Abstract
Neuropilin-1 (Nrp1) is a multifunctional protein, identified principally as a receptor for the class 3 semaphorins and members of the vascular endothelial growth factor (VEGF) family, but it is capable of other interactions. It is a marker of regulatory T cells (Tr), which often carry Nrp1 and latency-associated peptide (LAP)-TGF-β1 (the latent form). The signaling TGF-β1 receptors bind only active TGF-β1, and we hypothesized that Nrp1 binds the latent form. Indeed, we found that Nrp1 is a high-affinity receptor for latent and active TGF-β1. Free LAP, LAP-TGF-β1, and active TGF-β1 all competed with VEGF165 for binding to Nrp1. LAP has a basic, arginine-rich C-terminal motif similar to VEGF and peptides that bind to the b1 domain of Nrp1. A C-terminal LAP peptide (QSSRHRR) bound to Nrp1 and inhibited the binding of VEGF and LAP-TGF-β1. We also analyzed the effects of Nrp1/LAP-TGF-β1 coexpression on T cell function. Compared with Nrp1– cells, sorted Nrp1+ T cells had a much greater capacity to capture LAP-TGF-β1. Sorted Nrp1– T cells captured soluble Nrp1-Fc, and this increased their ability to capture LAP-TGF-β1. Conventional CD4+CD25–Nrp1– T cells coated with Nrp1-Fc/LAP-TGF-β1 acquired strong Tr activity. Moreover, LAP-TGF-β was activated by Nrp1-Fc and also by a peptide of the b2 domain of Nrp1 (RKFK; similar to a thrombospondin-1 peptide). Breast cancer cells, which express Nrp1, also captured and activated LAP-TGF-β1 in a Nrp1-dependent manner. Thus, Nrp1 is a receptor for TGF-β1, activates its latent form, and is relevant to Tr activity and tumor biology.
Keywords: binding motif, VEGF, LAP-TGF-β1, signal transduction, CD4+CD25–, T lymphocytes, suppressor cells
INTRODUCTION
Neuropilin-1 (Nrp1) is a multifunctional protein involved in axonal guidance through its ability to bind the chemorepulsive class 3 semaphorin (SEMA3) proteins and angiogenesis through its interactions with vascular endothelial growth factor (VEGF) family members (including VEGF165) and their receptors (VEGFR1 and VEGFR2) [1, 2]. It is a 130- to 140-kD type-1 membrane glycoprotein with a short cytoplasmic domain that has no apparent signaling motif but interacts with the post-synaptic density 95/Discs large/zona occludens (PDZ) protein denoted Nrp1-interacting protein, C (also denoted GIPC) [3]. The extracellular component consists of a1/a2, b1/b2, and c (meprin, A5 protein, protein tyrosine phosphatase μ) domains. The a1/a2 domains are involved in SEMA3 binding, and the b1/b2 domains are involved in SEMA3 and VEGF binding [4]. Nrp1 has affinity for other molecules including heparin, the peptide Tuftsin (TKPR), integrin β1, cMet, hepatocyte growth factor (HGF), and some heparin-binding growth factors [1, 2, 4,5,6,7,8,9,10]. Of note, Nrp1 or the homologous Nrp2 (45% homology with Nrp1) is frequently expressed by malignant tumor cells and contributes to their malignant potential [1, 2, 7, 8].
In the immune system, Nrp1 is expressed by dendritic cells (DCs) [11] and regulatory T cells (Tr cells) [12]. In DCs, it is found at the immunologic synapse, but no definitive function has yet been attached to its expression by Tr cells. Interestingly, Tr cells frequently bear latency-associated peptide (LAP)-TGF-β1, i.e., the latent form of this cytokine, and this appears to contribute to their suppressive activity in some settings [13]. The classical TGF-β1 receptors (TGF-βRI, -RII, and -RIII) bind only the active (mature) form of TGF-β1, and other receptors must be involved. Indeed, arginine-glycine-aspartic acid (RGD)-binding integrins [14] and the mannose-6-phosphate/insulin-like growth factor II receptor (M6P/IGFII-R) [15] can bind LAP-TGF-β1. However, these receptors appear be more relevant to DCs or macrophages and to our knowledge, have not been reported on Tr cells. We hypothesized that Nrp1 might be a receptor for LAP-TGF-β1, and indeed, we found that Nrp1-Fc (but not Fc) bound LAP-TGF-β1 and active TGF-β1 at high affinity. Interestingly, LAP-TGF-β competed with VEGF for binding to Nrp1-Fc and appeared to bind at the same site. Nrp1+ T cells, unlike Nrp1– T cells, captured LAP efficiently. Conventional CD4+CD25–Nrp1– T cells could be coated with Nrp1-Fc, which promoted the capture of LAP-TGF-β1 and imparted regulatory activity to these cells. Importantly, we found that Nrp1 activates LAP-TGF-β1, and this can occur on the membrane of Nrp1+ tumor cells. Thus, our study reveals that Nrp1 is a TGF-β1 receptor, which is relevant to immune regulation and tumor biology.
MATERIALS AND METHODS
Mice
Experiments were performed with the T cells of CD-1 and C57BL/6 mice (Charles River, Wilmington, MA, USA).
Nrp1-Fc, Fc, and peptides
Recombinant rat Nrp1, conjugated to an Fc fragment of human IgG1 (Nrp1-Fc), was purchased from R&D Systems (Minneapolis, MN, USA). There is 98% homology between mouse and rat Nrp1 and 93% homology between mouse and human Nrp1. A construct of identical sequence but containing only the spacer peptide and human IgG1-Fc fragment (R&D Systems) was used as a control (referred to as Fc). Peptides were synthesized by the Sheldon Biotechnology Centre (Montreal, Canada) and were >95% pure. The LAP C-terminal peptide was used in the N-acetylated QSSRHRR-OH form.
TGF-β1 components, VEGF165, and antibodies
All recombinant TGF-β component proteins and human recombinant VEGF165 were purchased from R&D Systems. Mouse anti-TGF-β-antibody was purchased from BD Biosciences (San Jose, CA, USA). Mouse anti-human VEGF165 and mouse anti-human LAP antibodies were from R&D Systems. Polyclonal rabbit anti-rat Nrp1 developed against the Nrp814–827 peptide was purchased from Oncogen (Seattle, WA, USA). We also used a mouse anti-Nrp1 mAb (R&D Systems, clone 130603) binding to a different epitope than rabbit anti-rat Nrp1.
Cell enrichment and culture
Mouse splenic T cells were isolated as described [16]. Nrp1+ T lymphocytes were isolated by positive magnetic sorting. In brief, cells were treated with rabbit anti-rat/mouse Nrp1 antibody (Oncogen) and biotinylated goat anti-rabbit antibody (BD PharMingen, San Diego, CA, USA). The mixture was then incubated with magnetic particles attaching to the biotinylated antibodies [streptavidin ferrofluid (R&D Systems)], and the labeled cells were separated in a magnetic field. Sorted cells were greater than 95% Nrp1-positive. CD4+ spleen cells were isolated by negative sorting using the EasySep CD4 enrichment kit from StemCell Technology (Canada), according to the manufacturer’s instructions. Further separation steps involved collection of CD4+CD25+ and CD4+CD25– using biotinylated anti-CD25 antibody and the same protocol as above.
T cells were cultured in AIM V serum-free medium (Invitrogen Canada, Ontario), supplemented with 50 μM 2-ME, 1 mM sodium pyruvate, and 2 mM HEPES. The mouse T cell line HT-2 (ATCC, Manassas, VA, USA) was grown in DMEM + 5% FBS medium containing 10 ng/ml IL-2 and 2-ME, and this was replaced by the same medium containing 5 ng/ml IL-4 instead of IL-2 for the TGF-β assay.
Binding to protein G-sepharose columns
This was performed with Protein G-sepharose microcolumns (HiTrap Protein G HP by Amersham Pharmacia Biotech, Uppsala, Sweden). Retained proteins were eluted in glycine buffer, pH 3.5, and subjected to SDS electrophoresis under reducing conditions. Immunoblots were developed in specific antibodies and avidin-HRP.
ELISA-binding assays
Nrp1-Fc or other proteins were directly immobilized to Nunc Maxisorb plates (Nalge Nunc International, Rochester, NY, USA), according the manufacturer’s instructions. Soluble ligands were incubated in the precoated plate for 2 h at room temperature or at 4°C overnight. Nonspecific binding was determined in wells treated with the blocking solution and specific antibody and subtracted from the OD for total binding to give the values of specific binding. The binding plate was treated with anti-TGF-β1, anti-LAP, or control antibodies, diluted to a final concentration 500 ng/ml in 0.1% BSA + 0.05% Tween 20, followed by the secondary biotinylated rabbit anti-mouse antibodies and HRP reaction. The assays were performed in duplicates. In some cases, we also determined the concentration of unbound (soluble) components by ELISA. Calculations and regression analysis were performed in GraphPad Prism software (GraphPad Software, Inc., San Diego, CA, USA). When necessary, the fits for alternative equations were compared.
Tr cell assays
Splenic sorted CD4+CD25– T cells were incubated with Nrp1-Fc (or not; 8×106 cells; final concentration, Nrp1-Fc was 5 nM in AIM V serum-free medium) at 37°C for 40 min. Unbound ligands were washed off. As a second step, Nrp1-Fc-pulsed cells (or control cells) were treated with LAP-TGF-β1 (or not) as above and washed. As a control, Nrp1-Fc was replaced by Fc. Tr (suppressor, Ts) cell assays were performed by adding these treated cells to CD4+CD25– T cells (effector/responder cells, Te) and measuring the decline in soluble CD3-stimulated proliferation (and IL-2 secretion), as described by Piccirillo et al. [17], except that assays were performed in serum-free medium. In brief, 5 × 105 CD4+CD25– cells were plated over 2 × 105 irradiated, T cell-depleted spleen cells, and increasing numbers of Tr cells were added. Soluble anti-CD3 antibody was added at 2 μg/ml. In some wells, a mAb (1D11) that neutralizes active TGF-β was added to determine whether suppression was mediated by this cytokine. Proliferation was determined with a MTT assay as described [18]. The percent suppression of responses was calculated as follows: Percent suppression = [1–(OD of Te+Ts)/(OD of Te)] × 100.
Flow cytometry
These analyses were performed as we have described previously [16] on a FACSCalibur flow cytometer.
Phospho-(p)-Smad2 staining
The cells were fixed in 4% formaldehyde and permeabilized in Triton X-100 before staining. Anti (human, mouse)-p-Smad2 rabbit IgG (Chemicon International, El Segundo, CA, USA) was used in a 1:500 dilution. Secondary antibody was goat anti-rabbit, FITC-conjugated IgG (Sigma Chemical Co., St. Louis, MO, USA), diluted 1:100.
Statistical analysis
Statistical analyses were performed with the GraphPad Prism 3.0 program (GraphPad Software Inc.). In each experiment, the significance of differences between experimental and control results was determined by Student’s t-test or ANOVA. P < 0.05 was considered significant.
RESULTS
TGF-β1, free LAP, and LAP-TGF-β1 bind to Nrp1
Protein G captured Nrp1-Fc or control Fc but not other components as a result of its Fc-binding capacity (not shown). We found that free β1-LAP, LAP-TGF-β1, and active TGF-β1 (like VEGF) all bound to the Nrp1-Fc-coated beads but not to control Fc-coated beads as determined by immunoblotting (Fig. 1A). Nrp1-Fc failed to bind IFN-γ or IL-2 (not shown).
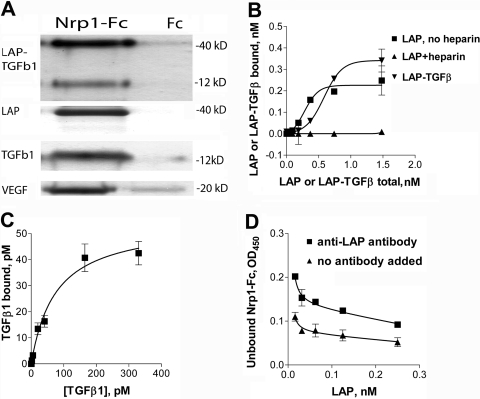
Fig. 1.
Nrp1 binds TGF-β1 components. (A) LAP-TGF-β1, LAP (β1), TGF-β1, and VEGF bound to Nrp1-Fc (but not control Fc) and were retained on protein G-sepharose beads. Bound proteins were recovered and immunoblots performed with specific antibodies. Molecular weight markers are indicated. (B) To demonstrate binding by ELISA, Nrp1-Fc-coated plates were incubated with increasing concentrations of the ligands. LAP (alone but not in the presence of 2 μg/ml heparin) and LAP-TGF-β1 bound at high affinity to Nrp1-Fc (see text). Several control proteins, including IFN-γ and IL-2, did not bind (not shown). (C) Active TGF-β1 bound to immobilized Nrp1-Fc. (D) Soluble Nrp1-Fc bound to plate-bound LAP, and this was inhibited by an anti-LAP antibody. The data in A–D are representative of three or more independent experiments.
Binding was also observed in ELISA cell-free assays. Plates coated with Nrp1-Fc retained active TGF-β1, free LAP, and LAP-TGF-β1 (Fig. 1, B and C). Heparin was not required for binding and prevented the binding of LAP but not TGF-β1. The cytokines IL-2 and IFN-γ did not bind to Nrp1-Fc (not shown). Active or latent TGF-β1 did not bind to immobilized Fc, and soluble Fc did not compete with soluble TGF-β1 for binding to immobilized Nrp1-Fc. No binding of any TGF-β1 components was noted when Nrp1-Fc was replaced by OVA, aprotinin, leupeptin, and a number of unrelated peptides (data not shown). To confirm specificity, we also performed blocking experiments with antibodies. Soluble LAP, when mixed with soluble Nrp1-Fc, competed with plate-bound LAP and decreased Nrp1-Fc retention on the plate (data not shown). Pretreatment of immobilized LAP with neutralizing concentrations of anti-LAP antibodies but not control antibody blocked the binding of soluble Nrp1-Fc to LAP (Fig. 1D).
Binding affinities were determined under equilibrium conditions by ELISA. This approach is sensitive and avoids the complexity of determining the kinetics of bivalent interactions. Affinity is expressed as EC50, an integrative equivalent of a Kd used when cooperativity between binding sites is observed (when binding sites do not interact, EC50=Kd). The affinity of LAP and LAP-TGF-β1 for Nrp1 was notably high: EC50 = 359 ± 80 and 338 ± 116 pM, respectively (mean±sem of seven or more experiments). Affinity for active TGF-β1 was even higher: Kd = 40 ± 8 pM (mean±sem of seven experiments). Strong positive cooperativity was observed for LAP (nH=2.9) and LAP-TGF-β1 (nH=3.7) binding to Nrp1-Fc (but not for TGF-β1 binding), suggesting that LAP binds to three or more interacting sites on the Nrp1-Fc molecule (Fig. 1B). To exclude possible effects of immobilization, we also measured reactant concentrations in soluble mixtures after filtration through Millipore filters with the molecular cut-off permitting separation of the unbound from the bound components. We also examined other variations of the assay (ELISA of the unbound instead of the retained ligands; immobilization of the ligand instead of the receptor protein). These alternative assays generated similar binding-affinity results (data not shown) to those reported above.
TGF-β1 and LAP components compete with VEGF165 for binding to Nrp1
Remarkably, LAP, LAP-TGF-β1, and active TGF-β1 diminished binding of VEGF165 to immobilized Nrp1-Fc (Fig. 2, A and B). LAP was a more effective competitor than active TGF-β1 with an IC50 = 0.28 nM versus 0.89 nM, respectively. In contrast, IFN-γ (Fig. 2B) and BSA (Fig. 2A) did not compete.
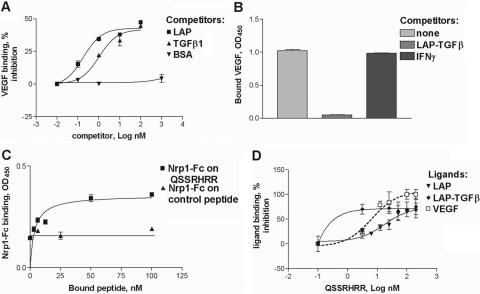
Fig. 2.
Mature TGF-β1 and LAP compete with VEGF for binding to Nrp1-Fc. (A) TGF-β1 and free LAP but not BSA-reduced binding of VEGF (2 nM) to the plate coated with 1 nM Nrp1-Fc. (B) Premixing LAP-TGF-β with VEGF (1 nM each) decreased the retention of VEGF by the Nrp1-Fc-coated plate. Under the same conditions, IFN-γ did not affect the VEGF retention. (C) Immobilized LAP C-terminal peptide, QSSRHRR, but not control peptide, bound soluble Nrp1-Fc. (D) Soluble QSSRHRR peptide reduced binding of LAP, LAP-TGF-β1, and VEGF to immobilized Nrp1-Fc (1 nM). The data in A–D are representative of two or three independent experiments.
Some ligands bind to Nrp1 through an arginine-rich C-terminal segment [5, 6]. In accord with this (Fig. 2, C and D), the C-terminal LAP-derived peptide QSSRHRR (but not an unrelated peptide) bound Nrp1-Fc, although with an affinity tenfold lower than that of LAP (EC50=4.18±0.99 nM), perhaps because of its monomeric form. In the soluble form, the QSSRHRR peptide inhibited the binding of VEGF165, LAP, and LAP-TGF-β1 to plate-bound Nrp1-Fc (Fig. 2D). Although VEGF binding could be blocked completely, the binding of LAP and LAP-TGF-β1 was markedly reduced but not completely inhibited. A modified peptide where the arginine was replaced by glutamic acid did not prevent binding (data not shown).
Coexpression of Nrp1 and LAP-TGF-β1 in T cell populations
Only 3–4% of splenic T cells expressed Nrp1 (Fig. 3A), as previously reported [19], and almost all were CD4+ and CD8– (data not shown). Some Nrp1-negative T cells incubated with soluble Nrp1-Fc captured this molecule on their membrane (Fig. 3B), and we made use of this finding in some of our subsequent experiments. The ability of T cells to capture Nrp1 has been reported previously [11], but the mechanism is unclear. It is not a result of FcRs, as FcR-blocking antibodies had no effect. Importantly, Nrp1-Fc incubation followed by LAP incubation increased the number of Nrp1+LAP+ T cells considerably (Fig. 1B).
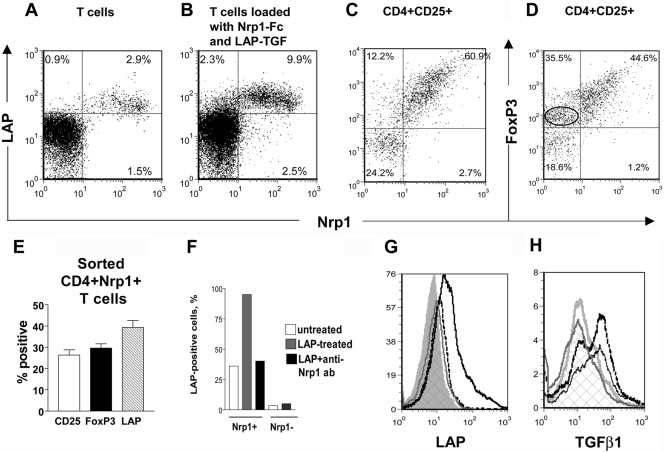
Fig. 3.
Coexpression of Nrp1 and TGF-β components analyzed by flow cytometry. (A) Approximately 3% of mouse splenic T cells coexpressed LAP and Nrp1 (percent-positive cells are indicated on the histograms). (B) Splenic T cells were incubated with Nrp1-Fc, washed, and incubated with LAP-TGF-β1. This approximately tripled the number Nrp1+LAP+ cells. (C and D) Splenic T cells were stained in four colors with antibodies against CD4, CD25, Nrp1, and LAP (C) or forkhead box P3 (FoxP3; D). The two-dimensional plots were gated on CD4+CD25+ cells. The number of Nrp1–FoxP3+ T cells is circled in D and shows that Nrp1+ cells generally express more FoxP3. (E) Tr-marker expression in CD4+Nrp1+ cells (percent±sem) in CD-1 mice. Similar results were obtained in C57BL/6 mice (not shown). (F) When Nrp1+ cells were incubated with LAP, the number of LAP+ cells increased from 40% to 95%, and this was blocked by an anti-Nrp1 mAb. Nrp1– T cells only minimally captured LAP. (G) Sorted CD4+Nrp1– T cells incubated with Nrp1-Fc and LAP-TGF-β1 capture LAP-TGF-β1 (LAP staining shown), and this was blocked by an anti-Nrp1 mAb. Silver peak, Isotype control; gray line/cross hatched peak, anti-LAP staining; solid black line, incubation with Nrp1-Fc and LAP-TGF-β1; dotted black line, incubation with Nrp1-Fc, then anti-Nrp1 mAb, and then LAP-TGF-β1. (H) T cells were incubated (or not) with Nrp1-Fc and then with active TGF-β1. Silver line, Isotype control; gray line/cross hatched peak, TGF-β1 staining; solid black line, T cell incubated with TGF-β1; dotted black line, T cells incubated with Nrp1-Fc and then TGF-β1. (A–H) The results are representative of two or more independent experiments. Cells were washed between treatments with Nrp1-Fc and TGF-β1 components.
We used four-color flow cytometry to analyze the coexpression of CD4, CD25, Nrp1, FoxP3, and LAP in splenic T cells. The expression of these markers did not completely overlap, indicating heterogenity in the Nrp1+ T cell population (Fig. 3, C–E; data not shown). Approximately 60% of CD4+CD25+ T cells coexpressed Nrp1 and LAP (Fig. 3C). There was a smaller number (10–12%) of CD4+CD25+Nrp1– T cells that also expressed LAP, suggesting that Nrp1 is not the only LAP receptor expressed by these T cells. Approximately 45% of CD4+CD25+ T cells coexpressed Nrp1 and FoxP3 (Fig. 3D). Interestingly, ∼35% of Nrp1– T cells also expressed FoxP3, but most of these cells stained at a lower intensity for FoxP3. We conclude that CD4+CD25+Nrp1+ express FoxP3 at higher levels than CD4+CD25+Nrp1– T cells.
We separated Nrp1+ and Nrp1– splenic T cells by magnetic sorting using rabbit polyclonal anti-Nrp1 antibody. In accord with the findings mentioned above, the sorted CD4+Nrp1+ splenic T cell population was enriched for cells expressing Tr markers, including LAP, FoxP3, and CD25 (Fig. 3E). It should be noted that this population overlaps with but is not identical to the CD4+CD25+ population analyzed in Figure 3, C and D, as approximately two-thirds of CD4+Nrp1+ T cells does not express CD25 (Fig. 3E). Thus, LAP was expressed by 40% of sorted CD4+Nrp1+ T cells, and FoxP3 and CD25 were expressed at a slightly lower frequency. When we incubated sorted CD4+Nrp1+ cells with free LAP, we observed that they captured LAP, such that ∼95% became LAP+, and this was blocked by an anti-Nrp1 mAb (Fig. 3F). In contrast, few sorted Nrp1– T cells captured LAP (Fig. 3F). Indeed, we found that the latter cells captured LAP poorly, unless they were precoated with Nrp1-Fc (Fig. 3G; in accord with the results of Fig. 3B with unsorted T cells), and LAP-TGF-β1 binding to Nrp1-Fc-coated T cells was inhibited by anti-Nrp1 mAb (Fig. 3G). These flow cytometry results, especially the blockade of binding by anti-Nrp1 antibodies, support the conclusion that Nrp1 is a LAP-TGF-β1 receptor. Unsorted T cells incubated with active TGF-β1 captured this cytokine (they have receptors for the active form), but even in this case, preincubation of cells with Nrp1-Fc moderately increased the number of TGF-β+ cells (Fig. 3H), consistent with the ability of Nrp1 to also bind the mature cytokine.
Nrp1 activates LAP-TGF-β1
When T cells were activated with anti-CD3 mAb, proliferation and IL-2 production were suppressed by active TGF-β1 (at ≤2 pM; not shown). Suppression was also observed with LAP-TGF-β1 but at much higher concentrations (50–200 pM), consistent with product information from the manufacturer and possibly because a fraction of this LAP-TGF-β1 is activated in culture. The suppression exerted by LAP-TGF-β1 was greatly increased when soluble Nrp1-Fc was added to cultures, revealing activation of LAP-TGF-β1 (Fig. 4A). Similarly, LAP-TGF-β1, in combination with Nrp1-Fc but not alone, suppressed the proliferation of TGF-β-sensitive HT-2 cells (Fig. 4B). In both cases, proliferation was restored by an anti-TGF-β mAb (1D11) that only binds the active form. Therefore, we conclude that Nrp1-Fc activates LAP-TGF-β1.
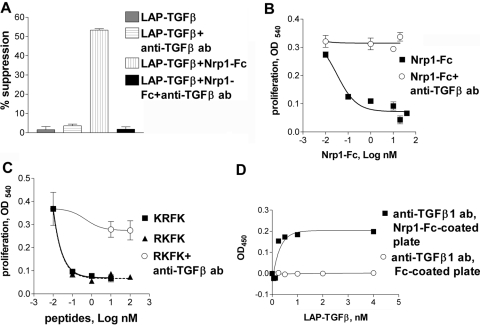
Fig. 4.
Nrp1-Fc and Nrp1 peptides activate LAP-TGF-β1. (A) Soluble Nrp1-Fc (0.4 nM) induced inhibition by LAP-TGF-β1 (0.2 nM) of the proliferation of T cells (CD3 mAb-stimulated), and this was abrogated by the 1D11 anti-TGF-β antibody (50 μg/ml) reactive only to active TGF-β. (B) LAP-TGF-β1 (2 nM) gained the ability to suppress the proliferation of HT-2 cells in the presence of Nrp1-Fc (5 nM), and this was abrogated by 1D11 mAb. In some wells, LAP-TGF-β1 and 1D11 were added without Nrp1-Fc (as a negative control), and as expected, this did not inhibit proliferation (not shown). (C) Nrp1 peptide RKFK and thrombospondin-1 (TSP-1) peptide KRFK were equally effective at activating LAP-TGF-β (2 nM), as determined in the HT-2 proliferation assay, and this was blocked by the 1D11 mAb. (D) Binding of LAP-TGF-β1 to immobilized Nrp1-Fc exposed a TGF-β1 epitope recognized by 1D11 mAb.
Nrp1 has a sequence (RKFK) in its b2 domain, closely similar to a sequence of TSP-1 (KRFK or RFK) that activates latent TGF-β1 [20,21,22]. In the HT-2 assay, we found that the RKFK and KRFK peptides (but not control peptides) were equally effective at activating TGF-β1 (Fig. 4C), and suppression was reversed by 1D11 mAb. The ability of Nrp1 to activate latent TGF-β1 was further confirmed by our finding that coincubating LAP-TGF-β1 with immobilized Nrp1-Fc in a cell-free system resulted in the exposure of an epitope recognized by1D11 (Fig. 4D). We hypothesize that this results in a change of conformation of LAP, where antigenic determinants of active TGF-β1 are exposed, but it remains attached and can interact with 1D11 mAb.
Nrp1-dependent activation of LAP-TGF-β1 by breast cancer cells
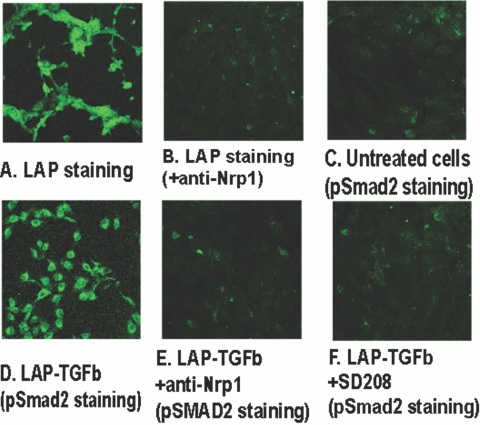
MDA-MB-231 human breast cancer cells are Nrp1+ (data not shown). These cells captured LAP-TGF-β1, which was markedly reduced by an anti-Nrp1 antibody (Fig. 5, A and B). The cells activated the latent cytokine as shown by increased p-Smad2 staining (Fig. 5, C and D), and this was almost completely blocked by an anti-Nrp1 antibody (Fig. 5E) or SD-208 (a TGF-βRI inhibitor; Fig. 5F). Mouse Lewis lung carcinoma cells similarly captured LAP-TGF-β1 in a Nrp1-dependent way (not shown).
Fig. 5.
MDA-MB-231 human Nrp1+ breast cancer cells bind and activate latent TGF-β1. (A) Incubation of the cells with LAP-TGF-β1 and fluorescent LAP staining showing retention of LAP. (B) Cells treated with anti-Nrp1 mAb-blocking antibody and LAP-TGF-β1 and stained for LAP. (C) p-Smad2 fluorescent staining of untreated cultured cells. (D) p-Smad2 staining of cells incubated with LAP-TGF-β1. (E) p-Smad2 staining of cells incubated with anti-Nrp1 antibody and LAP-TGF-β1. (F) p-Smad2 staining of cells incubated with SD-208 and LAP-TGF-β1. The results of A–F are representative of three separate experiments.
Conversion of conventional T cells into Tr cells in vitro
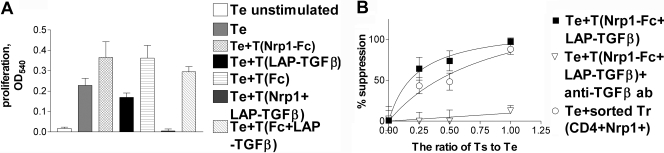
Conventional CD4+CD25–Nrp1– naïve (nonsuppressive), splenic T cells of C57BL/6 mice were incubated with Nrp1-Fc (or not), washed, and then incubated with LAP-TGF-β1 (or not). As a control, Nrp1-Fc was replaced by Fc. These treated cells (or untreated cells) were washed and then added to CD4+CD25– T lymphocytes (the Te cells) in serum-free culture medium containing soluble CD3 mAb and irradiated APCs, and proliferation was measured. The untreated CD4+CD25– cells and single-treated cells (with Nrp1-Fc or LAP-TGF-β1) were not suppressive or exerted only weak suppression, but the double-coated T cells (Nrp1-Fc/LAP-TGF-β1-treated) strongly suppressed proliferation of the Te cells (Fig. 6, A and B). IL-2 secretion was also suppressed (not shown). When Nrp1-Fc was substituted by Fc, no suppression was observed. Importantly, the suppression exerted by Nrp1-Fc/LAP-TGF-β1-treated cells was almost completely reversed by the 1D11 anti-TGF-β mAb. Interestingly, the suppression exerted by naturally occurring, sorted CD4+Nrp1+ T cells, isolated from the spleen and tested ex vivo, was comparable (although slightly less) with the suppression exerted by CD4+CD25–Nrp1– T cells coated with Nrp1-Fc/LAP-TGF-β1 (Fig. 6B).
Fig. 6.
Acquisition of Tr-like suppressive activity by conventional CD4+ T cells. We tested suppression in a conventional Tr assay, where CD4+CD25– T cells (the Te cells) are activated by soluble CD3 mAb. Putative Ts cells were added to Te cells, and decreased proliferation is used as an indicator of suppression. (A) Nrp1-Fc-treated CD4+CD25– T cells and Fc-treated cells did not suppress proliferation (data represent 1:1 Ts:Te ratio). LAP-TGF-β1-treated T cells were weakly suppressive. However, Nrp1-Fc/LAP-TGF-β1 double-treated T cells were strongly suppressive (1:1 Ts:Te ratio; see B for other ratios; P<0.0001 vs. all single-treated T cells). The suppressive effect was not seen when Nrp1-Fc was replaced by Fc. (B) Nrp1-Fc and LAP-TGF-β1 double-coated CD4+CD25– T cell-mediated suppression was TGF-β-dependent. The suppression was almost completely reversed by 1D11 anti-TGF-β mAb (50 μg/ml). Naturally occurring CD4+Nrp1+ T cells (sorted ex vivo) produced a comparable degree of suppression. The data are representative of three independent experiments.
DISCUSSION
We demonstrate that Nrp1 is a high-affinity receptor for the active and latent forms of TGF-β1. This is supported by several findings, including coprecipitation assays, cell-free ELISA-binding assays, competition with VEGF (a well-known Nrp1 ligand), use of blocking antibodies, flow cytometry data, and T cell functional assays. The binding affinities of Nrp1 for TGF-β1 components (LAP, LAP-TGF-β1, and TGF-β1) were notably high. These high affinities likely depend on prominent avidity effects as a result of the homodimeric structure of Nrp1-Fc and the target ligands, as shown, for example, for VEGFR2 [23]. It appears that Nrp1 frequently exists as a dimer or oligomer on cell membranes [1], and therefore, similar high-affinity binding can probably occur in vivo. LAP binding to Nrp1, unlike VEGF [1, 2], is not enhanced by heparin but rather is blocked.
Crystal structure studies of the b1 and b2 domains of Nrp1 [5, 6] provide possible explanations of how Nrp1 binds several seemingly unrelated ligands. Lee et al. [5] described a cleft with a negative charge within the b1 domain and postulated that positively charged C-terminal tails of VEGF and SEMA3 bind to this site. Vander Kooi et al. [6] reported similar findings and demonstrated that Tuftsin (TKPR), a peptide mimic of the C-terminal of VEGF165 (KPRR), binds to this cleft of the b1 domain. The terminal arginine residue of Tuftsin appears to be particularly important in this interaction. von Wronski et al. [24] showed that Tuftsin competes with VEGF165 for binding to Nrp1. Of note, a viral VEGF-like protein that binds Nrp1 has a C-terminal TRPPRRRR sequence [25]. In contrast, a splice variant of VEGF165 with a SLTRKD C-terminal sequence fails to bind Nrp1 [26]. In addition, other ligands or peptides that bind to Nrp1 are characterized by a basic structure and often two or more arginine residues [27]. As LAP has a highly basic, arginine-rich C-terminal motif (RHRR), we hypothesized that it binds to Nrp1 through this domain and tested some peptide interactions. A LAP C-terminal peptide (QSSRHRR) bound Nrp1, although with lower affinity than LAP, and blocked the binding of VEGF and LAP-TGF-β1. Unlike VEGF, the binding of LAP was not blocked completely. Taken together, these observations are indicative of participation of the C-terminal LAP peptide in the binding LAP to Nrp1 but do not exclude the possibility that there are other binding sites, especially as LAP binding was not blocked completely.
In the case of active TGF-β1, there is no similar C-terminal sequence, and binding presumably occurs through other interactions that have not yet been characterized. However, crystal structure studies of TGF-β3 and TGF-βRII complexes [28, 29] show that TGF-β3 has two arginine-containing, finger-like structures that interact with the RII receptor. In particular, the RII-binding site has an electronegative area, where D32 interacts with R94 of a TGF-β3 finger [28, 29]. TGF-β1 differs from TGF-β3 by only one amino acid in that segment and is presumably capable of similar interactions. In preliminary studies, we found that a basic peptide encompassing R94 of TGF-β1 (peptide VGRKPKV) interfered with the binding of mature TGF-β and VEGF to Nrp1 (data not shown). In view of this, we speculate that the TGF-β finger structure that binds to RII might also bind to the electronegative pocket of the b1 domain of Nrp1. It is interesting that this peptide did not have a C-terminal arginine residue, but in some cases, this may not be required as shown, for example, by the binding of cyclic arginine-containing peptides to Nrp1 [27]. Interestingly, the sequence 94RKPK of TGF-β1 has been identified as a binding site to LAP, and remarkably, the RKPK peptide activated LAP-TGF-β1 [30]. This peptide has obvious similarity with the basic peptides of Nrp1 (RKFK) and TSP-1 (KRFK), which also activate LAP-TGF-β1, as discussed below.
Flow cytometry analysis revealed that only 3–4% of splenic T cells were Nrp1+, and almost all were CD4+ cells, and they frequently coexpressed Tr markers (CD25, FoxP3, and LAP). However, these markers do not completely overlap, and there is obvious heterogenitiy in the Nrp1+ T cell population. By multicolor flow cytometry, we observed that 60% of CD4+CD25+ T cells coexpress Nrp1 and LAP. In addition, 10–12% of CD4+CD25+Nrp1– T cells also expressed LAP, suggesting that Nrp1 is not the only LAP receptor on the membrane of these T cells. Approximately 45% of CD4+CD25+ T cells coexpressed Nrp1 and FoxP3, and interestingly, the Nrp1+ cells expressed FoxP3 cells at a higher level than the Nrp1– T cells. This finding is similar to the report of Battaglia et al. [18], who studied human Nrp1+ Tr cells. Thus, Nrp1 expression appears to correlate with high FoxP3 expression in mice and humans. Approximately 40% of sorted CD4+Nrp1+ cells expressed LAP, but when these cells were incubated with soluble LAP, ∼95% became LAP+, revealing a high capacity to capture this component. The binding of LAP was blocked by an anti-Nrp1 antibody, showing this was a Nrp1-dependent interaction. In contrast, Nrp1– T cells only had a minimal ability to capture LAP, but this was considerably increased by precoating these cells with Nrp1-Fc. It is noteworthy that the blockade of other known receptors of LAP such as RGD-binding integrins and M6P/IGFII-R did not reduce the binding of LAP in our experiments (data not shown). Taken together, our results indicate that Nrp1 is the major LAP receptor of T cells, although it is not the only LAP receptor. The nature of other receptors has not been elucidated.
No definite role has yet been ascribed to Nrp1 in Tr cell-suppressive activity. Indeed, antibodies against Nrp1 did not alter the suppressive activity of natural Tr cells [12]. However, we hypothesized that Nrp1 might play a role by capturing LAP-TGF-β1 and contributing to its activation on the cell membrane. There is a precedent for this, as other molecules can activate latent TGF-β1 on the membrane of some cells [31]. First, we found that the coincubation of soluble Nrp1-Fc and LAP-TGF-β1 resulted in activation of the cytokine, as measured by suppression of HT-2 cells or normal T cells. Although the mechanism of TGF-β1 activation remains to be elucidated, it is striking that Nrp1 possesses a short basic sequence (RKFK) in its b2 domain, closely similar to a sequence of TSP-1 (KRFK or RFK) that binds LAP in its N-terminal portion and activates latent TGF-β1 by inducing a conformational change [20,21,22]. Importantly, the RKFK sequence of Nrp1 has been linked to heparin binding [6] and cell aggregation [32] and is evidently exposed for molecular interactions. We compared the RKFK and KRFK peptides and found that they are equally effective at activating latent TGF-β1. Although further studies are required, this suggests that the RKFK sequence of Nrp1 represents another binding site for LAP and by analogy, with TSP-1 [20,21,22], provides an important clue as to how it might activate LAP-TGF-β1. Indeed, our binding studies suggest more than one site for interaction of LAP with Nrp1. Interestingly, the RKFK sequence is present in Nrp1 and Nrp2 of the rat, mouse, and human [32].
Most malignant tumor cells express Nrp1 and/or Nrp2 [1], and it was of interest to see whether this influences LAP-TGF-β1 capture. We investigated this question with MDA-MB-231 human breast cancer cells, which are strongly Nrp1+. These cells captured LAP-TGF-β1, and LAP staining was greatly reduced by an anti-Nrp1 antibody, although some weak staining persisted, perhaps because of another receptor(s). Importantly, LAP-TGF-β1 was activated, as demonstrated by increased nuclear p-Smad2 expression, and this was almost completely blocked by an anti-Nrp1 antibody. The generation of p-Smad2 is strong evidence of the induction TGF-β1 signaling, and this is a Nrp1-dependent process. As expected, a TGF-βRI (ALK5) inhibitor, SD-208, also blocked signaling in this pathway.
To demonstrate an effect on T cell activity, we coated conventional (nonsuppressive) CD4+CD25–Nrp1– cells with Nrp1-Fc and LAP-TGF-β1 and tested their activity in a Tr cell assay (inhibition of CD3-mediated T cell proliferation). Of note, Nrp1-Fc binds to the membrane of CD4+ and CD8+ T cells, as well as tumor cell lines (data not shown). The receptors responsible for this interaction have not been characterized. However, as Nrp1 binds to several molecules commonly expressed on cell membranes (e.g., VEGF receptors, cMET, β1 integrin, galectin-1), it seems likely that several receptors are capturing soluble Nrp1. Single-treated CD4+CD25–Nrp1– cells (with Nrp1-Fc or LAP-TGF-β1) exerted minimal or no suppression, and double-coated T cells were strongly suppressive. Furthermore, this suppression was almost completely reversed by the 1D11 anti-TGF-β mAb, indicating that activated TGF-β1 was involved. We conclude that Nrp1 captured and activated TGF-β1 and through this mechanism, converted Te cells into Tr cells. As TGF-β1 is secreted primarily in the LAP-bound, latent form, which is present in plasma and tissues, it seems likely that Nrp1+ T cells will acquire LAP-TGF-β1 in vivo, although not all were LAP+. We speculate that Nrp1 allows T cells to capture and maintain TGF-β1 in an active form on their membrane and exert Tr-like suppression. This could be because these T cells are induced by TGF-β1 to differentiate into Tr cells [31] or because membrane-bound TGF-β1 directly exerts suppression, as suggested by some authors [13]. Under physiological conditions, the association of Nrp1 with the PDZ protein GIPC might be relevant. Nrp1 interacts with GIPC through a short C-terminal (cytoplasmic) sequence (SEA), and this segment is required for Nrp1-mediated angiogenesis [3]. GIPC is a PDZ scaffold protein [33] and can link G-protein signaling pathways [33]. It is also intriguing that Nrp1 and the TGF-βRIII [34] have a GIPC-binding domain, and it is plausible that they interact through this cytoplasmic protein. However, in our experiments, Nrp1-Fc does not have the GIPC-binding domain, demonstrating that GIPC is not required for the enhancement of LAP-TGF-β1-suppressive activity, presumably as signaling occurs through the classical TGF-βRs (RI, RII, RIII).
In our Tr cell assays, we artificially coated CD4+CD25– T cells with Nrp1-Fc, and the question arises whether these cells can express Nrp1 in vivo. With respect to this question, Bourbié-Vaudaine et al. [35] showed that Nrp1 is transferred from DCs (which express Nrp1) to CD4+ T cells during T cell/DC interactions by a process denoted trogocytosis. Interestingly, Nrp1 was not rapidly internalized, and Nrp1+ T cells could capture VEGF165 on their membrane. In this case, >60% of responding CD4+ T cells became Nrp1+, and although we have not studied this question, we hypothesize that when TGF-β1 is abundant, this could duplicate the conditions of our Tr assay.
In conclusion, we present evidence that Nrp1 is a receptor for the latent and active forms of TGF-β1, activates the latent form, and promotes Tr activity. Nrp1-dependent TGF-β1 activation also occurred on the membrane of breast cancer cells, and this is of great interest, as Nrp1 and TGF-β1 have been linked to cancer progression [1, 31]. Also of notable relevance to cancer, Nrp1 acts as a coreceptor in VEGF/VEGFR and HGF/cMet interactions and also binds fibroblast growth factor 2 and β1 integrin [1, 2, 7,8,9,10]. This astonishing variety of interactions suggests that Nrp1 has a general function of capturing growth factors on the membrane of normal or tumor cells and enhancing signaling through several pathways. Perhaps this occurs in some cases through the incorporation of Nrp1 into multicomponent signalosomes [36] involved in these pathways, with the participation of scaffold proteins such as GIPC. TGF-β1 is produced primarily in latent form (unlike other cytokines), and in this case, Nrp1 has the additional capacity of activating the latent cytokine. Our studies point to novel interactions between Nrp1 and TGF-β1, which are potentially relevant to several biological processes such as angiogenesis, immune regulation, and tumor biology.
Acknowledgments
This study was supported by Ontario Institute for Cancer Research of the Province of Ontario (Canada), the Juvenile Diabetes Foundation International, the Canadian Diabetes Association, the Krembil Foundation (Toronto, Canada), and the Li Ka Shing Knowledge Institute and Keenan Research Centre of St. Michael’s Hospital (Toronto, Canada).
References
- Staton C A, Kumar I, Reed M W, Brown N. Neuropilins in physiological and pathological angiogenesis. J Pathol. 2007;212:237–248. doi: 10.1002/path.2182. [DOI] [PubMed] [Google Scholar]
- Roskoski R., Jr Vascular endothelial growth factor (VEGF) signaling in tumor progression. Crit Rev Oncol Hematol. 2007;62:179–213. doi: 10.1016/j.critrevonc.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Wang L, Dutta S K, Kojima T, Xu X, Khosravi-Far R, Ekker S C, Mukhopadhyay D. Neuropilin-1 modulates p53/caspases axis to promote endothelial cell survival. PLoS ONE. 2007;2:e1161. doi: 10.1371/journal.pone.0001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H Q, Soker S, Feiner L, Alonso J L, Raper J A, Klagsbrun M. Neuropilin-1 mediates collapsin-1/semaphorin III inhibition of endothelial cell motility: functional competition of collapsin-1 and vascular endothelial growth factor-165. J Cell Biol. 1999;146:233–242. doi: 10.1083/jcb.146.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C C, Kreusch A, McMullan D, Ng K, Spraggon G. Crystal structure of the human neuropilin-1 b1 domain. Structure. 2003;11:99–108. doi: 10.1016/s0969-2126(02)00941-3. [DOI] [PubMed] [Google Scholar]
- Vander Kooi C W, Jusino M A, Perman B, Neau D B, Bellamy H D, Leahy D J. Structural basis for ligand and heparin binding to neuropilin B domains. Proc Natl Acad Sci USA. 2007;104:6152–6157. doi: 10.1073/pnas.0700043104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa M, Matsushita A, Korc M. Neuropilin-1 interacts with integrin β1 and modulates pancreatic cancer cell growth, survival and invasion. Cancer Biol Ther. 2007;6:1173–1180. doi: 10.4161/cbt.6.8.4363. [DOI] [PubMed] [Google Scholar]
- Matsushita A, Götze T, Korc M. Hepatocyte growth factor-mediated cell invasion in pancreatic cancer cells is dependent on neuropilin-1. Cancer Res. 2007;67:10309–10316. doi: 10.1158/0008-5472.CAN-07-3256. [DOI] [PubMed] [Google Scholar]
- Sulpice E, Plouet J, Berge M, Allanic D, Tobelem G, Merkulova-Rainon T. Neuropilin-1 and neuropilin-2 act as coreceptors, potentiating proangiogenic activity. Blood. 2008;111:2036–2045. doi: 10.1182/blood-2007-04-084269. [DOI] [PubMed] [Google Scholar]
- West D C, Rees C G, Duchesne L, Patey S J, Terry C J, Turnbull J E, Delehedde M, Heegaard C W, Allain F, Vanpouille C, Ron D, Fernig D G. Interactions of multiple heparin binding growth factors with neuropilin-1 and potentiation of the activity of fibroblast growth factor-2. J Biol Chem. 2005;280:13457–13464. doi: 10.1074/jbc.M410924200. [DOI] [PubMed] [Google Scholar]
- Tordjman R, Lepelletier Y, Lemarchandel V, Cambot M, Gaulard P, Hermine O, Roméo P H. A neuronal receptor, neuropilin-1, is essential for the initiation of the primary immune response. Nat Immunol. 2002;3:477–482. doi: 10.1038/ni789. [DOI] [PubMed] [Google Scholar]
- Bruder D, Probst-Kepper M, Westendorf A M, Geffers R, Beissert S, Loser K, von Boehmer H, Buer J, Hansen W. Neuropilin-1: a surface marker of regulatory T cells. Eur J Immunol. 2004;34:623–630. doi: 10.1002/eji.200324799. [DOI] [PubMed] [Google Scholar]
- Oida T, Xu L, Weiner H L, Kitani A, Strober W. TGF-β-mediated suppression by CD4+CD25+ T cells is facilitated by CTLA-4 signaling. J Immunol. 2006;177:2331–2339. doi: 10.4049/jimmunol.177.4.2331. [DOI] [PubMed] [Google Scholar]
- Travis M A, Reizis B, Melton A C, Masteller E, Tang Q, Proctor J M, Wang Y, Bernstein X, Huang X, Reichardt L F, Bluestone J A, Sheppard D. Loss of integrin α(v)β8 on dendritic cells causes autoimmunity and colitis in mice. Nature. 2007;449:361–365. doi: 10.1038/nature06110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godár S, Horejsi V, Weidle U H, Binder B R, Hansmann C, Stockinger H. 6P/IGFII-receptor complexes urokinase receptor and plasminogen for activation of transforming growth factor-β1. Eur J Immunol. 1999;29:1004–1013. doi: 10.1002/(SICI)1521-4141(199903)29:03<1004::AID-IMMU1004>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Glinka Y, Chang Y, Prud'homme G J. Protective regulatory T cell generation in autoimmune diabetes by DNA covaccination with islet antigens and a selective CTLA-4 ligand. Mol Ther. 2006;14:578–587. doi: 10.1016/j.ymthe.2006.03.021. [DOI] [PubMed] [Google Scholar]
- Piccirillo C A, Letterio J J, Thornton A M, McHugh R S, Mamura M, Mizuhara H, Shevach E M. CD4(+)CD25(+) regulatory T cells can mediate suppressor function in the absence of transforming growth factor β1 production and responsiveness. J Exp Med. 2002;196:237–246. doi: 10.1084/jem.20020590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia A, Buzzonetti A, Monego G, Peri L, Ferrandina G, Fanfani F, Scambia G, Fattorossi A. Neuropilin-1 expression identifies a subset of regulatory T cells in human lymph nodes that is modulated by preoperative chemoradiation therapy in cervical cancer. Immunology. 2008;123:129–138. doi: 10.1111/j.1365-2567.2007.02737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbel C, Lemarchandel V, Thomas-Vaslin V, Pelus A S, Agboton C, Roméo P H. Neuropilin 1 and CD25 co-regulation during early murine thymic differentiation. Dev Comp Immunol. 2007;31:1082–1094. doi: 10.1016/j.dci.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Schultz-Cherry S, Chen H, Mosher D F, Misenheimer T M, Krutzsch H C, Roberts D D, Murphy-Ullrich J E. Regulation of transforming growth factor-β activation by discrete sequences of thrombospondin 1. J Biol Chem. 1995;270:7304–7310. doi: 10.1074/jbc.270.13.7304. [DOI] [PubMed] [Google Scholar]
- Crawford S E, Stellmach V, Murphy-Ullrich J E, Ribeiro S M, Lawler J, Hynes R O, Boivin G P, Bouck N. Thrombospondin-1 is a major activator of TGF-β1 in vivo. Cell. 1998;93:1159–1170. doi: 10.1016/s0092-8674(00)81460-9. [DOI] [PubMed] [Google Scholar]
- Ribeiro S M, Poczatek M, Schultz-Cherry S, Villain M, Murphy-Ullrich J E. The activation sequence of thrombospondin-1 interacts with the latency-associated peptide to regulate activation of latent transforming growth factor-β. J Biol Chem. 1999;274:13586–13593. doi: 10.1074/jbc.274.19.13586. [DOI] [PubMed] [Google Scholar]
- Fuh G, Li B, Crowley C, Cunningham B, Wells J A. Requirements for binding and signaling of the kinase domain receptor for vascular endothelial growth factor. J Biol Chem. 1998;273:11197–11204. doi: 10.1074/jbc.273.18.11197. [DOI] [PubMed] [Google Scholar]
- von Wronski M A, Raju N, Pillai R, Bogdan N J, Marinelli E R, Nanjappan P, Ramalingam K, Arunachalam T, Eaton S, Linder K E, Yan F, Pochon S, Tweedle M F, Nunn A D. Tuftsin binds neuropilin-1 through a sequence similar to that encoded by exon 8 of vascular endothelial growth factor. J Biol Chem. 2006;281:5702–5710. doi: 10.1074/jbc.M511941200. [DOI] [PubMed] [Google Scholar]
- Wise L M, Wang Z, Grynpas M D. Vascular endothelial growth factor (VEGF)-like protein from orf virus NZ2 binds to VEGFR2 and neuropilin-1. Proc Natl Acad Sci USA. 1999;96:3071–3076. doi: 10.1073/pnas.96.6.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D O, MacMillan P P, Manjaly J G, Qiu Y, Hudson S J, Bevan H S, Hunter A J, Soothill P W, Read M, Donaldson L F, Harper S J. VEGF165b, an inhibitory splice variant of vascular endothelial growth factor, is down-regulated in renal cell carcinoma. Cancer Res. 2002;62:4123–4131. [PubMed] [Google Scholar]
- Hong T M, Chen Y L, Wu Y Y, Yuan A, Chao Y C, Chung Y C, Wu M H, Yang S C, Pan S H, Shih J Y, Chan W K, Yang P C. Targeting neuropilin 1 as an antitumor strategy in lung cancer. Clin Cancer Res. 2007;13:4759–4768. doi: 10.1158/1078-0432.CCR-07-0001. [DOI] [PubMed] [Google Scholar]
- Hart P J, Deep S, Taylor A B, Shu Z, Hinck C S, Hinck A P. Crystal structure of the human TβR2 ectodomain–TGF-β3 complex. Nat Struct Biol. 2002;9:203–208. doi: 10.1038/nsb766. [DOI] [PubMed] [Google Scholar]
- Groppe J, Hinck C S, Samavarchi-Tehrani P, Zubieta C, Schuermann J P, Taylor A B, Schwarz P M, Wrana J L, Hinck A P. Cooperative assembly of TGF-β superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol Cell. 2008;29:157–168. doi: 10.1016/j.molcel.2007.11.039. [DOI] [PubMed] [Google Scholar]
- Young G D, Murphy-Ullrich J E. Molecular interactions that confer latency to transforming growth factor-β. J Biol Chem. 2004;279:38032–38039. doi: 10.1074/jbc.M405658200. [DOI] [PubMed] [Google Scholar]
- Prud'homme G J. Pathobiology of transforming growth factor β in cancer, fibrosis and immunologic disease, and therapeutic considerations. Lab Invest. 2007;87:1077–1091. doi: 10.1038/labinvest.3700669. [DOI] [PubMed] [Google Scholar]
- Shimizu M, Murakami Y, Suto F, Fujisawa H. Determination of cell adhesion sites of neuropilin-1. J Cell Biol. 2000;148:1283–1293. doi: 10.1083/jcb.148.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramow-Newerly M, Roy A A, Nunn C, Chidiac P. RGS proteins have a signaling complex: interactions between RGS proteins and GPCRs, effectors, and auxiliary proteins. Cell Signal. 2006;18:579–591. doi: 10.1016/j.cellsig.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Blobe G C, Liu X, Fang S J, How T, Lodish H F. A novel mechanism for regulating transforming growth factor β (TGF-β) signaling. Functional modulation of type III TGF-β receptor expression through interaction with the PDZ domain protein, GIPC. J Biol Chem. 2001;276:39608–39617. doi: 10.1074/jbc.M106831200. [DOI] [PubMed] [Google Scholar]
- Bourbié-Vaudaine S, Blanchard N, Hivroz C, Roméo P H. Dendritic cells can turn CD4+ T lymphocytes into vascular endothelial growth factor-carrying cells by intercellular neuropilin-1 transfer. J Immunol. 2006;177:1460–1469. doi: 10.4049/jimmunol.177.3.1460. [DOI] [PubMed] [Google Scholar]
- Hoeller D, Volarevic S, Dikic I. Compartmentalization of growth factor receptor signaling. Curr Opin Cell Biol. 2005;17:107–111. doi: 10.1016/j.ceb.2005.01.001. [DOI] [PubMed] [Google Scholar]