Abstract
In the United States it is not allowed to make public any patient-specific information without the patient's consent. This ruling has led to difficulty for those interested in sharing three-dimensional (3D) images of the head and brain since a patient's face might be recognized from a 3D rendering of the skin surface. Approaches employed to date have included brain stripping and total removal of the face anterior to a cut plane, each of which lose potentially important anatomical information about the skull surface, air sinuses, and orbits. This paper describes a new approach that involves a) definition of a plane anterior to which the face lies, and b) an adjustable level of deformation of the skin surface anterior to that plane. On the basis of a user performance study using forced choices, we conclude that approximately 30% of individuals are at risk of recognition from 3D renderings of unaltered images and that truncation of the face below the level of the nose does not preclude facial recognition. Removal of the face anterior to a cut plane may interfere with accurate registration and may delete important anatomical information. Our new method alters little of the underlying anatomy and does not prevent effective registration into a common coordinate system. Although the methods presented here were not fully effective (one subject was consistently recognized under the forced choice study design even at the maximum deformation level employed) this paper may point a way toward solution of a difficult problem that has received little attention in the literature.
Keywords: facial recognition, 3D rendering, mathematical morphology, MRI, ITK
1. Introduction
Publicly available image databases provide a valuable resource to the scientific community. Within the United States, however, it is illegal to make images publicly available if a patient might be personally recognized from those images. Prohibited image types specifically include “Full face photographic images and any comparable images” (Health Insurance Portability and Accountability Act [HIPAA] 2006). During a recent HIPAA summit, Steinberg noted that Magnetic Resonance (MR) images of the head may provide information comparable to that of photographic images since a patient might be recognized via rendering of the skin surface (Steinberg, 2006). This risk is of concern to many Institutional Review Boards, and may complicate the process of making image databases available to other investigators. This paper discusses methods of disguising facial features while simultaneously altering as little of the underlying anatomy as possible.
The risk of identifying an individual from a three-dimensional (3D) rendering of the skin surface of his/her MR image is difficult to quantify. Among clinicians, anecdotal reports abound. Understandably, however, such observations have usually not been reported in the literature. Several psychophysical studies have attempted to define which facial features are most important to recognition of an individual. Chelappa et al., (1995) concluded that the hair, eyes, mouth, and nasal shape are of significance. Sadr et al., also found eyebrows to be important (Sadr 2003). When images are incomplete, the upper portion of the face is more important than the lower portion (Shepherd et al. 1981). Coloring may also play a role (Yip 2001). It is important to note, however, that MR images do not include coloring or hair. The likelihood of indentifying an individual from a surface rendering of his/her MR is unknown.
One solution to the potential problem of subject identification is brain stripping, which provides an isolated brain segmentation without any of the associated bone, muscle, and skin. The disadvantage of this approach is that it loses essential information. Surface cerebrospinal fluid (CSF) is stripped, for example, precluding CSF volume analyses during aging or under conditions of pathology. Additional, potentially valuable information about the orbits, skull shape and ears are also lost. We therefore do not view brain stripping as an optimal solution to the problem of avoiding facial recognition.
A second solution is to define a coronal plane that cuts off the face, removing in entirety the forehead, the nose and the mouth. We have found however, that affine registration of such images is less good than registration of images that include the face. Moreover, information about the bony orbits, nasal cavities, and other underlying structures is inherently lost, and a study of these structures may be important to diseases such as hyperthyroidism, fibroid dysplasia, and other illnesses.
In general, we see it as desirable to provide a means of preventing facial recognition while simultaneously preserving as much of the underlying anatomy as possible. Almost no literature is available on the subject. This paper describes a fully automated method of deforming facial structures in MR images. We then analyze via an observer performance study how much facial deformation is required to prevent recognition of an individual when the face is rendered in three dimensions. We finally examine the efficacy of image registration under conditions of varying levels of deformation and under conditions in which the face is removed in entirety. Our results should be of interest to investigators interested in providing image databases of the head and brain to the general public.
2. Methods
Our image processing approach involves a) delineation of the skin-air interface, b) filling in this outline to create a solid, 3D “mask” that contains all of the non-background voxels, c) delineation of a coronal plane anterior to which the face lies, and d) deformation of the skin surface of the face. Please note that this approach will not alter any voxels belonging to the brain or to any anatomical structure posterior to the defined coronal plane. Dependent upon the level of deformation selected, the alteration will affect only the skin surface of the anterior face or may, with increasing deformation levels, affect progressively deeper facial structures. The remainder of the image data is retained unchanged.
2.1. Image acquisition
Photographic and MR images of ten healthy adult volunteers were used in an observer performance study. The study was approved by our Institutional Review Board, and all subjects provided signed consent. Any image shown in the current report is included with the individual's consent. Subjects included six males and four females, ranging in age from 22 to 74.
The face of each volunteer was digitally photographed from the front and from the left side. In the attempt to standardize the photographic images, all subjects were photographed against a solid white background. Individuals with glasses were asked to remove them, and subjects with long hair were asked to pull the hair back from the face during the photographic session.
T1 and T2 12-bit greyscale images were also obtained of each volunteer. Images were acquired on a Siemens head-only 3T system (Allegra, Siemens Medical System Inc.) with a head coil. The imaging parameters were: TR/TE/TH=15msec/7msec/1mm for T1 and TR/TE/TH= 7730msec/ 80msec/1mm for T2. The inplane resolution was 1×1mm2 with an interslice spacing of 1mm. The size of each image was approximately 250×250×250 (about 15×106 voxels). Four of the ten subjects underwent MR imaging that included the entire face. The remaining six volunteers underwent imaging that included only the upper portion of the face, excluding the mouth and chin.
2.2. Extraction of the skin surface from the MR image and creation of the 3D mask
The purpose of this step was to define a solid, 3D “mask” that included all non-background voxels. The program was written using the National Library of Medicine's Insight Toolkit (ITK).
Three consecutive filters were employed to remove the background noise.
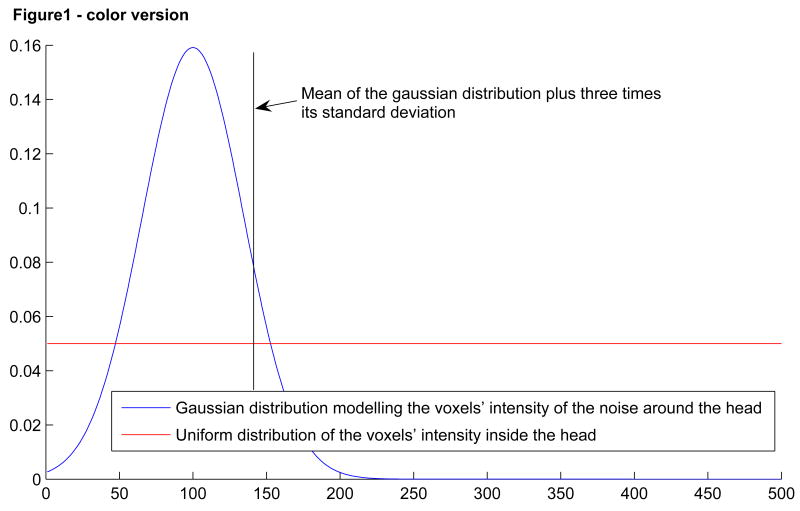
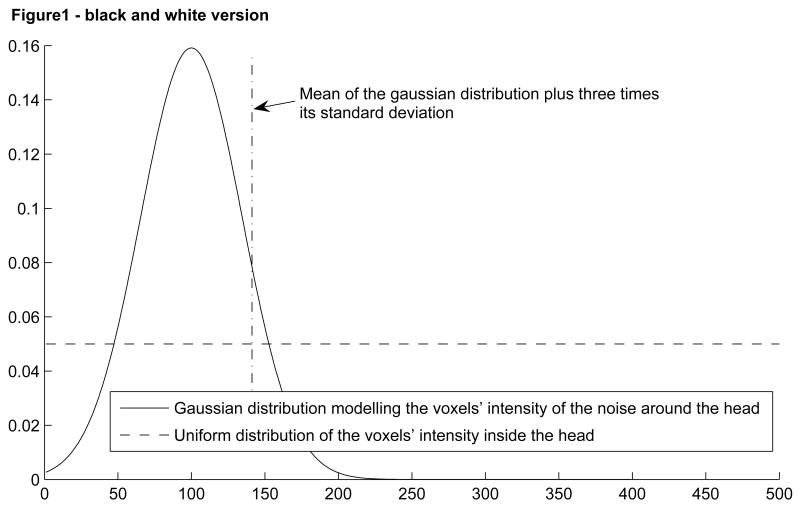
First, an intensity threshold was defined automatically. To find this threshold, we modeled the image intensities as a sum of two distributions: a Gaussian distribution representing the histogram of the voxel intensity of the noise surrounding the head and a uniform distribution modeling the histogram of the voxel intensity inside the head (Figure 1). The grayscale threshold was determined as the mean plus three times the standard deviation of the Gaussian curve. All pixels below this threshold were set to black. This threshold was not perfect and therefore some voxels were erased when they should not have been (voxels of image intensity lower than the threshold and lying inside the head) and others were not erased when they should have been (voxels of image intensity higher than the threshold and lying outside the head). To correct these errors, we applied two additional filters.
Figure 1.
Model of the voxels' intensity distribution and the threshold extracted
The second filter was a median filter (a rectangular kernel 5×5×3 with a radius of two voxels within each axial slice and one voxel between slices), used to darken small regions of noise outside the head and to brighten small dark regions within the head.
The final filter eliminated isolated bright points. For each point in the image, it counted the number of points linked to it that were brighter than the determined threshold in the first step. If the number of connected points was smaller than 10,0001, the image intensity at these points was set to 0. On the skin surface, bright points are adjacent to each other, and so such points were not erased.
Following this set of steps, only the contour of the head and points inside the head remained in the image. The filtered image was then binarized. Finally we filled in the contour of the head to obtain a mask (Figure 2). This final step of filling in the contour means that no “hole” should appear inside the head, even if a patient possesses a large, low density brain lesion.
Figure 2.


Axial (left) and sagittal (right) views of a binary facial mask.
To create a 3D model, the binary image was transformed into an isosurface using vtkContourFilter, a function of the Visualisation ToolKit (VTK) and was saved as a spatial object. The 3D rendering of the MRI could then be shown using the publicly available program SOViewer (Jomier et al). Figure 3 depicts a 3D rendering of the head whose mask is shown in Figure 2.
Figure 3.
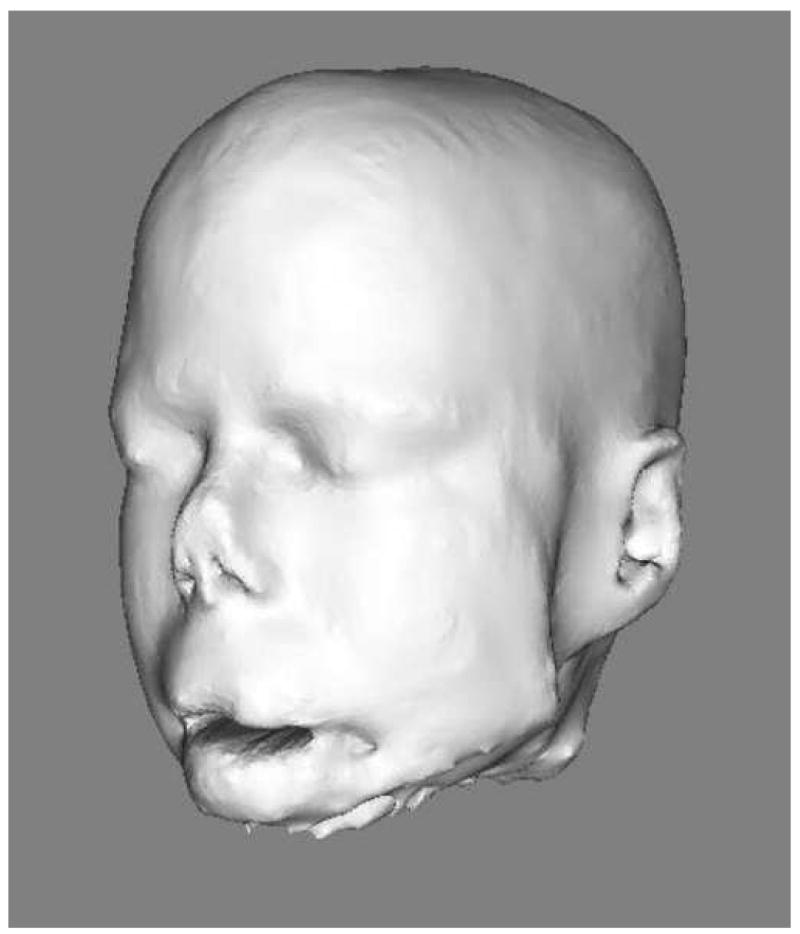
3D rendering of the head
2.3 Identification and deformation of the face
To deform the surface of the face without altering other structures requires identification of the face. We here employed a coronal plane that was initially manually defined in a reference image (Ir) so as to intersect the skin of the forehead, the nose, and the mouth. The reference image, together with its associated plane, was then registered with each image (I) we wished to deform (Figure 4). All pixels on the surface of the mask anterior to that plane were viewed as belonging to the face. The skin surface anterior to this plane was subsequently deformed whereas the remainder of the image was not. Although we could have deformed the surface of the entire head, we chose to alter only the face since facial features are generally most critical to recognition of an individual, since program execution was faster when only a portion of the skin surface was deformed, and since anatomical information was preserved for the other portions of the head.
Figure 4.
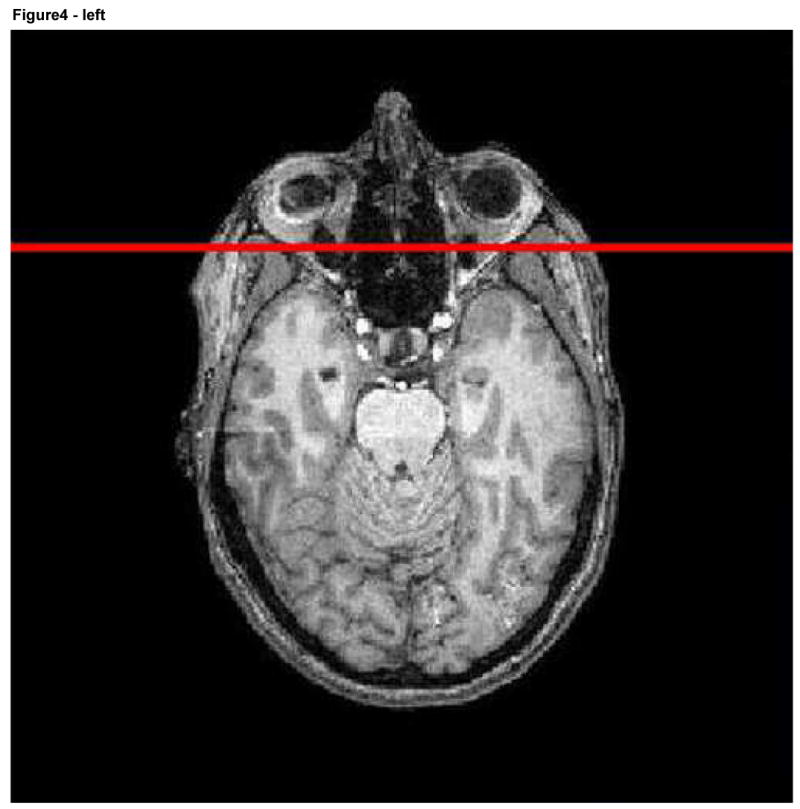
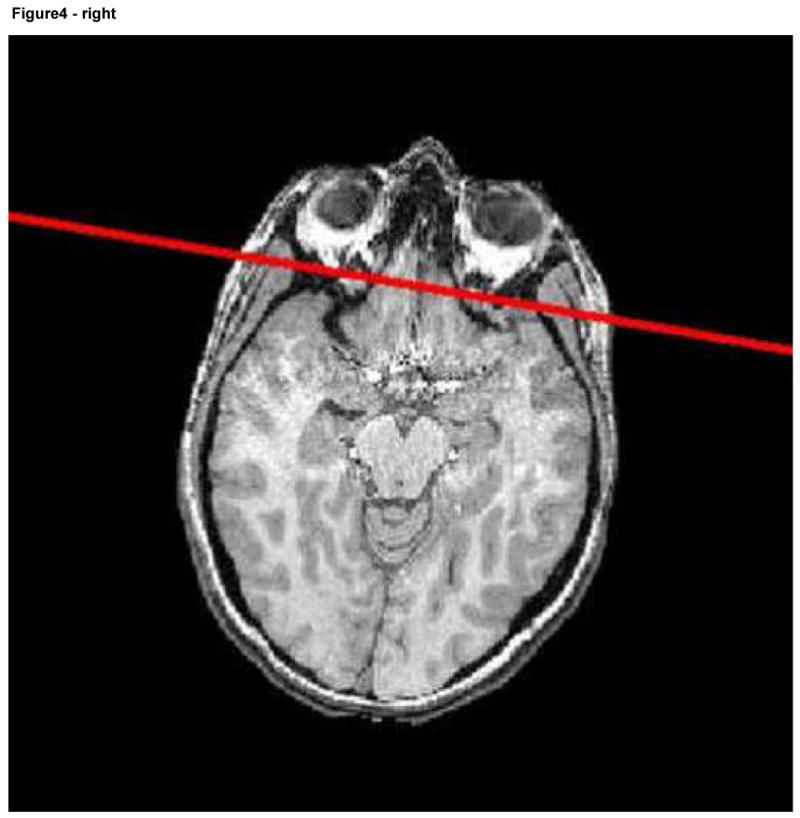
Reference image with the associated plane shown in red (left) and a second image including the same plane following registration.
The facial deformations themselves were produced by dilation and erosion (Serra, 1986), using a spherical kernel. Since dilation and erosion was applied to a binary mask, only the surface of the mask was altered. Kernel radius was selected by the user, with the same size kernel employed for both dilation and erosion. This approach allowed a graded amount of deformation dependent upon the size of the spherical kernel selected; the larger the radius the more information was removed. In all cases, an opening operation (first erosion, then dilation) was applied to the mask of the head to remove small bumps. Then a closing operation (first dilation, then erosion) was applied to remove small holes. This method smoothed the skin surface anterior to the defining plane while leaving the brain and the rest of the head intact. Since the kernel size used to deform the face in our experiment was small (radius between 4 and 12 voxels), the shape of the face was deformed only slightly. The voxels affected by the transformation were only on the surface of the face and belonged primarily to the skin, mouth and nose.
One potential problem was the assignment of image intensities to the new surface. Every point anterior to the plane defining the face might be changed. Image intensity values were assigned as outlined in Table 1.
Table 1.
Voxel grayscale value of the deformed mask surface
| Outside the deformed mask | Inside the deformed mask | |
|---|---|---|
| Inside the undeformed mask | Black | Unchanged |
| Outside the undeformed mask | Black | Gray value calculated with the neighborhood gray value |
For the cases “Inside the deformed mask” and “Outside the undeformed mask”, a new grayscale value must be calculated. During the deformation, grayscale information was saved for each voxel in the neighborhood. The grayscale value of the new point was picked randomly from the highest half of the preserved grayscale values to obtain realistic image intensity. Figures 5, 6, and 7 illustrate four levels of deformation for the same subject as shown in an axial slice, a sagittal slice, and in 3D.
Figure 5.
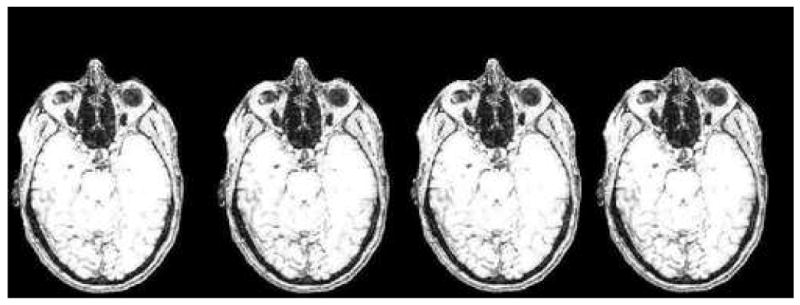
Axial slice of a progressively deformed image with deformation produced by different kernel sizes (from left to right: 0, 4, 8, 12 voxels). Note the progressive shortening of the nose. The brain, skull, orbital structures, deep nasal structures, and pharynx remain unaltered.
Figure 6.
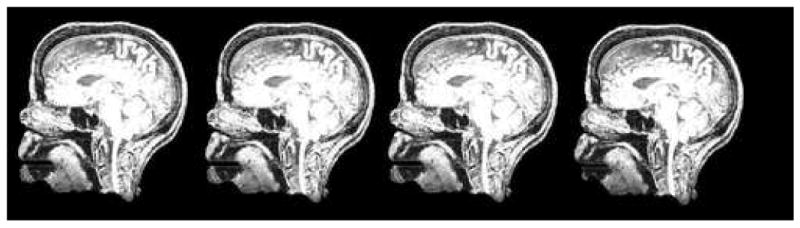
Sagittal slice of the same set of images shown in Figure 5.
Figure 7.
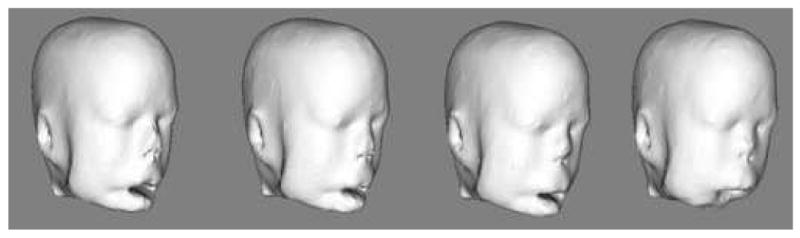
3D rendering of the same set of images shown in Figures 5 and 6.
2.4 Observer recognition study
The purpose of the observer recognition study was to determine how likely each of 10 different individuals was to be recognized from a 3D rendering of his/her facial features as defined from MR and as the underlying MR image was altered by progressive levels of facial deformation. The goal was to identify the lowest level of deformation that would preclude recognition of an individual subject. Four levels of facial deformation were applied to each original mask of the head including level 0 (undeformed), level 1 (spherical kernel with a 4 voxel radius), level 2 (8 voxel radius) and level 3 (12 voxel radius). Although additional and higher levels of deformation could have been included in this study we did not do so because of the additional time required for evaluation. Thirty-three volunteers participated in the observer recognition study.
The program employed in the observer recognition study is illustrated in Figure 8. Paired frontal and lateral facial photographs of the ten healthy imaged volunteers were displayed at the sides of the main window. Any photograph could be enlarged for closer inspection by clicking on it. The images had a resolution of 211 pixels by 200 pixels and 528 pixels by 500 pixels when enlarged. A central window displayed a 3D rendering of a head defined from an MRI of one of the healthy volunteers. This visualization could be rotated so as to be seen from any point of view by the observer. It could also be zoomed in and zoomed out to see more specific details. Since four levels of deformation were employed and images of ten subjects were analyzed, a total of forty 3D renderings were displayed consecutively by the program during each observer session. For each of these forty 3D visualizations, the observer was asked to identify the correctly associated pair of photographs. If the observer could not associate the 3D rendering with any photographic pair, he/she was asked to make a best guess among the ten possible choices.
Figure 8.
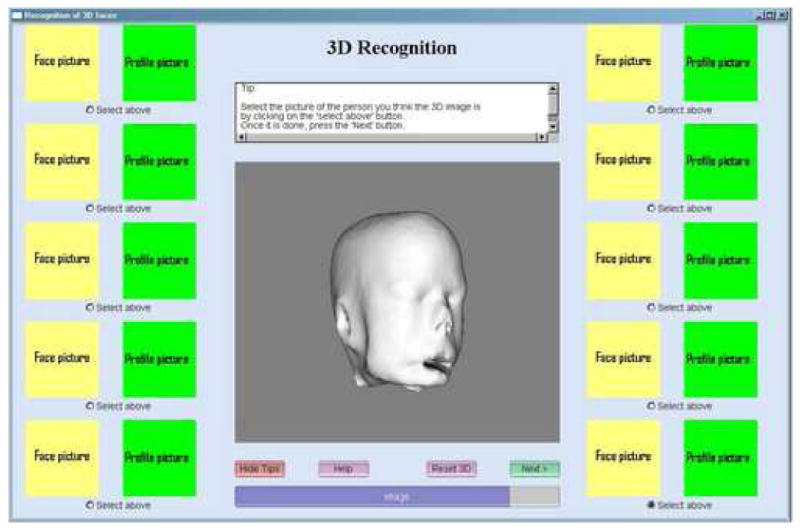
View of the program used in the observer recognition study. Photographs are not shown because we do not have permission from all participants to use their photographs in a publication.
This type of observer recognition study can introduce bias for a number of different reasons. In order to reduce bias, the following steps were employed:
The order of photographic presentation might affect the likelihood of selection of a particular pair of photographs. The order in which photographs were presented was therefore set to change randomly during each program run. For each individual observer, the photographs remained in the same position during the entire session. For the next observer running the program, however, the photographs were presented in a different order.
It is possible that knowing a person individually might aid an observer identify that person's 3D image. At the outset of the program, each observer was therefore asked to indicate (by clicking in a box) which of the 10 people he or she was acquainted with personally.
We assumed that it would be more difficult to associate a highly deformed image with the appropriate set of photographs than an undeformed image. If the undeformed images were presented first to each observer, the observer might note some feature that would allow him later to more readily associate a more deformed 3D image to the same photographic pair. The 10 most deformed images were therefore presented first to each observer, followed by the 10 next most deformed images, followed by the 10 least deformed images, and culminating in the 10 undeformed images.
It is possible that the order in which the 3D images were presented could affect the results. For each of the four deformation levels, the order in which the ten images were shown in 3D was therefore random.
Before the test, each observer was told to spend less than one minute on each 3D rendering. There was no mandatory time limit, but with this advice, it took between twenty and forty minutes to finish the test.
2.5. Statistical analysis
The two primary objectives of analysis were: a) to examine whether the selection was random for each imaged subject and at each deformation level, and b) to examine differences in the likelihood of facial recognition at different deformation levels.
To verify the hypothesis of random selection, we examined the observed probability of correct recognition for each imaged subject at each deformation level. A binomial test was used to determine whether the binomial proportion in each cell was equal to 1/10. The Bonferroni correction was used to adjust for type I error in the multiple tests where the significance level was set to be 0.05/40.
For the second objective, another generalized linear model with logistic link was used to fit this data. The outcome variable was correctness/incorrectness of matching the images with photographs and so was a dichotomous variable. The independent variable list included the 4 different deformation levels, whether the person in the photograph was known to the reader, and whether the subject's 3D image presented the entire face (4 subjects) or half face (6 subjects). A generalized estimating equation with the exchangeable working covariance was used to analyze the data.
2.6. Image registration
Many applications require registration of a test image to a healthy atlas. If appropriately anonymized images are made publicly available but the images cannot be readily registered into an atlas-based coordinate system, then the utility of the public image database could be severely compromised. Deforming or otherwise altering an image might well influence how well that image can be registered into another coordinate system, however. Since our approach to anonymizing image data minimizes deformation, we hypothesized that registration using our deformed images might be more accurate than registration under conditions in which the face has simply been removed.
We performed two different sets of registration evaluations that each compared our facial deformation method to a method based upon total face removal. In the first evaluation, all 40 images (these 40 images represented the 10 subjects at each of 4 deformation levels) were registered to the fuzzy McConnell T1 atlas (ICBM Atlas). An additional registration of each of the 10 subjects to the same atlas was also performed; in these cases, however, a plane was defined to separate the face from the head and all pixels anterior to that plane were set to black, thus removing the face entirely (Figure 9).
Figure 9.
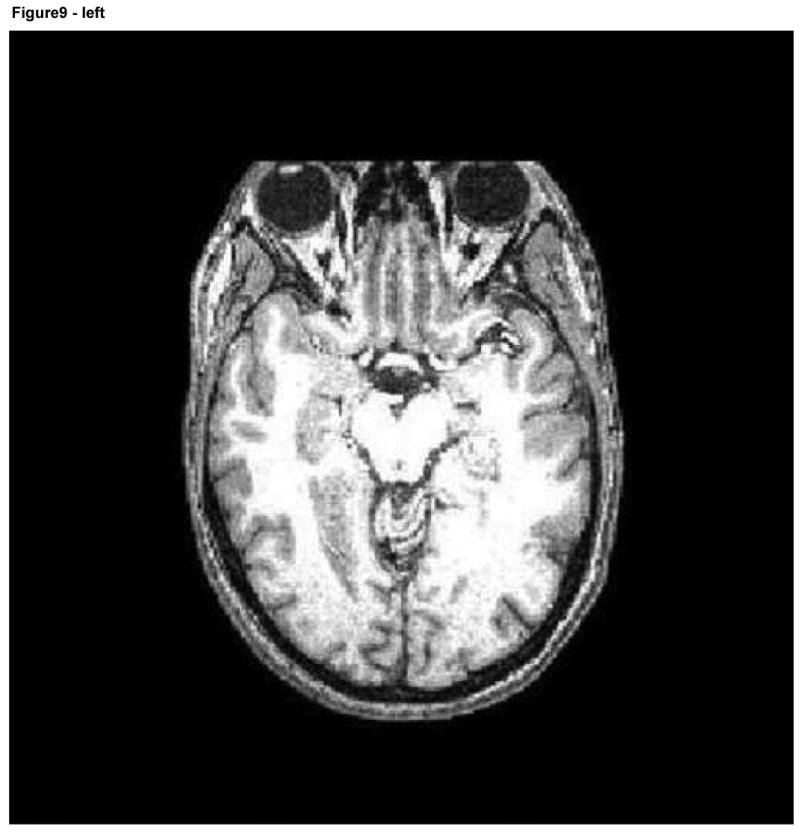
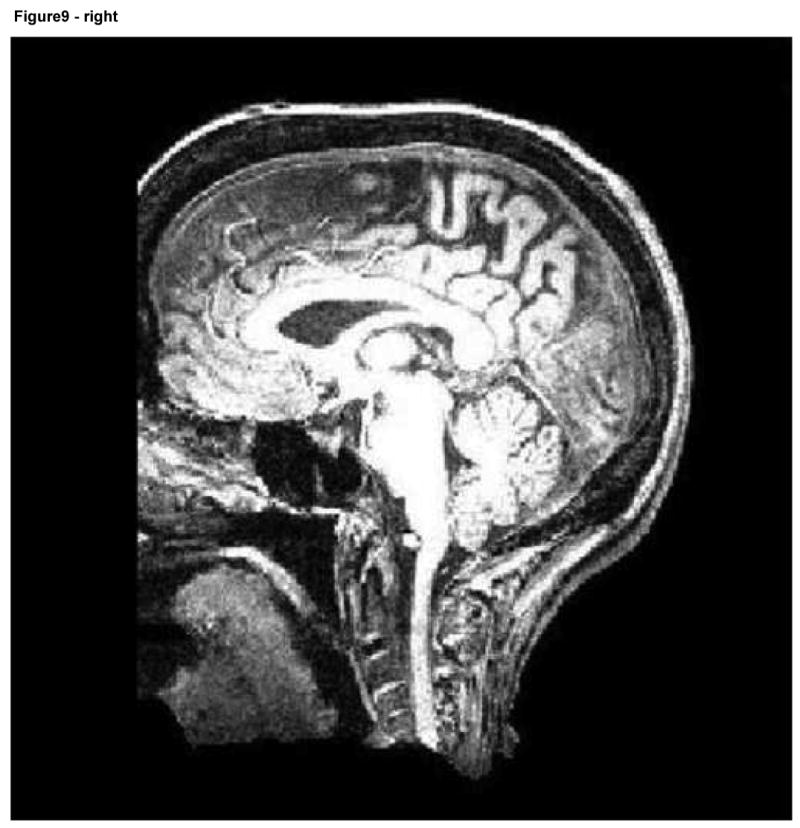
Axial and sagittal slices of an image following facial removal. Note the loss of information about the air sinuses, orbits, and anterior skull.
For the second registration evaluation, the same 50 images were registered to a single, sharp reference image of a different, subject. This approach was performed twice, using two different reference images to avoid bias. The rationale for evaluation against different target images (atlas and two sharp reference images) was that the fuzzy McConnell atlas represents a standard used by several groups but that other groups may sometimes employ a single, sharp image of an individual as the reference coordinate system.
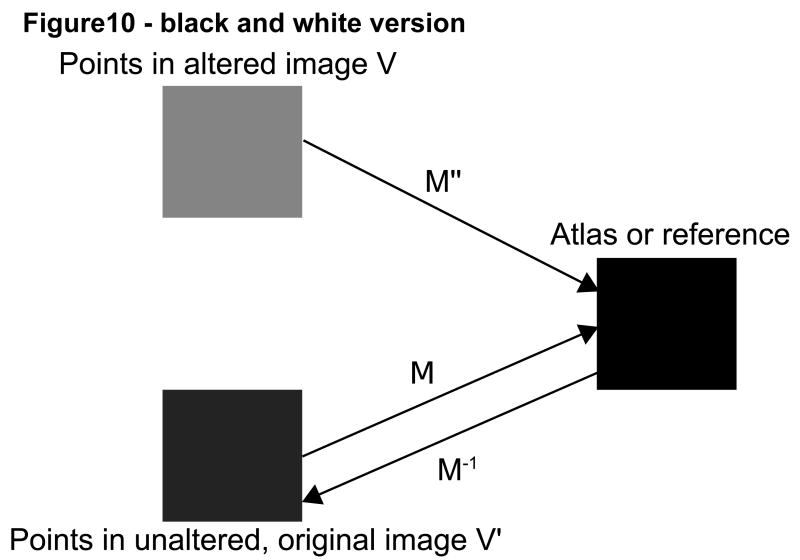
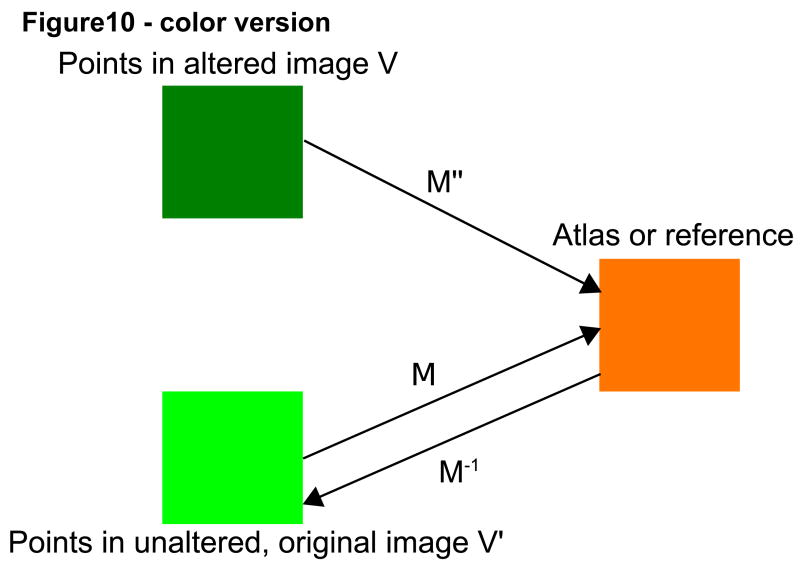
Registration was performed affinely, using the normalized mutual information method described by Rueckert and Schnabel [Rueckert; Schnabel et al, 2001]. In the absence of absolute ground truth, we viewed the registration matrix produced by registration of an unaltered test image to the target image as the “goldish standard of truth” for that particular subject. The results of registration with deformed images or with complete facial removal for the same subject were then compared to the “goldish standard” of registration of the same subject's unaltered image to the same target. The remainder of this section employs the term “altered image” to refer both to an image in which the skin surface has been deformed and to an image in which the face has been cut off. For each subject, 5000 randomly selected points were taken within each altered image and each point's position was compared following registration to the location of the same point following registration of the unaltered, original image. The closer the transformed points in each altered image were to the transformed points in the original image, the better the registration was judged to be. Figure 10 provides a graphical illustration of the methods employed.
Figure 10.
Graphical depiction of affine mappings between the original (V') and altered (V) images via registration to a common coordinate system (atlas or reference).
More specifically, 5000 randomly selected points (V) were selected in each altered image. We calculated the location of these points following registration to a target coordinate system, and then compared the location of these points to the locations of the same points in the unaltered image (V') after the unaltered, original image had been independently registered to the same target.
We then calculated the distance between the points in the altered image and their corresponding points in the unaltered one. If the matrices of registration were exactly the same for the altered and unaltered images, the distance should be zero. We calculated both the mean distance and the maximum distance between these point pairs.
3. Results
3.1. Facial recognition
Tables 2 and 3 summarize the results of the observer recognition study. This study had two objectives. The first was to determine how differing levels of facial deformation affected the probability of recognizing an individual subject and to assess how far such identification differed from random chance.
Table 2.
Summary of Facial Recognition Study from 33 Observers. A perfect recognition score given 33 raters would be 33 for each imaged subject at each level of deformation and 330 for the total value at each level of deformation.
| How many got it right out of 33 observers | Level of Deformation | |||
|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |
| Imaged Subject | ||||
| 0 | 6 | 3 | 1 | 2 |
| 1 | 7 | 7 | 8 | 2 |
| 2 | 4 | 4 | 3 | 5 |
| 3 | 6 | 3 | 3 | 0 |
| 4 | 3 | 2 | 2 | 4 |
| 5 | 10 | 5 | 0 | 3 |
| 6 | 2 | 6 | 6 | 4 |
| 7 | 9 | 15 | 6 | 2 |
| 8 | 7 | 8 | 8 | 3 |
| 9 | 17 | 24 | 15 | 14 |
| Total | 71 | 77 | 52 | 39 |
Table 3.
The predicted probability of correct recognition for each imaged subject at each deformation level. Each cell provides the selection probability (upper value) and the probability that this selection differed significantly from chance (lower value).
| Observed probability of recognition (two-sided p-value) according to LevelDeformation and ImageID | Level of Deformation | |||
|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |
| Imaged Subject | ||||
| 0 | 0.182
(0.137) |
0.091
(1) |
0.03
(0.25) |
0.061
(0.769) |
| 1 | 0.212
(0.042) |
0.212
(0.042) |
0.242
(0.014) |
0.061
(0.769) |
| 2 | 0.121
(0.567) |
0.121
(0.567) |
0.091
(1) |
0.152
(0.374) |
| 3 | 0.182
(0.137) |
0.091
(1) |
0.091
(1) |
0
(0.073) |
| 4 | 0.091
(1) |
0.061
(0.769) |
0.061
(0.769) |
0.121
(0.567) |
| 5 | 0.303
(0.001) |
0.152
(0.374) |
0
(0.073) |
0.091
(1) |
| 6 | 0.061
(0.769) |
0.182
(0.137) |
0.182
(0.137) |
0.121
(0.567) |
| 7 | 0.273
(0.004) |
0.455
(<0.0001) |
0.182
(0.137) |
0.061
(0.769) |
| 8 | 0.212
(0.042) |
0.242
(0.014) |
0.242
(0.014) |
0.091
(1) |
| 9 | 0.515
(<0.0001) |
0.727
(<0.0001) |
0.455
(<0.0001) |
0.424
(<0.0001) |
Results indicated that the hypothesis of random selection was only rejected for subject 5 at deformation level 0, subject 7 at deformation level 1, and subject 9 at all four deformation levels. Table 2 provides the number of correct choices made by the 33 raters at each deformation level for each of the 10 imaged subjects. The observed selection probability and the p-values are given in Table 3.
These results suggest that the majority of faces rendered from MR images appear to be difficult to recognize even without deformation. However, 3 of the 10 imaged subjects (subjects 5,7, and 9) could be recognized from 3D renderings of their MR images at no or at low levels of deformation, and this event should be avoided.
The second objective of this study was to establish differences in the ability to recognize subjects according to level of facial deformation and other parameters. Results indicate that the probability of correct recognition at deformation level 0-1 was significantly greater than the probability at deformation level 2-3. (p < 0.02). The small number of observations did not permit us to discriminate between levels 0 and 1 (p = 0.43) or between levels 2 and 3 (p = 0.08). In other words, the probability of recognizing an individual was approximately the same at levels 0 and 1 but was significantly more likely than at the higher deformation levels 3 and 4. These results suggest that deformations of level 2 or greater significantly reduce the chance that an individual's face can be recognized from 3D renderings of his/her MR image.
There was no significant difference in the correct association of 3D image with photographs if the subject was known to the observer (p =0.51). There was also no significant difference in the likelihood of facial recognition if the MR image included the entire face or truncated the face below the level of the nose (p =0.13).
The 3D images of one subject (case 9) were associated with the correct set of photographs more often than might be expected on the basis of chance even at deformation levels 2 and 3 (Table 3). Images of this volunteer's 3D renderings at deformation levels 2 and 3 are shown in Figure 11. As illustrated there, the degree of deformation is sufficiently marked to make it seemingly highly difficult to recognize the person as an individual. We address some of the issues inherent to the “forced choice” design of the observer study under the Discussion.
Figure 11.
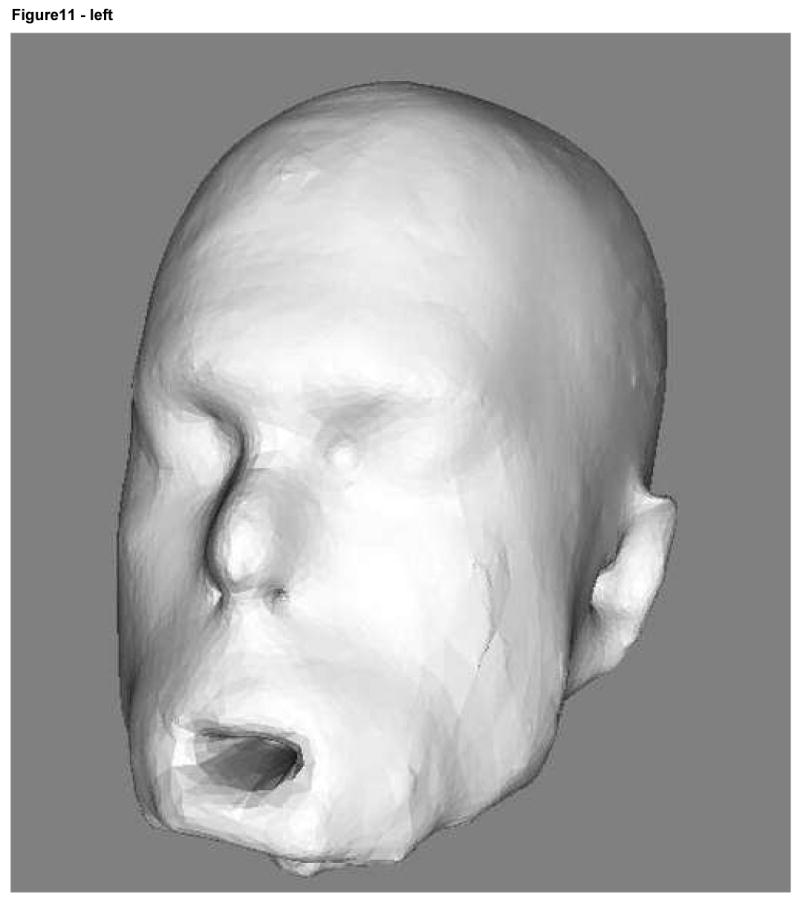
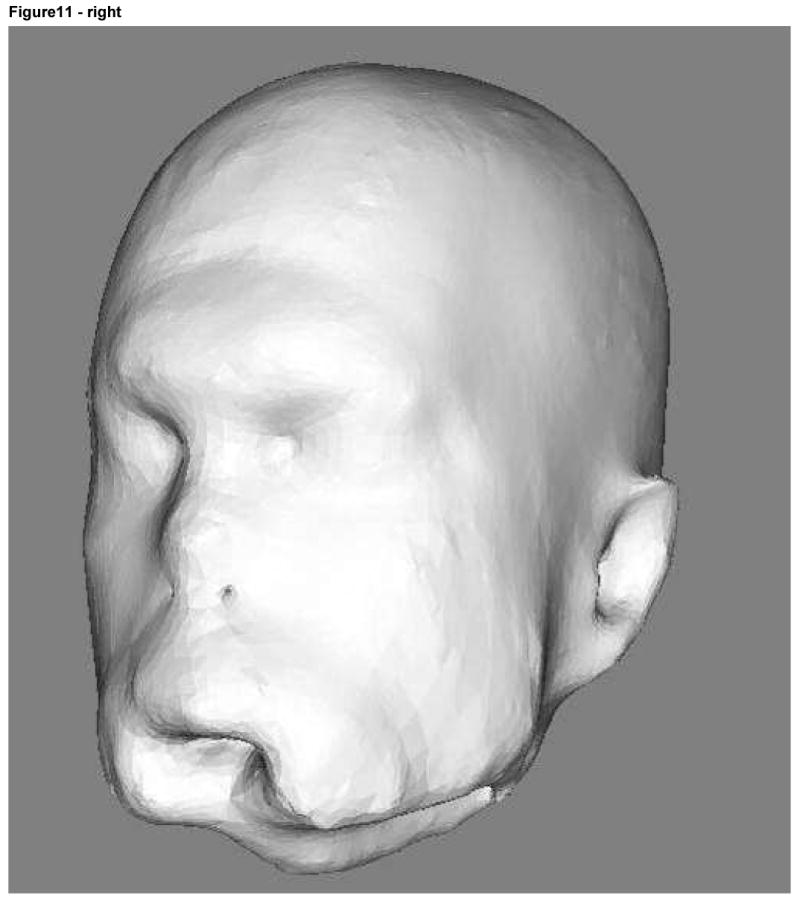
Subject 9 3D rendering at deformation level 2 (radius of 8 voxels) on the left and level 3 (radius of 12 voxels) on the right.
3.2. Registration
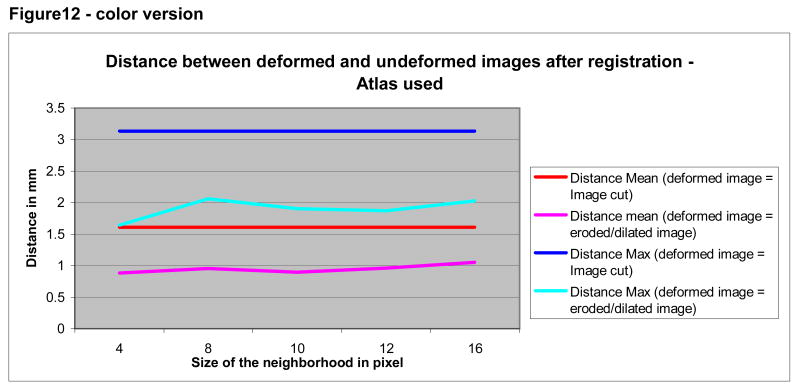
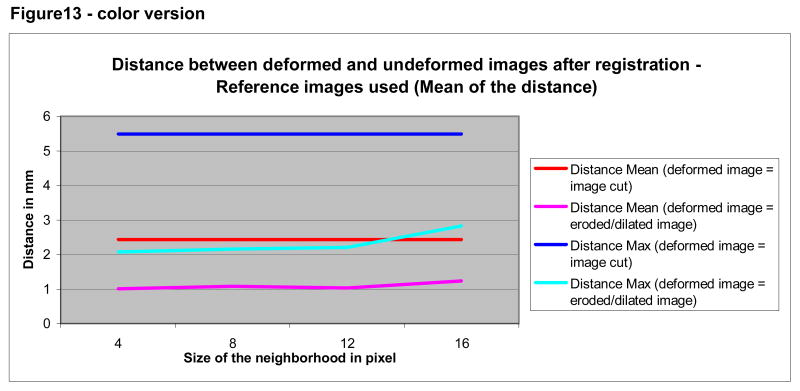
As shown by Figures 12 and 13, registration using a minimally deformed image produced more accurate results than those obtained following removal of the face. The mean and maximum distances as compared to “ideal registration” were small for both approaches However, the registration error for a deformed image was half the distance of that of a cut face (compare the blue curves, indicating to the maximum distance between the altered images and the undeformed images; compare the “red” curves indicating the mean distance).
Figure 12.
Mean and maximum distance between deformed and undeformed images according to method of deformation (erosion/dilation or removal of the face). Registration was to the ICBM atlas.
Figure 13.
Mean and maximum distance between deformed and undeformed images according to method of deformation (erosion/dilation or removal of the face). Registration was to reference images of two different individuals.
4. Discussion
For purposes of scientific analysis, it would be preferable to employ original, unmodified images from which no information has been removed. Ideally, each patient would provide consent to allow his/her images to be made publicly available, thus permitting widespread usage of unmodified images. Obtaining patient consent is not always feasible, however, and so, at least in the United States, it is often necessary to perform some form of image modification to prevent potential recognition of a patient's face.
This study is, to our knowledge, the first to address the risk of recognizing the facial features of an individual from a 3D surface rendering of his/her MR image of the head. MR images cannot provide information about the coloring of the skin, eyes, or hair, or about the configuration of hair and eyebrows--features often considered important when recognizing an individual (Chelappa 1995, Sadr 2003, Yip 2001). A three-dimensional (3D) rendering of the face from an MR image therefore inherently omits much of the information that human beings normally use to identify each another visually. On the basis of this preliminary study, we conclude that only about 30% of imaged patients may be at risk of identification on the basis of 3D renderings of the face. A 30% risk of identification is too high to permit indiscriminate publication of images of human patients, however.
Omission of the mouth and chin from the 3D image acquisition does not appear sufficient to protect a patient from facial identification. In our study, the chance of correctly identifying subjects with full face 3D image acquisitions did not differ significantly from the chance of identifying patients whose 3D image acquisitions omitted the mouth and chin (p=0.13). These results are consistent with the findings of Shepherd (1981), who concluded that when facial information is incomplete that the upper face is more important than the lower. Compatible with Shepherd's observations, our study also found that the presence of sharp and distinctive features (such as a large nose or an uncommon profile) appeared to be more important than inclusion of the entire face.
The most common means of intentionally obscuring the face in 2D photographs is to place a black bar over the eyes of the subject. This method is often used in articles and medical presentations that contain patient photographs. The approach produces an image in which the individual is presumed to be unrecognizable, thereby allowing publication of full face images while respecting the underlying HIPAA requirement of avoiding images that permit identification of a specific individual. This “black bar across the eyes” approach is obviously not appropriate to 3D images, however, since with rotation of a 3D image the subject's profile will remain unobscured and potentially recognizable. Similarly, recent work on the importance of using facial asymmetry to recognize individuals from video sequences (Mitra 2006) is not fully relevant to the task at hand since, even if a 3D facial rendering is altered to become perfectly symmetrical, the face may remain recognizable in profile.
The most reliable method of preventing facial recognition is to delete the face entirely. The two most common methods employed to date have been brain stripping, which deletes a great deal of anatomical information, and removal of the face anterior to an arbitrary plane, which deletes less anatomical information than brain stripping but is likely to interfere with any study of the orbits, skull, air sinuses, skull shape, and other structures that extend anterior to the brain itself (see Figure 9). This paper describes a novel, automated approach to the prevention of facial recognition that deforms the surface of the face, leaving underlying anatomical regions intact. We demonstrate that the registration results are superior to those using a simple deletion of the face—and we also demonstrate that our proposed method simultaneously preserves the underlying anatomy.
The approach described in the current report employs dilation and erosion over the skin surface so as to “blur” the facial features of each subject. As indicated by our results, a radius of 8 voxels appears sufficient to significantly reduce the likelihood of recognizing an individual. A variety of other approaches to blurring facial features could have equally well been employed to achieve a similar result.
Four limitations of the methods employed should be noted, however. First, the method inherently alters the superficial structures of the face. Although our approach permits the study of many anatomical regions whose analysis is precluded by brain stripping or by cutting off the face, images processed by our method should not be used to study such topics as the shapes of noses, the curvature of lips, or the smoothness of forehead skin.
Second, the approach should not be used on any patient with unique and recognizable deformities likely to make his/her images identifiable to the public. Such situations are extremely rare, however, since almost all diseases affect multiple individuals and almost any type of injury suffered by one individual will also have been suffered by many others.
Third, although our method allows an arbitrary amount of facial deformation there will always be a tradeoff between the amount of information lost and the difficulty with which the face can be recognized. Levels of deformation higher than those examined here could be used to reduce the face to a shapeless blob, but such levels of deformation would also be likely to affect the orbits and air sinuses, making these structures unsuitable for study.
Finally, the major limitation of the current study is that it did not perform perfectly in preventing patient identification under the forced-choice methods employed for evaluation of the approach. The 3D image of subject 9 was associated with the correct set of facial photographs more often than would have been expected by chance even at the maximum deformation level employed. Subject 9 had a long face, a noble nose, and a distinctive profile different from any of the other subjects imaged under the current study. Subject 9 could therefore be recognized as distinct from the other available choices when only a limited set of choices (10) were given among which to choose. In real life, each rater may have mental images of hundreds of acquaintances against which to compare a 3D rendering of an arbitrary image (and the rater may not be acquainted with the imaged subject at all). Although our study would ideally have included hundreds of photographs among which to choose, such a study would have been prohibitively time-consuming for each rater to complete. The study can be seen as representing the worst case scenario and the risk of recognition estimated in the study as the upper bound of the recognition rate. As shown by Figure 11, the degree of deformation employed would seem to make it unlikely that an individual could be recognized specifically. Nevertheless, under the terms of the current study subject 9 was associated with the correct set of photographs more often than would have been predicted by chance and so we cannot conclude with certainty that the proposed method is fully protective of patient identification.
There were three reasons why the study was designed as a forced choice study. First, this approach represents the worst-case scenario in which the rater performing the study is acquainted with the individual whose image is shown in 3D and under conditions in which the rater already knows/suspects that he will be shown an image of someone he knows. Second, this design allows the statistician to know with certainty what the likelihood is of correctly associating the correct set of images by random chance (1/10 for the case in which ten choices are available and the rater must make a choice from among the ten). In the absence of forced-choice and under conditions in which the rater is given the additional option of checking a box indicating that the 3D image cannot be matched to any photograph, different raters will have different thresholds of certainty and the likelihood of picking the correct image by random chance becomes much more difficult to determine. Finally, without forced choice the study becomes not only more difficult to analyze statistically but also less powerful in discriminating between responses at different deformation levels. The problem arises because raters may mentally match a 3D image with a person who was not on the actual subject list.
The primary contribution of this report may be to provide a set of questions not previously addressed by others. What is the best way to prevent patient recognition when making 3D MR images publicly available while still preserving as much as the underlying anatomy as possible? How can one preserve information and allow good registration between images while concomitantly preventing patient recognition? What is the best way to evaluate the success or failure of any such effort? This paper provides a first attempt to delineate many of the issues involved and to provide a useful solution.
Acknowledgments
This work was supported by R01EB000219, NIH-NIBIB
Footnotes
The size of our images was approximately 250×250×250 (about 15×106 voxels). The head occupies about a third of the voxels in the image (5×106 voxels). The connected voxel threshold should therefore be in the order of 106 voxels. Voxels representing noise outside the head are not connected to many others. The 10,000 voxel threshold was chosen relatively arbitrarily.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Chellappa R, Wilson CL, Sirohey S. Human and Machine Recognition of Faces: A Survey. Proceeding of the IEEE. 1995;83:705–740. [Google Scholar]
- 2.HIPAA Privacy Rules. 2006 Available at http://privacyruleandresearch.nih.gov/pr_08.asp.
- 3.Ibañez L, Schroeder W, Ng L, Cates J. The ITK Software Guide. Second. Kitware Inc; 2005. [Google Scholar]
- 4.ICBM Atlas, McConnell Brain Imaging Centre, Montréal Neurological Institute, McGill University, Montréal, Canada.
- 5.Jomier J, Guyon JP, Aylward S, et al. The Spatial Object Viewers Toolkit. http://public.kitware.com/SOViewer.
- 6.Mitra S, Savvides M, Kumar BV. Human Face Identification from Video based on Frequency Domain Asymmetry Representation using Hidden Markov Models. Lecture Notes in Computer Science. 2006;4105:26–33. [Google Scholar]
- 7.Otsu N. A Threshold Selection Method from Gray Level Histograms. IEEE Trans Syst Man Cyber SMC. 1979;9:62–66. [Google Scholar]
- 8.Rueckert D. Rview. Available: www.doc.ic.ac.uk/∼dr/software.
- 9.Sadr J, Jarudi I, Sinha P. The role of eyebrows in face recognition. Perception. 2003;32:285–293. doi: 10.1068/p5027. [DOI] [PubMed] [Google Scholar]
- 10.Schnabel JA, Rueckert D, Quist M, Blackall JM, Castellano-Smith AD, Hartkens T, Penney GP, Hall WA, Liu H, Truwit CL, Gerritsen FA, Hill DLG, Hawkes JD. A generic framework for non-rigid registration based on uniform multi-level free form deformations. Lecture Notes in Computer Science. 2001;2208:573–581. [Google Scholar]
- 11.Shepherd JW, Davies GM, Ellis HD. Studies of cue saliency. In: Davies GM, Ellis HD, Shepherd JW, editors. Perceiving and Remembering Faces. Academic Press; London, U.K: 2001. [Google Scholar]
- 12.Serra J. Image analysis and mathematical morphology. Academic Press; 1982. [Google Scholar]
- 13.Smith S. Fast robust automated brain extraction. Human Brain Mapping. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinberg D. Privacy and security in a federated research network. Thirteenth National HIPAA Summitt; 2006. Available at http://www.hipaasummit.com/past13/agenda/day2.html. [Google Scholar]
- 15.Yip A, Sinha P. Role of Color in Face Recognition. AI Memo 2001-035, CBCL Meme 212. 2001 [Google Scholar]