies, we report in the accompanying manuscript that expression in pancreatic beta cells of human Vps35 containing a mutation in the corresponding PRLYL motif (hVps35-R107W) affects the localization of the MPR and insulin (27). Based on these studies, we propose that the conserved PRLYL motif of yeast and mammalian Vps35p is engaged in mediating efficient interaction with their respective Vps26p counterparts.
Our identification of the R98 residue as an important structural feature is consistent with the previous two-hybrid analyses and in vitro pull-down studies that mapped regions of Vps35p and Vps26p that interact with one another (15,25). In particular, the two-hybrid data indicated that the interaction domain was within residues 1–172 of human Vps35 (containing the conserved PRLYL motif), while the hVps35-interacting domain on human Vps26p was mapped to loop L16 that contains two highly conserved acidic residues. Identification of a basic R residue in Vps35p important for association with Vps26p suggests the possibility that the two proteins may engage in ionic interactions between R98 and one or more acidic residues on loop L16of Vps26p. Consistent with an ionic interaction, Vps26p has been shown to dissociate from the rest of the retromer complex by the addition of 250 mM NaCl (26). Further experiments to test this hypothesis are in progress.
While our study was being reviewed, Gokool et al. also reported the identification of the PRLYL motif in Vps35p as playing an essential role in interaction with Vps26p (28). However, while Gokool et al. reported a loss of Vps35 membrane association because of a L99P mutation in the yeast protein and R107A or L108P mutations in the mammalian protein, we did not observe a decrease in membrane association of yeast or human Vps35 because of the R98W/R107W mutations. Thus, while Vps35/Vps26 association clearly relies on the PRLYL motif, differences in residue substitution or differences in methodology may account for whether membrane binding of the mutant Vps35 is observed. More work is needed to clarify whether membrane binding of the mutant Vps35 is required in order to achieve the dominant-negative phenotypes described in this report.
The structure of human Vps26p has recently been shown to resemble that of the arrestin family (29), a group of proteins that act as cargo-selective adaptors during clathrin-mediated endocytosis (29,30). Therefore, it is tempting to speculate that Vps26p may aid Vps35p in cargo selectivity. Indeed, it is interesting that R98 (this report) is relatively proximate to another highly conserved residue, D123, that has previously been implicated in cargo recognition by Vps35p (12).
The structural elements of Vps35p that mediate association with Vps29p appear to be more extensive than those mediating association with Vps26p. In the case of the mammalian retromer, two-hybrid analysis and experiments measuring in vitro association of Escherichia-coli-expressed Vps35p and Vps29p indicate that the N-terminal conserved region of Vps35p is not sufficient to associate with Vps29 (15,25). These analyses were only able to narrow down the interacting region to residues 307–796 of mammalian Vps35, suggesting that amino acids from distant regions of the primary sequence may converge in space to form an interacting patch. Our analysis of yeast Vps35p indicated that, with the full complement of retromer subunits present, neither R98W nor Q733* mutations in Vps35p caused much of a reduction in Vps29p coimmunoprecipitation. With other retromer subunits absent, both mutations caused a modest reduction in Vps35p/Vps29p coimmunoprecipitation. Combining our results with those of previous analyses, it seems likely that Vps29p association with Vps35p is sensitive to mutations that perturb the global three-dimensional structure of Vps35p. A precise structural definition of the Vps35/Vps29 interface will have to await X-ray crystallographic analysis of Vps35p and subsequent testing of the role of surface residues in the interaction.
The precise mechanism by which the dominant-negative Vps35 proteins affected retromer function is unknown. Some evidence suggests that retromer complexes may undergo self-assembly into larger arrays on the membrane, a process driven by the Vps5/SNX1 component (9,31). Expression of mutant forms of Vps35p that possess some but not all of the structural features necessary for interaction with other subunits might then result in incomplete and non-functional retromer complexes interspersed with functional retromer complexes. This would likely prevent some normal function of the vesicle or tubule coat from occurring. This deficiency in coat function, which could relate to problems with higher-order organization on the membrane, membrane curvature or disassembly, would prevent efficient late endosome-to-TGN trafficking of cargo.
An interesting aspect of this study is the ability of the dominant-negative Vps35-R98W mutant (expressed in the presence of wild-type Vps35p) to trigger degradation of Vps26p, even as it fails to interact efficiently with Vps26p. Vps35-R98W does not destabilize Vps26p when wild-type Vps35p is absent (Figure 7B; data not shown). Furthermore, Vps26-HA expressed in the complete absence of the other four retromer subunits is actually quite stable and equal to that in a strain containing them (S. F. N., unpublished). Thus, the instability of Vps26p is probably not simply because of a lack of association with Vps35p or other retromer subunits. Immunoprecipitating Vps26-HA from a strain expressing Vps35-13xMyc at normal levels and overexpressing Vps35-R98W results in the coprecipitation of the same amount of Vps35-13xMyc as when Vps35-R98W is not expressed. Importantly, under these same conditions, Vps35-R98W is not coprecipitated. However, Vps35-R98W clearly can interact with other retromer subunits such as Vps29p (Figures 7 and 8). One possibility is that high-level expression of Vps35-R98W acts as a sink for Vps29p causing a shortage of Vps29p for association with wild-type Vps35p and Vps26p. This could result in a large proportion of Vps26p being present in incomplete retromer complexes. These Vps35-R98W/Vps29p complexes apparently do not effectively engage the Vps35p/Vps26-containing complexes within the context of a large array. How this renders Vps26p subject to degradation without destabilizing wild-type Vps35p is not known. Vps26p is thought to associate with the retromer in a more transient or dynamic fashion than other subunits (26), and this might enable it to be selectively degraded. The membrane-associated pool of Vps26-HA was most destabilized compared with the control (Figure 6). Thus, it is possible that Vps35-R98W caused Vps26-HA to dissociate from the membrane and, in turn, caused its rapid degradation in the cytosol.
In an accompanying paper, we show that expression of hVps35 in yeast also causes a dominant-negative phenotype that actually requires the R107 residue (of the PRLYL motif) (27). Further, we show that hVps35 in yeast is predominantly membrane associated and binds to yeast Vps26p in a manner dependent on its R107 residue. The simplest explanation is that the PRLYL motif allows association of hVps35 with yeast Vps26p, while hVps35 has altered many of the structural domains required to properly bind other yeast retromer subunits. It is likely that, as in this report, complete pentameric retromer complexes with wild-type subunits become interspersed with those that are incomplete and defective. And as with expression of yeast Vps35p-R98W in an otherwise wild-type strain, expression of hVps35 in an otherwise wild-type strain destabilizes Vps26p. These data support the model presented herein that Vps26p is susceptible to degradation when cycling on membranes in higher-order complexes containing a mixture of functional and non-functional retromer complexes.
In summary, we have identified two structural features of yeast Vps35p important for its role in the retromer complex, one of which involves a conserved PRLYL motif engaged in binding to Vps26p. An accompanying paper further demonstrates that the PRLYL motif is also essential for the function of mammalian Vps35 (27). These results will help guide future studies aimed at more precisely defining structure-function relationships of Vps35p.
Materials and Methods
General methods, antibodies and yeast strains
The production of yeast medias, the genetic manipulation of yeast strains and all general molecular biology methods were performed as described previously (32) or as otherwise noted. Rabbit anti-CPY polyclonal antibodies and the mouse anti-HA 12CA5 monoclonal antibody were gifts from Tom Stevens (University of Oregon, Eugene, OR, USA). Rabbit anti-HA antibodies were from Covance. Mouse anti-myc monoclonal antibody 9E10, mouse anti-CPY and rabbit anti-glucose 6-phosphate dehydrogenase antibodies were from Santa Cruz Biotechnology, Inc., Molecular Probes and Sigma-Aldrich, respectively. Rabbit polyclonal antibodies against yeast alkaline phosphatase and Cps1p have been previously described (2,33,34). Rabbit polyclonal antibodies against an N-terminal region of Vps35p, termed anti-Vps35-N, were raised against a fusion protein consisting of residues 1–357 of Vps35p. Likewise, rabbit polyclonal antibodies against a C-terminal region of Vps35, termed anti-Vps35-C, were generated against a fusion protein consisting of Vps35p residues 647–883. Yeast strains used in this study are described in Table 2.
Table 2.
Saccharomyces cerevisiae strains and plasmids used in this study
| Strain | Description | Origin or reference |
|---|---|---|
| SNY36-9A | MATa leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 pho8Δ::ADE2 | (39) |
| LSY2 | SNY36-9A pep4Δ::TRP1 | (34) |
| SNY80 | SNY36-9A vps35Δ::HIS3 | (21) |
| PBY15 | SNY36-9A vps35Δ::HIS3 pep4-ΔH3 | (12) |
| SNY175 | SNY36-9A pep4Δ::TRP1 VPS17::3xHA | This study |
| SNY183 | SNY36-9A pep4Δ::TRP1 VPS29::3xHA | This study |
| SNY204 | SNY36-9A vps26Δ::NatR | This study |
| SNY205 | SNY36-9A pep4Δ::TRP1 vps26Δ::NatR | This study |
| AHY40 | SNY36-9A vps5Δ::HIS3 | (10) |
| PBY35 | SNY36-9A pep4Δ::TRP1 vps5Δ::HIS3 | This study |
| WVY12 | SNY36-9A SNX3::3xHA | (13) |
| ZHY1 | WVY12pep4Δ::TRP1 | This study |
| SNY197 | SNY36-9A pep4Δ::TRP1 VPS35::13xMyc-KanR | This study |
| SNY200 | SNY36-9A pep4Δ::TRP1 SNX3::3xHA VPS35::13xMyc-KanR | This study |
| SNY201-5 | SNY36-9A pep4Δ::TRP1 VPS29::3xHA VPS35::13xMyc-KanR | This study |
| SNY206 | SNY36-9A pep4Δ::TRP1 VPS17::3xHA VPS35::13xMyc-KanR | This study |
| SNY208-9D | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 pho8Δ::ADE2 pep4Δ::TRP1 vps26Δ::LEU2 VPS35::13xMyc-KanR | This study |
| SNY209 | SNY36-9A pep4Δ::TRP1 vps5Δ::HIS3 VPS35::13xMyc-KanR | This study |
| SNY193 | SNY36-9A pep4Δ::TRP1 VPS17::3xHA vps35Δ::KanR | This study |
| SNY195 | SNY36-9A pep4Δ::TRP1 VPS29::3xHA vps35Δ::KanR | This study |
| AHY42 | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 Iys2-801 suc2-Δ9 pho8Δ:: ADE2 pep4Δ::TRP1 vps5Δ::HIS3 | (10) |
| SNY202b | AHY42 vps35Δ::KanR | This study |
| SNY207-1C | MATα leu2-3, 112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 pho8Δ:: ADE2 pep4Δ::TRP1 vps26Δ::LEU2 vps35Δ::KanR | This study |
| SNY215 | SNY36-9A pep4-ΔH3 vps29Δ::KanR vps5Δ vps26Δ::NatR vps17Δ::KanR vps35Δ::HIS3 + pSN432 (2μ-TRP1-VPS29::3xHA) | This study |
| SNY216 | SNY36-9A pep4-ΔH3 vps29Δ::KanR vps5Δ vps26Δ::NatR vps17Δ::KanR vps35Δ::HIS3 + pSN433 (2μ-TRP1-VPS26::3xHA) | This study |
DNA manipulations
VPS35 plasmids were constructed by first subcloning the 3.9-kbp MscI fragment containing VPS35 into the Smal site of pRS316 (CEN-URA3). The EcoRI-SacI fragment from the resulting plasmid, pLS8, was subcloned into the EcoRI/SacI sites of YEp352 (2μ-URA3) resulting in pAH60. To randomly mutagenize VPS35, the VPS35 insert of pAH60 and ~100 bp of each region flanking the polylinker was polymerase chain reaction (PCR) amplified under mutagenic conditions using the GeneMorph Random Mutagenesis Kit (Stratagene). The4.1-kbp PCR fragments were cotransformed with linearized YEp352 into yeast strain SNY36-9A (Table 2). Plasmids were rescued from transformants that exhibited CPY sorting defects and were subjected to DNA sequence analysis resulting in 14 distinct dominant-negative alleles (Table 1).
To introduce VPS35 and the derived mutant alleles into a centromeric plasmid, the inserts from pAH60 and derived mutant plasmids were excised as EcoRI–EagI fragments and were subcloned into the same sites of pRS314 (CEN-TRP1). To generate VPS35 solely containing the R98W mutation, the 1.5-kbp EcoRI-AatII fragment from a pAH60 derivative containing the vps35-hp8 allele was swapped for the corresponding EcoRI-AatII fragment in pLS8 to generate pSN358. To generate an R98W, Q733* double mutant allele, the 2.5-kbp AatII-XbaI fragment from a pAH60 derivative containing the hp11 allele was swapped for the corresponding AatII–XbaI fragment in pSN358. The 2μ versions of these single and double mutant alleles were generated by subcloning them into plasmids YEp351 (2μ-LEU2) and YEp352. To place the expression of VPS35 under the control of the TDH3 promoter, the3.4-kbp BseRI-PvuII VPS35 ORF fragment from pLS13 (12) was blunted with T4 DMA polymerase and was ligated into the SmaI site of pSN359, a pRS316 derivative containing the TDH3 promoter and terminator separated by SmaI and other polylinker sites.
Plasmids pVps26-HA and pAH31 consist of pRS316 containing the VPS26::HA and VPS5::HA alleles, respectively (10,26). The 2.0-kbp PvuII fragment from pVPS26-HA containing the VPS26::HA allele was subcloned into the PvuII sites of pRS313 (CEN-HIS3) and pRS424 (2μ-TRP1) to generate plasmids pSN420 and pSN433, respectively. To generate VPS29::3xHA plasmids, the VPS29::3xHA allele was amplified by PCR from SNY183 (Table 2) yeast genomic DMA. This fragment was digested with XbaI/Bg/II, and the resulting 0.956-kbp fragment corresponding to the 3′ region of the VPS29::3xHA allele was ligated, along with the 0.985-kbp HindIII/XbaI fragment from pEMY29-1 (8) corresponding to the 5′ region of VPS29, into the HindIII/BamHI sites of pRS316 through a three-way ligation giving rise to pSN431. The VPS29::3xHA allele from pSN431 was subcloned as a 2.0-kbp EagI-Sa/I fragment into the EagI/Sa/I sites of pRS424 giving rise to pSN432. A VPS35 allele with a single HA tag at the 3′ end of the ORF was constructed by introducing a Bg/II site just upstream of the VPS35stop codon in pLS8. An oligo duplex encoding an HA tag was ligated into the Bg/II site, resulting in plasmid pAH58.
Assessment of retromer subunit stability, protein processing and CPY sorting
The procedure for immunoprecipitation of CPY was performed using a rabbit antibody against CPY as described previously (35). Likewise, immunoprecipitations to assess A-ALP and Cps1p processing and the degradation of retromer subunits were performed using the basic procedure previously described for A-ALP (3) with variations on the chase times and antibodies used. In the case of Cps1, the immunoprecipitated protein was deglycosylated using Endoglyosidase H (Roche Diagnostics) according to a published procedure (36) prior to running on the gel. Radioactively labeled proteins were quantified from gels using the Molecular Imager FX phosphorimager and associated Quantity One software (Bio-Rad Laboratories). For calculation of the half-time of A-ALP processing, the log of the percentage of unprocessed precursor at each time-point was plotted as a function of time, and the plots were analyzed by linear regression analysis. The half-time of retromer subunit turnover was calculated similarly except the log of the percent protein remaining at a given time-point (as compared with the 0-min time-point) was plotted versus time.
Colony blots of secreted CPY were performed as previously described (37). Briefly, cells were applied to the surface of agar plates, each was overlaid with a nitrocellulose filter and incubated for 14 h at 30°C. Cells were then rinsed off the filters using dH2O and the filters immunoblotted using a mouse anti-CPY antibody followed by an anti-mouse alkaline phosphatase-coupled secondary antibody and chemiluminescent development using the Lumiphos reagent (Pierce Biotechnology).
Membrane fractionation and generation of yeast whole cell extracts
Six optical density (OD)600 units of yeast cells harvested from log-phase cultures were incubated in 1 mL of 0.1 M Tris, pH 9.4, and 10 mM DTT at23°C for 10 min. The cells were pelleted and spheroplasted in 1.2 mL of minimal selective media containing 1 M sorbitol, 20 mM Tris, pH 7.5, and 24 μg of Zymolyase 100T (U.S. Biological). The spheroplasts in media containing 1 M sorbitol were pulse labeled with [35S]methionine/cysteine at 30°C for the indicated time and then chased with unlabeled amino acids for the indicated time before stopping the chase through the addition of NaN3 to 10 mM and transfer to ice. After pelleting, the spheroplasts were resuspended in 600 μL of ice-cold lysis buffer (20 mM HEPES-KOH, 50 mM potassium acetate, 2 mM ethylenediaminetetraacetic acid (EDTA), 0.2 M sorbitol) containing protease inhibitors (0.5 mM phenylmethylsulphonyl fluoride, 1 μg/mL leupeptin and 1 μg/rnL pepstatin A) and were dounce homogenized through 10 strokes witha 1-mL dounce. Unlysed cells were pelleted bycentrifugation at 600 × g for 5 min at 4°C. The supernatant was then centrifuged at 150 000 × g for 1 h at 4°C to generate a cytosol and membrane fraction. Eighty microliters of 8 M urea, 5% sodium dodecyl sulfate (SDS) was added to both the pellet and the supernatant followed by heating at 100°C for 5 min. Twenty microliters of the resuspended pellet and 170 μL of the supernatant were then diluted to 1 mL so as to both contain: 0.15× lysis buffer, 0.02% SDS, 160 mM urea, 90 mM Tris, pH 8.0, 0.1 % Triton-X-100 and 2 mM EDTA. Immunoprecipitation of each fraction was carried out as described previously (3) using a rabbit anti-HA antibody for HA-tagged subunits or rabbit anti-Vps35-N antibody for wild-type and mutant Vps35p alleles.
Whole cell protein extracts were generated by incubating 10 OD600 units of yeast cells in 100 μL of 50 mM Tris, pH 6.8, containing 8 M urea, 5% SDS and 5% 2-mercaptoethanol for 10 min at 70°C. Cells were then lysed by agitation with glass beads and cellular debris pelleted by centrifugation at 16 100 × g for 5 min. Four percent of the supernatant was loaded on an SDS–PAGE gel per lane.
Coimmunoprecipitation of retromer subunits
To assess Coimmunoprecipitation of retromer subunits from yeast cell extracts, 6 OD600 units of cells were spheroplasted as described above. Spheroplasts were then lysed in 500 μL ice-cold buffer C-DTT (25 mM HEPES–KOH, 125 mM potassium acetate, 2.5 mM magnesium acetate and 0.1% Triton-X-100) containing protease inhibitors. After incubating on ice for 15 min, the lysate was centrifuged at 16 100 × g for 5 min, and the supernatant was precleared by adding 50 μL (bed volume) of Sepharose CL-4B, rotating for 30 min at 4°C, sedimenting the sepharose and transferring the supernatant to a fresh tube. The immunoprecipitation antibody was then added followed by rotating at 4°C for 90 min, addition of protein A–Sepharose and incubation for an additional 60 min. The supernatant was then transferred to a fresh tube, and the protein A–Sepharose was washed three times with 1 mL volumes of buffer C. The supernatant and bead-associated proteins were separated by SDS–PAGE, transferred to polyvinylidene fluoride membranes and were probed with the appropriate primary antibody. After incubation with enzyme-conjugated anti-rabbit or anti-mouse secondary antibodies, chemiluminescent detection of the proteins of interest was carried out using the LumiPhos substrate for alkaline phosphatase or the SuperSignal substrate for horseradish peroxidase (both from Pierce). The blots were imaged using a Fuji LAS-1000 charge-coupled device camera and ImageReader LAS-1000 1.2 software (Fuji Photo Film Co.). Images were quantified using ImageGuage 4.22 and formatted using Adobe Photoshop 7.
Acknowledgments
We are grateful to Tom Stevens for supplying antibodies and strain WVY12 and to Matt Seaman for providing the Vps26-HA construct. This work was supported by grants from the National Institutes of Health: R01GM053449 awarded to S. F. N and DK48280 awarded to P. A.
References
- 1.Costaguta G, Stefan CJ, Bensen ES, Emr SD, Payne GS. Yeast Gga coat proteins function with clathrin in Golgi to endosome transport. Mol Biol Cell. 2001;12:1885–1896. doi: 10.1091/mbc.12.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foote C, Nothwehr SF. The clathrin adaptor complex 1 directly binds to a sorting signal in Ste13p to reduce the rate of its trafficking to the late endosome of yeast. J Cell Biol. 2006;173:615–626. doi: 10.1083/jcb.200510161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nothwehr SF, Roberts CJ, Stevens TH. Membrane protein retention in the yeast Golgi apparatus: dipeptidyl aminopeptidase A is retained by a cytoplasmic signal containing aromatic residues. J Cell Biol. 1993;121:1197–1209. doi: 10.1083/jcb.121.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant NJ, Stevens TH. Two separate signals act independently to localize a yeast late Golgi membrane protein through a combination of retrieval and retention. J Cell Biol. 1997;136:287–297. doi: 10.1083/jcb.136.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper AA, Stevens TH. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J Cell Biol. 1996;133:529–541. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cereghino JL, Marcusson EG, Emr SD. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPS gene products regulate receptor stability, function, and localization. Mol Biol Cell. 1995;6:1089–1102. doi: 10.1091/mbc.6.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilcox CA, Redding K, Wright R, Fuller RS. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol Biol Cell. 1992;3:1353–1371. doi: 10.1091/mbc.3.12.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seaman MNJ, Marcusson EG, Cereghino JL, Emr SD. Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the Vps29, Vps30, and Vps35 gene products. J Cell Biol. 1997;137:79–92. doi: 10.1083/jcb.137.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seaman MNJ, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nothwehr SF, Hindes AE. The yeast VPS5/GRD2 gene encodes a sorting nexin-1-like protein required for localizing membrane proteins to the late Golgi. J Cell Sci. 1997;110:1063–1072. doi: 10.1242/jcs.110.9.1063. [DOI] [PubMed] [Google Scholar]
- 11.Horazdovsky BF, Davies BA, Seaman MNJ, McLaughlin SA, Yoon S, Emr SD. A sorting nexin-1 homologue, Vps5p, forms a complex with Vps17p and is required for recycling the vacuolar protein-sorting receptor. Mol Biol Cell. 1997;8:1529–1541. doi: 10.1091/mbc.8.8.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nothwehr SF, Bruinsma P, Strawn LS. Distinct domains within Vps35p mediate the retrieval of two different cargo proteins from the yeast prevacuolar/endosomal compartment. Mol Biol Cell. 1999;10:875–890. doi: 10.1091/mbc.10.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voos W, Stevens TH. Retrieval of resident late-Golgi membrane proteins from the prevacuolar compartment of Saccharomyces cerevisiae is dependent on the function of Grd19p. J Cell Biol. 1998;140:577–590. doi: 10.1083/jcb.140.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strochlic TI, Setty TG, Sitaram A, Burd CG. Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J Cell Biol. 2007;177:115–125. doi: 10.1083/jcb.200609161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haft CR, Sierra MDL, Bafford R, Lesniak MA, Barr VA, Taylor SI. Human orthologs of yeast vacuolar protein sorting proteins Vps26, 29, and 35: assembly into multimeric complexes. Mol Biol Cell. 2000;11:4105–1116. doi: 10.1091/mbc.11.12.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurten RC, Cadena DL, Gill GN. Enhanced degradation of EGF receptors by a sorting nexin, SNX1. Science. 1996;272:1008–1010. doi: 10.1126/science.272.5264.1008. [DOI] [PubMed] [Google Scholar]
- 17.Arighi CN, Hartnell LM, Aguilar RC, Haft CR, Bonifacino JS. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J Cell Biol. 2004;165:123–133. doi: 10.1083/jcb.200312055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seaman MN. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J Cell Biol. 2004;165:111–122. doi: 10.1083/jcb.200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verges M, Luton F, Gruber C, Tiemann F, Reinders LG, Huang L, Burlingame AL, Haft CR, Mostov KE. The mammalian retromer regulates transcytosis of the polymeric immunoglobulin receptor. Nat Cell Biol. 2004;6:763–769. doi: 10.1038/ncb1153. [DOI] [PubMed] [Google Scholar]
- 20.Rojas R, Satoshi K, Haft CR, Bonifacino JS. Interchangeable but essential function of SNX1 and SNX2 in the association of retromer with endosomes and the trafficking of mannose 6-phosphate receptors. Mol Cell Biol. 2007;27:1112–1124. doi: 10.1128/MCB.00156-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nothwehr SF, Ha S-A, Bruinsma P. Sorting of yeast membrane proteins into an endosome-to-Golgi pathway involves direct interaction of their cytosolic domains with Vps35p. J Cell Biol. 2000;151:297–309. doi: 10.1083/jcb.151.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burda P, Padilla SM, Sarkar S, Emr SD. Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 Ptdlns 3-kinase. J Cell Sci. 2002;115:3889–3900. doi: 10.1242/jcs.00090. [DOI] [PubMed] [Google Scholar]
- 23.Cozier GE, Carlton J, McGregor AH, Gleeson PA, Teasdale RD, Mellor H, Cullen PJ. The phox homology (PX) domain-dependent, 3-phosphoinositide-mediated association of sorting nexin-1 with an early sorting endosomal compartment is required for its ability to regulate epidermal growth factor receptor degradation. J Biol Chem. 2002;277:48730–48736. doi: 10.1074/jbc.M206986200. [DOI] [PubMed] [Google Scholar]
- 24.Carlton J, Bujny M, Peter BJ, Oorschot VM, Rutherford A, Mellor H, Klumperman J, McMahon HT, Cullen PJ. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high-curvature membranes and 3-phosphoinositides. Curr Biol. 2004;14:1791–1800. doi: 10.1016/j.cub.2004.09.077. [DOI] [PubMed] [Google Scholar]
- 25.Collins BM, Skinner CF, Watson PJ, Seaman MN, Owen DJ. Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly. Nat Struct Mol Biol. 2005;12:594–602. doi: 10.1038/nsmb954. [DOI] [PubMed] [Google Scholar]
- 26.Reddy JV, Seaman MNJ. Vps26p, a component of retromer, directs the interactions of Vps35p in endosome-to-Golgi retrieval. Mol Biol Cell. 2001;12:3242–3256. doi: 10.1091/mbc.12.10.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao X, Nothwehr SF, Lara-Lemus R, Zhang B, Peter H, Arvan P. Mammalian Vps35 behaves as a dominant negative protein in yeast: mutation of a conserved PRLYL motif alters protein function and localization. Traffic. 2007 doi: 10.1111/j.1600-0854.2007.00658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gokool S, Tattersall D, Reddy JV, Seaman MN. Identification of a conserved motif required for Vps35p/Vps26p interaction and assembly of the retromer complex. Biochem J. 2007 doi: 10.1042/BJ20070555. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi H, Rojas R, Bonifacino JS, Hurley JH. The retromer subunit Vps26 has an arrestin fold and binds Vps35 through its c-terminal domain. Nat Struct Mol Biol. 2006;12:540–548. doi: 10.1038/nsmb1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller WE, Lefkowitz RJ. Expanding roles for β-arrestins as scaffolds and adapters in GPCR signaling and trafficking. Curr Opin Cell Biol. 2001;13:139–145. doi: 10.1016/s0955-0674(00)00190-3. [DOI] [PubMed] [Google Scholar]
- 31.Kurten RC, Eddington AD, Chowdhury P, Smith RD, Davidson AD, Shank BB. Self-assembly and binding of a sorting nexin to sorting endosomes. J Cell Sci. 2001;114:1743–1756. doi: 10.1242/jcs.114.9.1743. [DOI] [PubMed] [Google Scholar]
- 32.Ausebel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Struhl K, Smith JA. Current Protocols in Molecular Biology. USA: John Wiley & Sons, Inc; 2000. [Google Scholar]
- 33.Nothwehr SF, Bryant NJ, Stevens TH. The newly identified yeast GRD genes are required for retention of late-Golgi membrane proteins. Mol Cell Biol. 1996;16:2700–2707. doi: 10.1128/mcb.16.6.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spelbrink RG, Nothwehr SF. The yeast GRD20 gene is required for protein sorting in the trans-Golgi network/endosomal system and for polarization of the actin cytoskeleton. Mol Biol Cell. 1999;10:4263–4281. doi: 10.1091/mbc.10.12.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vater CA, Raymond CK, Ekena K, Howald-Stevenson I, Stevens TH. The VPS1 protein, a homolog of dynamin required for vacuolar protein sorting in Saccharomyces cerevisiae, is a GTPase with two functionally separable domains. J Cell Biol. 1992;119:773–786. doi: 10.1083/jcb.119.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orlean P, Kuranda MJ, Albright CF. Analysis of glycoproteins from Saccharomyces cerevisiae. Methods Enzymol. 1991;194:682–697. doi: 10.1016/0076-6879(91)94050-m. [DOI] [PubMed] [Google Scholar]
- 37.Roberts CJ, Raymond CK, Yamashiro CT, Stevens TH. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- 38.Ammerer G, Hunter CP, Rothman JH, Saari GC, Vails LA, Stevens TH. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol Cell Biol. 1986;6:2490–2499. doi: 10.1128/mcb.6.7.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nothwehr SF, Conibear E, Stevens TH. Golgi and vacuolar membrane proteins reach the vacuole in vps1 mutant yeast cells via the plasma membrane. J Cell Biol. 1995;129:35–46. doi: 10.1083/jcb.129.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]




