Abstract
Homologues of the human major histocompatibility complex (MHC) HLA-A, -B, -E, -F, and -G loci are present in all the Catarrhini (Old World primates, apes, and humans), and some of their allelic lineages have survived several speciation events. Analysis of 26 MHC class I cDNAs from seven different genera of New World primates revealed that the Callitrichinae (tamarins and marmosets) are an exception to these rules of MHC stability. In gene trees of primate MHC class I genes, sequences from the Callitrichinae cluster in a genus-specific fashion, whereas in the other genera of New World primates, as in the Catarrhini, they cluster in a transgeneric way. The genus-specific clustering of the Callitrichinae cDNAs indicates that there is no orthology between MHC class I loci in genera of this phyletic group. Additionally, the Callitrichinae genera exhibit limited variability of their MHC class I genes, in contrast to the high variability displayed by all other primates. Each Callitrichinae genus, therefore, expresses its own set of MHC class I genes, suggesting that an unusually high rate of turnover of loci occurs in this subfamily. The limited variability of MHC class I genes in the Callitrichinae is likely the result of the recent origin of these loci.
One of the hallmarks of the major histocompatibility complex (MHC) is the high polymorphism and intralocus variability of its loci (1). At the human classical MHC class I locus HLA-B, more than 149 alleles have been sequenced, with pairwise differences that range from 1 to 49 aa (2). Despite this enormous diversity, the MHC class I loci and some of their allelic lineages are very stable. Orthologues of the human MHC class I loci HLA-A, -B, -E, -F, and -G are remarkably well preserved throughout the infraorder Catarrhini (Old World primates, apes, and humans; refs. 3–8). In addition to the evolutionary stability of the loci, sharing of allelic lineages has been observed between different species of primates (9–11). This stability, however, appears to be time-dependent and framed within particular phylogenetic hierarchies. Indeed, MHC class I genes have evolved independently in each class of the subphylum vertebrates (12–16), and in mammals, there is no documented example of orthologous relationships of MHC class I genes between the different orders. In addition, this disruption in the orthology pattern of MHC class I genes has been observed in two marsupial species from the same order that diverged 48 million years ago (17). Thus, a process of duplication, followed by differential expansion of loci, characterizes the long-term evolution of the MHC class I region.
The cotton-top tamarin (Saguinus oedipus), a New World primate, is, in many ways, an exception to the rules of polymorphism, variability, and stability that characterize the MHC class I genes of primates (18). Cotton-top tamarins express MHC class I genes with limited polymorphism and variability. In more than 100 wild and captive cotton-top tamarins analyzed thus far, only 11 different MHC class I molecules have been described (19, 20). These molecules, encoded by at least three loci, display high sequence similarity and show no locus-specific motifs that distinguish the products of one locus from those of another. Furthermore, the MHC class I genes that are expressed in this New World primate are not orthologous to any of the classical MHC class I loci of the Catarrhini (A, B, or C loci), but instead, they are most similar to the human nonclassical HLA-G (21). Interestingly, the MHC class II DRB genes in New World primates appear to have an independent origin from their Catarrhini counterparts (22, 23), indicating that a discontinuity in the orthology relationships has also occurred in DRB genes after the divergence of the Platyrrhini (New World primates) and the Catarrhini.
Recently, we described the presence of an MHC class I processed pseudogene (MHC-PS1) in the genome of several species of the genus Saguinus (24). Gene trees revealed that none of the expressed MHC class I genes in the cotton-top tamarin clustered with this processed pseudogene. Interestingly, MHC-PS1 was closely related to the cotton-top tamarin MHC class I pseudogene So-N1 (19), a defective gene from which no transcripts have been found. Thus, MHC-PS1 likely represents a remnant of a once-functional MHC class I gene in New World primates that has been inactivated in the lineage leading to the tamarins. To investigate the evolutionary origin of the unusual tamarin MHC class I genes, we have cloned and sequenced 26 MHC class I cDNAs from seven genera of New World primates.
MATERIALS AND METHODS
Samples and Cell Culture.
Whole blood was obtained by venipuncture from one saddle-backed tamarin (Saguinus fuscicollis), one common marmoset (Callithrix jacchus), one golden lion tamarin (Leontopithecus rosalia), one owl monkey (Aotus trivirgatus), one squirrel monkey (Saimiri sciureus), one spider monkey (Ateles belzebuth), and one saki monkey (Pithecia pithecia). The infraorder Platyrrhini (New World primates) is divided in two families, Atelidae and Cebidae (25). Atelidae is divided into two subfamilies, Atelinae and Pitheciinae; in turn, Atelinae is divided into tribes Alouattini (Alouatta) and Ateline (Ateles, Brachyteles, and Lagothrix), and Pitheciinae is divided into tribes Callicebine (Callicebus) and Pitheciini (Pithecia, Chiropotes, and Cacajao). The family Cebidae is divided into four subfamilies, Cebinae (Cebus), Saimiriinae (Saimiri), Aotinae (Aotus), and Callitrichinae (Saguinus, Leontopithecus, Callimico, Callithrix, and Cebuella). The animals were originally housed at the Oak Ridge Associated Universities (Oak Ridge, TN; S. fuscicollis), the National Zoo (Washington DC; L. rosalia), the New England Regional Primate Research Center (Southborough, MA; C. jacchus, A. trivirgatus, and S. sciureus), Zoológico Santacruz (Mesitas del Colegio, Colombia; A. belzebuth), and Roger Williams Park Zoo (Providence, RI; P. pithecia). Peripheral blood lymphocytes were separated from whole blood using Ficoll/diatrizoate gradient centrifugation. These cells were transformed with Epstein–Barr virus by culturing peripheral blood lypmhocytes with supernatants of the cell line B958. Peripheral blood lypmhocytes were also cultured with concanavalin-A (5 μg/ml; Sigma) and interleukin-2 (20 units/ml; Roche). Both transformed and concanavalin-A-activated lymphocytes were cultured at 1 × 106 cell/ml in RPMI 1640 medium (GIBCO), supplemented with 10% fetal bovine serum (Sterile Systems), 2 mM l-glutamine, 5 × 10−5 M 2-mercaptoethanol, 20 mM Hepes, 50 units/ml penicillin, and 50 μg/ml streptomycin (Sigma).
RNA Extraction, cDNA synthesis, Cloning, and Sequencing.
Total cellular RNA was extracted from 5 × 106 lymphocytes using RNAzol (Tel-Test). One microgram of total RNA was used to synthesize cDNA using the Access reverse transcription–PCR kit (Promega). The cDNA synthesis reaction contained 50 mM Tris (pH 8.3), 5 mM MgCl2, 0.2 mM each deoxyribonucleotide, 1 μM each PCR primers 3′UTRI and 5′LPHIII (19), 1 mM MgSO4, 5 units of avian myeloblastosis virus reverse transcriptase, and 5 units of TfI DNA polymerase. For the first-strand cDNA synthesis, the reaction mixture was incubated at 48°C for 45 min, followed by heating at 94°C for 2 min. The second-strand synthesis and PCR amplification were carried out for 40 cycles as follows: denaturing at 94°C for 30 sec, annealing at 60°C for 1 min, and extension at 68°C for 2 min. After amplification, PCR products were ligated into the pCR II vector (Invitrogen), and at least three identical copies of each cDNA were sequenced using fluorescent dye-labeled dideoxy terminators (Applied Biosystems) in a model 373 automated DNA sequencer (Applied Biosystems). On average, 30 cDNA clones per animal were sequenced.
Evolutionary Analysis.
Gene trees were constructed using the neighbor-joining method (26), based on the number of nucleotide substitutions per site (d), estimated by the method of Jukes and Cantor (27). In the estimation of d, sites were excluded from the analysis when the alignment indicated a gap. The reliability of clustering patterns in the tree was tested by the standard error test for internal branches (28). The number of synonymous substitutions per synonymous site (dS) and the number of nonsynonymous substitutions per nonsynonymous site (dN) were estimated by the method of Nei and Gojobori (29), and SEMs for d, dS, and dN were estimated by the method of Nei and Jin (30).
Radiolabeling, Immunoprecipitation, and One-Dimensional Isoelectric Focusing (1-D IEF).
MHC class I glycoproteins from Callitrichinae (S. fuscicollis, C. jacchus, and L. rosalia) Epstein–Barr virus-transformed peripheral blood lymphocytes were labeled with [35S]methionine, immunoprecipitated, and subjected to 1-D IEF, as described before (31). To immunoprecipitate Callitrichinae MHC class I products, we used mouse and rabbit antibodies directed against human MHC class I products. The BB7.7 antibody-producing cell line was obtained from the American Type Culture Collection (ATCC; Rockville, MD). Rabbit antiserum directed against human β2-microglobulin (Dako) and denatured human MHC class I molecules (provided by H. Ploegh, Netherlands Cancer Institute, Amsterdam, The Netherlands) were also used. Full-length Callitrichinae MHC class I cDNAs were subcloned into the CDM8 expression vector (32) and COS cells (SV40-transformed simian CV-1 cells; ATCC) were transfected using the diethylaminoethyl/dextran method (33). Expressed MHC class I molecules were precipitated using the rabbit antiserum directed against denatured MHC class I heavy chains and focused using 1-D IEF.
RESULTS
Low Variability of Callitrichinae MHC Class I Genes.
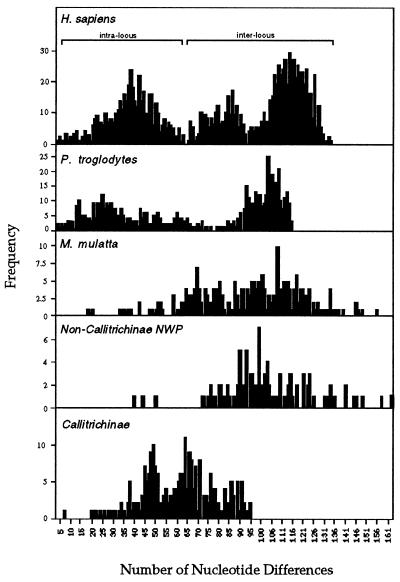
We cloned and sequenced 26 MHC class I cDNAs from seven species of New World primates. These primates include representatives from five of the six subfamilies of the infraorder Platyrrhini (25). Comparison of the New world primate MHC class I cDNAs revealed an unusually limited diversity in species from the subfamily Callitrichinae (S. fuscicollis, C. jacchus, and L. rosalia) that is not seen in other New World primate species. Indeed, the frequency distribution of nucleotide differences of MHC class I sequences of New World primates (Fig. 1) showed that inter- and intralocus, as well as interspecies, comparisons in Callitrichinae have a distribution that corresponds to the intralocus distribution seen in HLA sequence comparisons. In contrast, in the other species of New World primates (S. sciureus, A. trivirgatus, A. belzebuth, and P. pithecia), the frequency distribution of differences corresponds to interlocus comparisons of HLA alleles. Additionally, the average number of nucleotide substitutions in pairwise comparisons of MHC class I cDNAs of New World primates (Table 1) shows that cDNAs from genera of the subfamily Callitrichinae display a much more limited variability than do their counterparts in other New World primate genera. This indicates that the limited variability seen in the cotton-top tamarin MHC class I genes (21) is a general feature of all Callitrichinae. Analysis of the pattern of nucleotide substitutions in the Platyrrhini MHC class I cDNAs (Table 2) revealed an elevated rate of nonsynonymous substitutions (dN > dS) in the peptide-binding region (PBR) of these molecules. This indicates that positive Darwinian selection is acting to diversify the PBR of the Platyrrhini MHC class I genes (34). Thus, despite the limited sequence variability of MHC class I molecules in the Callitrichinae, natural selection promotes the diversification of the PBR of these proteins.
Figure 1.
Frequency distribution of numbers of pairwise nucleotide differences in primate MHC class I sequences. The frequency histogram was obtained from all possible pairwise comparisons of full-length sequences between 50 HLA-A, -B, and -C alleles, 25 chimpanzee (Pan troglodytes) Patr-A, Patr-B, and Patr-C alleles, 20 rhesus monkey (Macaca mulatta) Mamu-A and Mamu-B alleles, 15 non-Callitrichinae New World primate sequences, and 22 Callitrichinae sequences. Distribution of inter- and intralocus nucleotide differences in human are indicated. NWP, New World primate.
Table 1.
Mean numbers of nucleotide substitutions per 100 sites (d ± SEM), per 100 synonymous sites (dS ± SEM), and per 100 nonsynonymous sites (dN ± SEM) in pairwise comparisons of MHC class I alleles from Callitrichinae and non-Callitrichinae New World primates
| d | dS | dN | |
|---|---|---|---|
| Within-species comparisons | |||
| Callitrichinae | |||
| S. oedipus | 4.1 ± 0.3 | 4.4 ± 0.7 | 4.0 ± 0.4 |
| S. fuscicollis | 4.6 ± 0.5 | 5.3 ± 1.1 | 4.4 ± 0.6 |
| C. jacchus | 5.6 ± 0.5* | 7.3 ± 1.2 | 5.0 ± 0.5 |
| Non-Callitrichinae | |||
| A. trivirgatus | 8.3 ± 0.8† | 11.4 ± 1.9† | 7.4 ± 0.9† |
| P. pithecia | 8.7 ± 0.6† | 10.1 ± 1.4† | 8.3 ± 0.7† |
| A. belzebuth | 12.5 ± 0.8† | 17.2 ± 2.0† | 11.0 ± 0.9† |
| Between-species comparisons | |||
| Callitrichinae | |||
| S. oedipus vs. S. fuscicollis | 4.8 ± 0.3 | 5.4 ± 0.7 | 4.7 ± 0.4 |
| S. oedipus vs. C. jacchus | 6.2 ± 0.5‡ | 8.4 ± 1.2 | 5.6 ± 0.5 |
| S. fuscicollis vs. C. jacchus | 6.4 ± 0.5§ | 9.0 ± 1.2 | 5.6 ± 0.5 |
| Non-Callitrichinae | |||
| A. trivirgatus vs. P. pithecia | 10.5 ± 0.6§ | 12.3 ± 1.2§ | 9.9 ± 0.7§ |
| A. trivirgatus vs. A. belzebuth | 10.7 ± 0.6§ | 14.5 ± 1.4§ | 9.5 ± 0.7§ |
| A. pithecia vs. A. belzebuth | 11.0 ± 0.6§ | 14.4 ± 1.4§ | 10.0 ± 0.6§ |
Test of the hypothesis that values of d, dS, or dN for within-species comparisons equal corresponding values for S. oedipus: *P < 0.05;
P < 0.001. Test of the hypothesis that values of d, dS, or dN for between-species comparisons equal corresponding values for S. oedipus. vs. S. fuscicollis:
P < 0.01;
P < 0.001.
Table 2.
Mean number of nucleotide substitutions per 100 synonymous sites (dS ± SEM) and per 100 nonsynonymous sites (dN ± SEM)
| Comparison | PBR (n = 57)
|
α1 − α2 remainder (n = 125)
|
α3 (n = 92)
|
|||
|---|---|---|---|---|---|---|
| dS | dN | dS | dN | dS | dN | |
| Saoe-G | 4.3 ± 1.9 | 16.3 ± 1.9* | 5.0 ± 1.3 | 2.0 ± 0.4† | 4.0 ± 1.7 | 2.4 ± 0.7 |
| Safu-G | 2.7 ± 1.9 | 14.5 ± 2.5* | 7.0 ± 2.0 | 2.1 ± 0.6* | 4.2 ± 1.9 | 1.5 ± 0.6 |
| Caja-G | 10.0 ± 3.7 | 18.0 ± 2.8† | 8.2 ± 2.2 | 3.3 ± 0.8† | 4.8 ± 2.0 | 2.0 ± 0.7 |
| Lero-G | 0.0 ± 0.0 | 3.9 ± 1.8† | 4.3 ± 2.2 | 1.1 ± 0.6 | 11.9 ± 4.6 | 1.9 ± 1.0† |
| Sasc-G | 14.0 ± 6.1 | 24.2 ± 4.9 | 4.4 ± 2.2 | 2.2 ± 0.9 | 13.2 ± 4.8 | 5.4 ± 1.6 |
| Aotr-G | 8.3 ± 3.2 | 17.5 ± 2.9† | 10.3 ± 2.5 | 4.7 ± 0.9† | 11.2 ± 3.5 | 2.7 ± 0.8* |
| Atbe-G | 15.5 ± 5.4 | 38.3 ± 5.2* | 14.0 ± 3.4 | 8.5 ± 1.5 | 15.3 ± 4.2 | 5.0 ± 1.3* |
| Pipi-G | 8.9 ± 3.3 | 21.5 ± 2.9* | 4.9 ± 1.6 | 5.1 ± 0.9 | 16.7 ± 3.9 | 4.5 ± 1.0* |
n, number of codons compared. Mean dS is significantly different from mean dN for the same region at the 5% (†) or 1% (∗) level.
Most of the MHC Class I Genes in New World Primates Are Related to HLA-G.
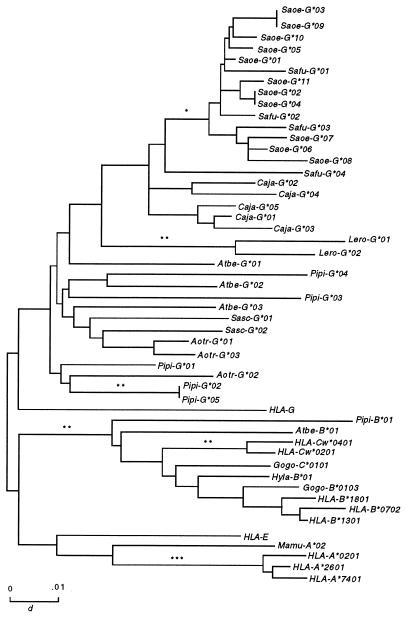
To investigate the evolutionary relationships of the MHC class I cDNA isolated, we constructed gene trees from exon 4 to exon 8 (Fig. 2). This region contains the majority of the locus-defining motifs (35), and consequently, tree construction with this region leads to a better understanding of the phylogenetic relationships among different loci. The gene tree showed that most of the Platyrrhini MHC class I loci were related to the human nonclassical HLA-G locus, as was previously shown in the cotton-top tamarin (21). Thus, because all the Catarrhini express classical MHC class I loci that are orthologous to HLA-A and -B (36), this finding indicates that different MHC class I loci were expanded in the Platyrrhini and Catarrhini. Surprisingly, two cDNAs, Pipi-B*01 (Pithecia pithecia) and Atbe-B*01 (Ateles belzebuth), clustered significantly with the primate MHC class I B and C alleles. Because the C locus appeared in hominoids (apes and humans) after their divergence from Old World monkeys (3), it its likely that these two New World primate cDNAs are orthologous to HLA-B. This suggests that the family Atelidae may represent an intermediate stage between the G locus expansion in the Platyrrhini and the A, B, and C loci expansion in the Catarrhini.
Figure 2.
Gene trees of exons 4–8 of primate MHC class I alleles showing the genus-specific distribution of MHC class I sequences in the Callitrichinae. Test of significance of internal branches: ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001. Saoe, Saguinos oedipus; Safu, Saguinos fuscicollis; caja, callithrix jacchus; Lero, Leontopithecos rosalia; Sasc; Saimiri sciureus; Aotr, Aotus trivirgatus; Atbe, Ateles belzebuth; Pipi, Pithecia pithecia; Gogo, Gorilla gorilla; Hyla, Hylobates lar.
Absence of Orthology Among the Callitrichinae MHC Class I Loci.
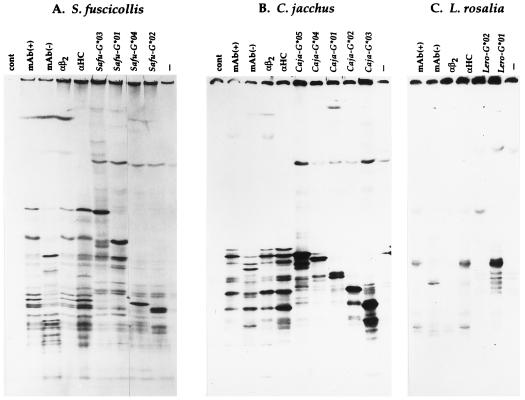
The most unusual feature of the gene tree of exons 4–8 is the clustering of Callitrichinae alleles in a genus-specific fashion (Fig. 2). Callitrichinae MHC class I loci are more similar to loci of the same genus than they are to those of a different genus. The MHC class I cDNAs of the saddle-backed tamarin (Safu-G) were closely related to those of the cotton-top tamarin (Saoe-G), suggesting that they expressed the same MHC class I loci. Interestingly, the common marmosets’ MHC class I cDNAs (Caja-G) did not cluster with the cotton-top tamarins’ cDNAs, but instead, they formed a separate clade. The genus-specific clustering was also observed in the golden lion tamarin’s sequences (Lero-G). This suggests that there is no orthologous relationships between MHC class I loci in the different genera of Callitrichinae. Nevertheless, MHC class I cDNAs of genera Aotus, Saimiri, Ateles, and Pithecia cluster in a transgeneric fashion, indicating that these primates share their MHC class I loci, i.e., they have orthologous MHC class I loci. Because common marmosets and cotton-top tamarins probably shared a common ancestor 5–10 million years ago (37), it was surprising that the MHC class I genes in these two species are not orthologous. Chimpanzees and humans, who last shared a common ancestor at approximately the same time that cotton-top tamarins and common marmosets did (38), express MHC class I genes from almost identical HLA-A, -B, and -C loci (9, 39). To evaluate whether the Callitrichinae MHC class I cDNAs isolated were the actual expressed genes or whether they were nonfunctional genes, we transfected COS cells with the MHC class I cDNAs sequenced from each Callitrichinae species (Fig. 3). Each transfected cDNA produced a band on the 1-D IEF gel that had the same isoelectric point as MHC class I molecules expressed by the Epstein–Barr virus-transformed cell line from which the RNA was extracted. This suggests that we had, indeed, isolated MHC class I molecules expressed by the Callitrichinae cell lines. Thus, each Callitrichinae genus expresses their own set of MHC class I genes, with no orthology among the various genera.
Figure 3.
1-D IEF of Callitrichinae MHC class I molecules. Transfections of Callitrichinae MHC class I PCR-generated cDNAs from S. fuscicollis (A), C. jacchus (B), and L. rosalia (C). Biosynthetically labeled MHC class I molecules were precipitated with an antiserum directed against human β2-microglobulin (αβ2), an antiserum generated against denatured MHC class I H chains (αHC), or the monoclonal antibody BB7.7 [mAb(+)]. Transfected MHC class I molecules were precipitated from denatured lysates with the antiserum generated against denatured human H chains. Precipitates were treated with neuraminidase in all cases except one [mAb(−)]. These precipitates were then subjected to 1-D IEF.
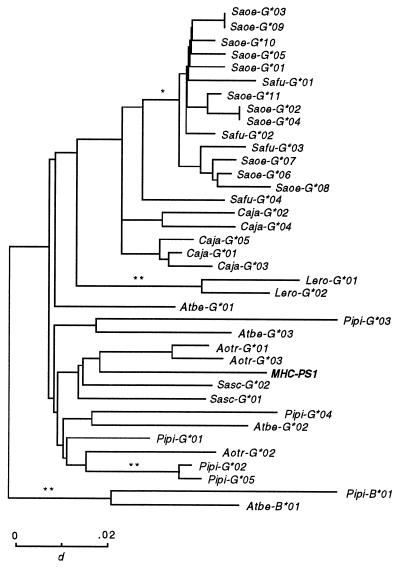
The Tamarin MHC-PS1 Is Related to Two Owl Monkey Expressed MHC Class I Genes.
To investigate the phylogenetic relationships of the tamarin MHC class I processed pseudogene (MHC-PS1), we constructed a gene tree of exons 4–8, including the New World primate MHC class I alleles and the MHC-PS1-processed pseudogene (Fig. 4). Interestingly, MHC-PS1 did not cluster with any of the cotton-top tamarins’ MHC class I alleles, but it clustered with two expressed MHC class I alleles from the owl monkey (Aotus trivirgatus). This finding is interesting because MHC-PS1 was present only in the genome of species from the genus Saguinus, and no functional counterparts were found among the expressed MHC class I genes in the tamarin. This indicates that the gene that gave rise to MHC-PS1 is no longer functional in the Saguinus species and, likely, is orthologous to the expressed owl monkey MHC class I genes.
Figure 4.
Gene tree of exons 4–8 of New World primate MHC class I sequences. The MHC class I processed pseudogene MHC-PS1 clusters with the expressed owl monkey MHC class I alleles. Test of significance of internal branches: ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001.
DISCUSSION
Analysis of the MHC class I genes of New World primates revealed an unusual evolutionary history of this gene family in tamarins and marmosets. The genera of the subfamily Callitrichinae express MHC class I genes with limited variability, compared with their counterparts in other genera of New World primates and in the Catarrhini. Moreover, each Callitrichinae genus seems to express a different set of MHC class I loci, with no apparent orthology among them. This remarkable feature contrasts with the stability of the Catarrhini or even the non-Callitrichinae New World primate MHC class I loci. Orthologues of HLA-A, -B, -E, -F, and -G have been conserved for more than 35 million years, throughout the evolution of Old World primates, apes, and humans. In addition, the transgeneric distribution of MHC class I loci in the non-Callitrichinae suggests that some of the Platyrrhini have also preserved their loci during several speciation events. Both the limited variability of the Callitrichinae MHC class I alleles and the apparent independent origin of the MHC class I loci of the genera Saguinus, Callithrix, and Leontopithecus may be explained by a very high rate of turnover of MHC class I loci. A rapid process of duplication, followed by differential fixation of loci, may account for the lack of orthology among the MHC class I genes of the Callitrichinae genera. Although the MHC class I loci evolve by a continual process of duplication and subsequent inactivation of loci (40), this process appears to be highly accelerated in the Callitrichinae. Consequently, the recently duplicated MHC class I genes have not had enough time to generate the variability that is seen in other primate taxa, and a pattern of limited inter- and intralocus variability should eventually arise. The inability to assign allelic relationships and to differentiate loci in tamarins and marmosets support the fairly recent origin of MHC class I genes in these genera. Recent divergence of MHC class I genes has also been reported in the koala (17) and in the horse (41). The limited variability and the lack of orthologous relationships seen in the Callitrichinae MHC class I loci is, therefore, consistent with a rapid process of duplication and differential expansion of loci. Our data indicate that, in approximately 5 million years, a complete turnover of MHC class I genes has occurred in three genera of the subfamily Callitrichinae.
The tamarin MHC class I processed pseudogene MHC-PS1 clustered with sequences from the owl monkey and was not closely related to any of the expressed MHC class I genes in the Saguinus species. This suggests that these owl monkey genes may be the functional counterparts of the MHC-PS1 processed pseudogene, supporting the view that there is a high rate of turnover of MHC class I loci in the Callitrichinae. Thus, MHC-PS1 probably represents a genomic remnant of a once active MHC class I gene that likely was orthologous to one of the currently expressed MHC class I loci in non-Callitrichinae New World primates.
A common feature of the subfamily Callitrichinae is that most of their births are fraternal twins. During gestation, placental fusion occurs resulting in a cross-circulation of genetically distinct hematopoietic elements (42). Therefore, these primates are born as bone marrow chimeras. The limited variability of the MHC class I loci in Callitrichinae might facilitate the coexistence of genetically different lymphocytes in the same individual. However, the higher rate of nonsynonymous over synonymous substitutions in the PBR indicates that positive selection acts by diversifying the PBR in Callitrichinae MHC class I genes. Therefore, it does not appear to be a cause–effect relationship between the limited variability of MHC class I genes and the establishment of bone marrow chimerism in the Callitrichinae.
As can be inferred from the lack of orthology of MHC class I genes between major vertebrate taxa, this region evolves by gene duplication and differential expansion of loci. In primates, ancestral duplication of MHC class I genes lead to the expansion of A and B loci in the Catarrhini and G-related loci in the Platyrrhini. Interestingly, homologues of HLA-B were found in two species of New World primates (Pithecia pithecia and Ateles belzebuth) from the family Atelidae. Because the other New World primates studied are from the family Cebidae and no locus that is orthologous to HLA-B was found, it is possible that the Atelidae represents a transitional stage between the complete expansion of G locus in the Cebidae and B locus in the Catarrhini. An ancestral duplication and differential expansion of MHC class II genes has also been proposed for Platyrrhini, indicating that the disruption of the orthology relationships is a general feature of the MHC in New World primates.
The MHC class I region evolves by cycles of turnover of loci in which each cycle may coincide with the adaptive radiation of a major phyletic group. The Callitrichinae genera appear to have an unusually high rate of turnover of MHC class I loci, in that they express their own set of MHC class I loci, with no orthology among loci from the different genera. The recent origin of MHC class I genes in this taxon, therefore, may account for the limited inter- and intralocus variability displayed by the MHC class I genes in Callitrichinae.
Acknowledgments
We thank Drs. Prabhat Seghal, David Lee Parritz, Neal Clapp, Lisa Forman, Anne Savage, and Haydy Monsalve for their help in obtaining blood samples. We also thank Jon Boyson for critical review of this manuscript. This work was supported by National Institutes of Health Grants RR00167 and DK44886 (to D.I.W.) and GM43940 (to A.L.H.).
ABBREVIATIONS
- MHC
major histocompatibility complex
- 1D IEF
one-dimensional isoelectric focusing
- PBR
peptide-binding region
Footnotes
References
- 1.Klein J. Natural History of the Major Histocompatibility Complex. New York: Wiley; 1986. [Google Scholar]
- 2.Parham P, Ohta T. Science. 1996;272:67–74. doi: 10.1126/science.272.5258.67. [DOI] [PubMed] [Google Scholar]
- 3.Boyson J E, Shufflebotham C, Cadavid L F, Urvater J A, Knapp L A, Hughes A L, Watins D I. J Immunol. 1996;156:4656–4665. [PubMed] [Google Scholar]
- 4.Boyson J E, McAdam S N, Gallimore A, Golos T G, Liu X, Gotch F M, Hughes A L, Watkins D I. Immunogenetics. 1995;41:59–68. doi: 10.1007/BF00182314. [DOI] [PubMed] [Google Scholar]
- 5.Otting N, Bontrop R E. Immunogenetics. 1993;38:141–145. doi: 10.1007/BF00190901. [DOI] [PubMed] [Google Scholar]
- 6.Corell A, Morales P, Martínez-Laso J, Martín-Villa J M, Varela P, Paz-Artal E, Allende L M, Rodríguez C, Arniz-Villena A. Hum Immunol. 1994;41:52–55. doi: 10.1016/0198-8859(94)90084-1. [DOI] [PubMed] [Google Scholar]
- 7.Cadavid L F, Watkins D I. In: Molecular Biology and Evolution of Blood Group and MHC Antigens in Primates. Blencher A, Klein J, Socha W W, editors. Berlin: Springer; 1997. pp. 339–357. [Google Scholar]
- 8.Chen Z W, McAdam S N, Hughes A L, Dogon A L, Letvin N L, Watkins D I. J Immunol. 1992;148:2547–2554. [PubMed] [Google Scholar]
- 9.Lawlor D A, Ward F E, Ennis P D, Jackson A P, Parham P. Nature (London) 1988;335:268–271. doi: 10.1038/335268a0. [DOI] [PubMed] [Google Scholar]
- 10.Lawlor D A, Warren E, Ward F E, Parham P. Immunol Rev. 1990;113:147–185. doi: 10.1111/j.1600-065x.1990.tb00040.x. [DOI] [PubMed] [Google Scholar]
- 11.McAdam S N, Boyson J E, Liu X, Garber T L, Hughes A L, Bontrop R E, Watkins D I. J Immunol. 1995;154:6421–6429. [PubMed] [Google Scholar]
- 12.van Erp S H M, Dixon B, Figueroa F, Egberts E, Stet R J M. Immunogenetics. 1996;44:49–61. doi: 10.1007/BF02602656. [DOI] [PubMed] [Google Scholar]
- 13.Dixon B, van Erp S H M, Rodrigues P N S, Egberts E, Stet R J M. Dev Comp Immunol. 1995;19:109–133. doi: 10.1016/0145-305x(94)00056-l. [DOI] [PubMed] [Google Scholar]
- 14.Flajnik M F, Canel C, Kramer J, Kasahara M. Proc Natl Acad Sci USA. 1991;88:537–541. doi: 10.1073/pnas.88.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grossberger D, Parham P. Immunogenetics. 1992;36:166–174. doi: 10.1007/BF00661093. [DOI] [PubMed] [Google Scholar]
- 16.Kaufman J, Andersen R, Avila D, Engberg J, Lambris J, Salomonsen J, Welinder K, Skjødt K. J Immunol. 1992;148:1532–1546. [PubMed] [Google Scholar]
- 17.Houlden B A, Greville W D, Sherwin W B. Mol Biol Evol. 1996;13:1119–1127. doi: 10.1093/oxfordjournals.molbev.a025674. [DOI] [PubMed] [Google Scholar]
- 18.Watkins D I, Hodi F S, Letvin N L. Proc Natl Acad Sci USA. 1988;85:7714–7718. doi: 10.1073/pnas.85.20.7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watkins D I, Garber T L, Chen Z W, Toukatly G, Hughes A L, Letvin N L. Immunogenetics. 1991;33:79–89. doi: 10.1007/BF00210819. [DOI] [PubMed] [Google Scholar]
- 20.Gyllensten U, Bergstrom T, Josefsson A, Sundvall M, Savage A, Blumer E S, Giraldo L H, Soto L H, Watkins D I. Immunogenetics. 1994;40:167–176. doi: 10.1007/BF00167076. [DOI] [PubMed] [Google Scholar]
- 21.Watkins D I, Chen Z W, Hughes A L, Evans M G, Tedder T F, Letvin N L. Nature (London) 1990;346:60–63. doi: 10.1038/346060a0. [DOI] [PubMed] [Google Scholar]
- 22.Trtková K, Mayer W, O’Huigin C, Klein J. Mol Phylogenet Evol. 1995;4:408–418. doi: 10.1006/mpev.1995.1038. [DOI] [PubMed] [Google Scholar]
- 23.Satta Y, Mayer W E, Klein J. J Mol Evol. 1996;42:648–657. doi: 10.1007/BF02338798. [DOI] [PubMed] [Google Scholar]
- 24.Cadavid L F, Hughes A L, Watkins D I. J Immunol. 1996;157:2403–2409. [PubMed] [Google Scholar]
- 25.Schneider H, Schneider M P C, Sampaio I, Harada M L, Stanhope M, Czelusniak J, Goodman M. Mol Phylogenet Evol. 1993;2:225–242. doi: 10.1006/mpev.1993.1022. [DOI] [PubMed] [Google Scholar]
- 26.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 27.Jukes T H, Cantor R C. In: Mammalian Protein Metabolism. Munro H N, editor. New York: Academic; 1969. pp. 21–132. [Google Scholar]
- 28.Rzhetsky A, Nei M. Mol Biol Evol. 1992;9:945–967. [Google Scholar]
- 29.Nei M, Gojobori T. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 30.Nei M, Jin L. Mol Biol Evol. 1989;6:290–300. doi: 10.1093/oxfordjournals.molbev.a040547. [DOI] [PubMed] [Google Scholar]
- 31.Watkins D I, Kannagi M, Stone M E, Letvin N L. Eur J Immunol. 1988;18:1425–1432. doi: 10.1002/eji.1830180919. [DOI] [PubMed] [Google Scholar]
- 32.Seed B. Nature (London) 1997;329:840–842. [Google Scholar]
- 33.Watkins D I, Letvin N I, Hughes A L, Tedder T F. J Immunol. 1990;144:1136–1143. [PubMed] [Google Scholar]
- 34.Hughes A L, Nei M. Nature (London) 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 35.Parham P, Lawlor D A, Lomen C E, Ennis P D. J Immunol. 1989;142:3937–3950. [PubMed] [Google Scholar]
- 36.Watkins D I. Crit Rev Immunol. 1995;15:1–29. doi: 10.1615/critrevimmunol.v15.i1.10. [DOI] [PubMed] [Google Scholar]
- 37.Hershkovitz P. Living New World Monkeys (Platyrrhini) with an Introduction to Primates. Vol. 1. Chicago: University of Chicago Press; 1977. [Google Scholar]
- 38.Goodman M. In: Primatology Today. Ehara A, Kimura T, Takenaka O, Iwamoto M, editors. New York: Elsevier; 1991. pp. 11–18. [Google Scholar]
- 39.Mayer W E, Jonker M, Klein D, Ivanyi P, van Seventer G, Klein J. EMBO J. 1988;7:2765–2774. doi: 10.1002/j.1460-2075.1988.tb03131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes A L, Nei M. Mol Biol Evol. 1989;6:559–579. doi: 10.1093/oxfordjournals.molbev.a040573. [DOI] [PubMed] [Google Scholar]
- 41.Ellis S A, Martin A J, Holmes E C, Morrison W I. Eur J Immunogenet. 1995;22:249–260. doi: 10.1111/j.1744-313x.1995.tb00239.x. [DOI] [PubMed] [Google Scholar]
- 42.Benirschke K, Anderson J M, Brownhill L C. Science. 1962;138:513–515. doi: 10.1126/science.138.3539.513. [DOI] [PubMed] [Google Scholar]