Abstract
The phase I enzyme known as cytochrome P450 1B1 (CYP1B1) is involved in the metabolism of many endogenous and exogenous compounds, including carcinogens. CYP1B1 is overexpressed in a wide variety of human diseases ranging from diabetes to malignancies, such as invasive breast cancer. Because of its microsomal location in the cell, CYP1B1 could not be measured directly by existing methods but only assessed indirectly via the determination of the catalytic products. We report here a rapid, sensitive piezoimmunosensor for detection of CYP1B1 using single-chain fragment variable antibodies (scFv) as recognition elements and a quartz crystal microbalance (QCM) as the transducer. Three anti-CYP1B1 scFvs (designated B-66, D-23, and L-21) were biotiny-lated and used to capture and specifically detect CYP1B1 from samples in solution. ScFvs are smaller than most commonly used antibodies and can be coated onto QCM surfaces at much higher density to improve sensor sensitivity and specificity. The scFv-QCM biosensors showed excellent sensitivity (detection limit, 2.2 ± 0.9 nM) and specificity with a dissociation constant Kd = (1.54 ± 0.59) × 10−7 M. CYP1B1 were quantitatively detected in normal and malignant cell lysates (e.g., human T47D breast cancer cell microsomes). Results demonstrate that an anti-CYP1B1 scFv-QCM immunosensor could be used to detect P450 enzymes in biological samples.
The cytochrome P450 gene superfamily encodes multifunctional phase I enzymes that are involved in the metabolism of many drugs and dietary substances and in the synthesis of steroid hormones and other extracellular lipid signaling molecules.1 Not surprisingly, cytochrome P450s (CYPs) have been implicated to play important roles in a wide variety of human diseases ranging from diabetes to malignancies.2 Because of their important functions in normal physiology and disease, it would be desirable to measure CYPs. To date, CYPs have been visualized by immunocytochemistry and localized within the microsomal (endoplasmic reticulum) compartment of the cell. However, attempts to quantitate the concentration of CYPs have been unsuccessful with existing methods because of the location in the complex microsomal membrane, which consists of a mixture of proteins and lipids containing the embedded CYPs. Thus, individual CYPs have not been measured directly by existing methods but only assessed indirectly via the determination of their catalytic products.
In the present study, we have developed a novel analytical approach, which was based on a piezoimmunosensor, to detect and quantitate individual CYPs in cellular extracts. As a prototype, we selected one member of the CYP family, CYP1B1, because it has been implicated in the estrogen carcinogenesis of breast cancer, in which it is overexpressed.3,4 Several studies have also targeted CYP1B1 as a universal tumor antigen, using specific cytotoxic T cells for cancer immunotherapy.5 Thus, CYP1B1 is a potential tumor biomarker as well as a target for immunotherapy,6 making its quantitation desirable.
The established method for quantifying cytochrome P450 enzymes is carbon monoxide-difference spectrometry,7 which can be applied to P450s with the detection limit of 10 pmol of P450/mL.8 This method, however, does not allow the detection of a specific P450 isoform such as CYP1B1, especially in tissue or complex samples, due to the fact that all CYP isoforms produce an identical UV absorption peak. Also, several different pigments in cells, such as mitochondrial flavorproteins, hemoproteins, and the endoplasmic reticulum cytochrome b5 significantly influence the cytochrome P450 spectrum. Thus, problems of turbidity and interfering chromophores make it impossible to accurately quantify a specific cytochrome P450 by CO-difference spectrometry. This has led to the development of immunological methods to assess CYP1B1 expression. Murray et al.9 produced polyclonal anti-CYP1B1 antibody against a synthetic peptide corresponding to amino acids 332–345 of CYP1B1, which are not shared with CYP1A1 and CYP1A2. Additionally, monoclonal antibodies against peptides corresponding to amino acids 422–436 and 437–451 of CYP1B1 were prepared for immunohistochemical studies of formalin-fixed, paraffin-embedded breast tissue.10 These studies were important for the localization of CYP1B1 in breast tumors, but they only allowed a semiquantitative assessment of CYP1B1 expression.
To achieve both specific identification and quantitation of CYP1B1, immunoassay methods using monoclonal antibodies (MAbs) were initially explored. The commonly used MAb-based immunoassays include enzyme-linked immunosorbent assay (ELISA), radioimmunoassy, and Western blotting.8 The main limitations of MAb-based immunoassays are high cross-reactivity and nonspecific adsorption. Cross-reaction would prevail when the MAbs target epitopes (antigenic sites) that are common to other cytochrome P450s. Also, the large size and structural complexity of MAbs increase their susceptibility to nonspecific adsorption and contaminant trapping. Steric hindrance is another problem encountered when antibodies are used to detect P450s in cell samples. Cytochrome P450s are microsomal membrane-bound enzymes that can be buried and undetected by MAbs used in immunoassays.
In this paper. we described a novel method for the detection of cytochrome P450 (CYP1B1) based on single-chain fragment variable (scFv) antibody and quartz crystal microbalance (QCM) transducers. QCM is an acoustic sensor based on a piezoelectric crystal. The characteristics of the sensor have been described by Sauerbrey11 and are represented by the following equation Δf = −2Δmnf02/[A(µqρq)1/2] where n is the overtone number, µq is the shear modulus of the quartz (2.947 × 1011 g/(cm·s2)), and ρq is the density of the quartz (2.648 g/cm3)). The sensor is extremely sensitive and allows for noninvasive on-line measurements of adsorption and biophysical changes. QCM can be easily automated or combined with flow injection systems to extend their capability for continuous and repeated assays.12 Recently, we have demonstrated that the decreased molecular size and homogeneity of scFvs offer significant advantages over polyclonal and monoclonal antibodies as recognition elements for immunochemical detection of antigens.13,14 Due to the small size of scFvs (MW ~27 kDa), they can be coupled, at high density, to a transducer surface to greatly improve sensor sensitivity and to significantly reduce antibody nonspecific adsorption and contaminant trapping. Thus, the detection of CYP1B1 and characterization of the binding abilities to specific antibodies or their fragments can be realized using scFv-QCM technology that not only can identify and quantify an antigen such as CYP1B1 but also can provide important information about the kinetics and thermodynamics of CYP1B1–scFv interactions.
Immunological recognition is based on the spatial complementarities of groups of amino acids within the antigenic site or epitope of an antigen with those in the paratope or antigen-binding site of the antibody. In the case of macromolecules, each antibody recognizes a specific epitope that generally constitutes a fraction of the total surface on an antigen. Multiple molecules or closely related antigens may have the same or similar epitopes. In such a case, the antibodies specific for one antigenic site would cross-react with closely related, but not identical sites on other antigens. In an effort to overcome false positives, a second antigen-specific antibody is used to detect the same antigen. The chances are that two or more closely related molecules would not share the same antigenic sites at different locations on the same molecule. In this study, three scFvs specific for different antigenic sites on CYP1B1 were used in QCM assays to specifically detect CYP1B1 and to reduce assay false positive results. Positive detection requires simultaneous binding on at least two of the scFv-modified QCMs. Mouse and rat antibodies contain two naturally occurring lysines, which are located near the C-terminus of every antibody light-chain variable region. The free amines located on these two lysines, on other lysines that may be present at other locations in an scFv, or at the scFv amino terminus, can be readily biotinylated using commercially available biotinylation reagents, without destroying the scFv antigen-binding activity or specificity. Three anti-CYP1B1 scFv antibodies designated B-66, L-21, and D-23 were biotinylated at a ratio of ~2 biotins to 1 scFv. The biotinylated scFv, immobilized onto neutravidin-coated gold QCM surfaces, were used to quantify CYP1B1 in breast cancer microsomes, cancer cell lysates (Hela, HCC, 4T1), and normal cell lysates. The scFv-QCM biosensors showed excellent sensitivity [detection limit, 2.2 ± 0.9 nM (n = 3)] and specificity (confirmed by utilizing different negative control antigens). The binding affinity for the scFv–CYP1B1 interaction was also determined.
MATERIALS AND METHODS
Reagents
Bovine serum albumin and rabbit IgG were purchased from Sigma Inc. Phosphate-buffered saline (PBS) and fetal bovine serum (FBS) were obtained from Gibco. All other chemicals were purchased from Aldrich (reagent grade) and used as received.
Production, Characterization, and Biotinylation of anti-CYP1B1 scFvs
Construction, size, and diversity of the rodent phage displayed scFv antibody library used to select for anti-CYP1B1 scFv antibodies have been described.15 Phage antibody selections and E-tagged soluble scFv antibody assays were published earlier.15 ScFv recombinant antibodies were cloned into the pCANTAB5E phagemid expression vector. Each scFv antibody using pCANTAB5E displays a tag known as the E-tag. An anti-E tag monoclonal antibody is used to detect E-tagged scFv bound to an antigen in an assay and is also used to affinity purify E-tagged scFv expressed by Escherichia coli. Two rounds of phage antibody selection were performed using biotinylated CYP1B1 and streptavidin magnetic beads. Phage-displayed scFvs bound to CYP1B1 were eluted using 100 mM triethylamine. The pH of the eluted phage antibodies was neutralized using 1 M Tris buffer, pH 7.4–8. Eluted phage antibodies were used to infect E. coli TG1 cells. Infected TG1 cells were plated onto 2xYT AG (2xYT bacterial culture medium containing 100 µg/mL ampicillin and 2% glucose) agar plates and grown overnight at 30 °C. Colonies were M13KO7 helper phage rescued for a second round of selection on CYP1B1. Colonies stemming from a second round of selection on CYP1B1 were individually picked into microtiter plates containing 2xYT AI (2xYT medium containing 100 µg/mL ampicillin and 1 mM IPTG) to induce soluble E-tagged scFv expression. Bacterial clones (designated I-20, D-23, L-21, and B-66) were identified by ELISA as producing CYP1B1-specific scFv and were scaled up and purified using the RPAS purification kit (G.E. Healthcare) according to the manufacturer’s instructions. All purified scFvs were biotinylated at a ratio of ~2:1 (2 biotins/1 ScFv) using biotinamidocaproate-NHS.
Normal and Tumor Cell Lines
Human ECV22 endothelial cells, human umbilical vein endothelial cells (Huvec), mouse liver cells, and African Green Monkey Cos7 kidney fibroblast cells were used as sources for normal cells. Human cervical carcinoma HeLa cells, human hepatocellular carcinoma HCC cells, and 4T1 mouse mammary tumor cells were used as a cancer cell source. All cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA).
Cell Cultures and Cell Lysate Preparation
All cell lines were cultured as specified by ATCC protocols. The cells were washed with PBS three times and then resuspended in a lysis buffer (1 mM sodium phosphate, 1 mM EDTA, and 1 mM Tris-HCl, pH 7.5 plus protease inhibitors), and kept on ice for 30 min to lyse cells. The cell suspensions were freeze-thawed five times using dry ice and room-temperature water. The cell lysates were centrifuged at 3000 rpm for 10 min. Supernatants from cell lysates were transferred to microcentrifuge tubes for use in this study.
Preparation of T47D Human Breast Cancer Cell Microsomes
Confluent T47D cells were grown in the presence of 10 nM TCDD in 0.1% DMSO for 48 h to induce CYP1B1 expression. T47D cells were then prepared according to Spink et al. to prepare microsomes containing CYP1B1.16
Protein Concentration Assay
The protein concentration in cell lysates was determined using a BCA protein assay kit according to the protocol provided by the company (Pierce, Rockford, IL). BSA was used as a protein standard.
CO-Difference Spectrometry
The protocol reported by Omura and Sato7 was used to obtain visible spectra of the CO–CYP1B1complex. Briefly, 2 mL of solution (e.g., 100 µL of CYP1B1 sample and 1900 µL of spectral buffer (50 mM K3PO4, 0.1 mM EDTA, pH 7.4, 20% glycerol)) was prepared and kept on ice until measurements were made. Carbon monoxide was bubbled through the sample cuvette using a disposable glass pipet for 1 min, with a total of 60–100 bubbles. Then, a pinch of solid sodium dithionite was added to both sample and reference cuvettes, inverting each 5 or 6 times. Visible spectra were obtained for both reference and sample cuvettes between 400 and 500 nm.
Preparation of CYP1B1 Standard Samples
Recombinant purified human CYP1A1 and CYP1B1 proteins were purified as previously described.17 It was found that sodium carbonate could strip extrinsic proteins from microsomal membrane without affecting transmembrane and lipid-anchored proteins such as the CYP proteins.18 Therefore, sodium carbonate was used to remove extrinsic proteins from microsomal membranes to enable CYP1B1 quantification via spectrophotometric analysis and biosensor assays. Experiments were also performed to characterize the effects of detergents and lipids on the binding activity of the CYP1B1 scFvs immobilized on QCM surfaces. Microsomes from E. coli in which CYP1B1 was absent served as negative controls.
Immobilization Procedure
A nonpolished gold piezoelectric quartz surface was cleaned with concentrated sulfuric and nitric acid mixture (1:1 v/v), biograde water (i.e., 2-µm prefiltered 18-MΩ deionized water treated with UV radiation), and ethanol in series three times and dried using nitrogen. The fresh-washed gold quartz crystal surface was immersed in neutravidin (1 mg/mL) in PBS for 6–8 h at 4 °C and then rinsed with PBS and biograde water to remove unbound neutravidin. The neutravidin-coated Au QCM surface was immersed in the biotinylated scFv solution (~0.5 mg/mL) for 3–4 h, blocked with 0.1% BSA in PBS for 0.5 h, washed with PBS and biograde water, and then dried under nitrogen.
QCM Measurements
A 1000Å thickness, 0.23 cm2 AT-cut 10 MHz nonpolished gold quartz crystal (International Crystal Co., Inc.) was mounted in a Kel-F cell. The biotinylated scFv/neutravidin-coated gold surface was placed in a Faraday cage containing 1 mL of PBS that was continuously stirred during measurement. A network/spectrum/impedance analyzer (Agilent 4395A) was used to measure the change of frequency and damping resistance caused by the analyte addition.
RESULTS AND DISCUSSION
ScFv-QCM Sensor Sensitivity
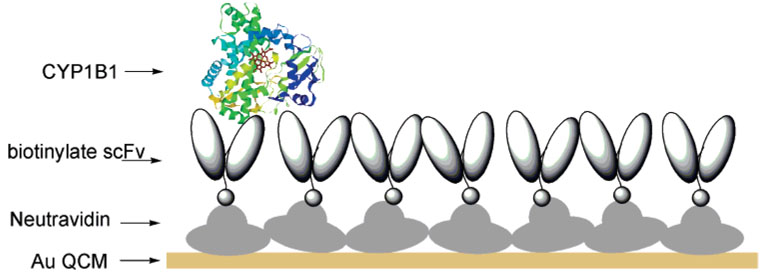
ScFv-QCM sensor sensitivity and specificity are dependent upon scFv affinity (binding strength), orientation, and sensor surface coverage. In this study, biotinylated scFv antibodies (designated D-23, B-66, L-21), specific for different CYP1B1 antigenic sites, were immobilized onto a neutravidin-coated gold QCM surface (Figure 1). Samples containing different CYP1B1 concentrations were sequentially added to one QCM sensor to produce the calibration curve.19,20 The concentration of biotinylated scFv was kept high to ensure that scFv antigen-binding sites would not become saturated upon sequential addition of CYP1B1.
Figure 1.
Schematic representation of scFv-QCM-based CYP1B1 measurements.
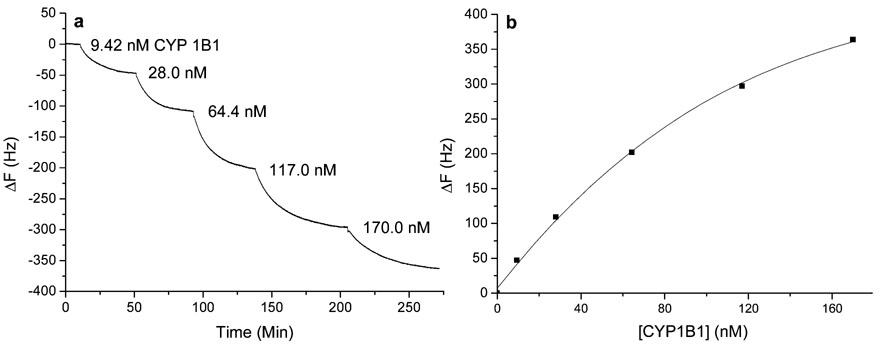
Figure 2a is a representative dose–response curve when samples containing purified CYP1B1 were sequentially added to scFv B-66 modified QCM sensor. A linear relationship was obtained when CYP1B1 concentrations ranging from ~9 to 120 nM were plotted against frequency changes for all scFv-QCMs. The representative calibration curve (i.e., ΔF vs C curve) for scFv B-66 QCM sensor is shown in Figure 2b. All scFvs used to detect CYP1B1 exhibited similar binding constants [Kd = (1.54 ± 0.59) × 10−7 M] and detection limits (2.2 ± 0.9 nM). The standard deviation was obtained on individual calibration curves. All calibration curves gave similar linear ranges and detection limits. The anti-CYP1B1 scFv piezoimmunosensors exhibited nanomolar sensitivity.
Figure 2.
The scFv B-66 QCM dose response curves. (a) Frequency change vs time when purified CYP1B1 samples were consecutively added to 1 mL of PBS; (b) Frequency change vs purified CYP1B1 concentration curve.
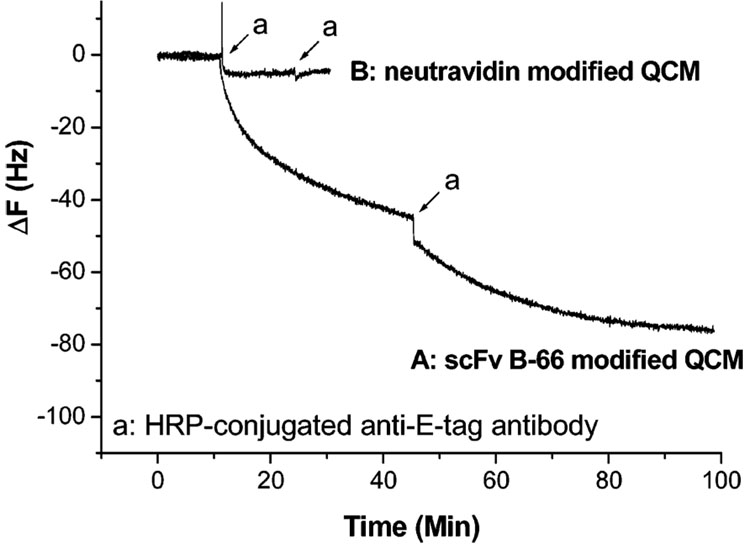
The scFv immobilization on gold QCM surfaces was further characterized by an immunoassay using the horseradish peroxidase-conjugated, anti-E tag monoclonal antibody to detect biotinylated E-tagged scFv immobilized onto neutravidin-coated QCM surfaces. After an addition of peroxidase-conjugated anti-E tag antibody to the scFv-modified electrode, the sample was incubated at room temperature for 1 h and then the surface was rinsed with PBS. When the H2O2/ABTS peroxidase substrate (1.8 µL of 30% H2O2/mL of ABTS) was added to the QCM surface, a green color was observed, indicating that the peroxidase-conjugated anti-E tag monoclonal antibody bound to the biotinylated scFv immobilized onto the neutravidin-coated QCM surface. The addition of anti-E-tag antibody to the biotinylated scFv B-66 on the neutravidin-coated QCM surface produced a decrease in frequency of 80 Hz. This was not observed when the anti-E-tag antibody was applied to the QCM electrode bearing neutravidin only (Figure 3). These results indicated that biotinylated scFvs were successfully immobilized onto the neutravidin-coated QCM surface.
Figure 3.
QCM sensor responses to consecutive additions of 20 µL of HRP-conjugated anti-E-tag antibodies. Curve A: biotinylated scFv B-66 on neutravidin-coated QCM surface. Curve B (negative control): neutravidin-coated QCM surface. Final concentration of HRP-conjugated anti-E-tag antibodies was 1.60 × 10−6 M.
ScFv-QCM Sensor Specificity
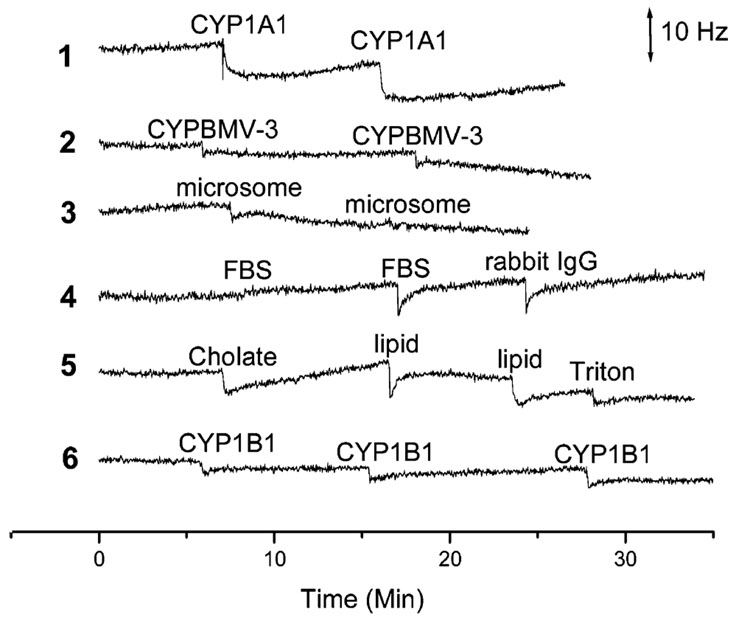
QCM is a mass sensor, and any contaminating molecule that nonspecifically binds to the QCM surface can adversely interfere with a QCM-based assay. The amount of nonspecific adsorption may be small, but can have a significant effect on assay sensitivity if the concentration of the antigen in a sample is very low. CYP1B1 is a detergent-soluble eukaryotic P450 enzyme that is found associated with lipids and other proteins in cell microsomal membranes. Anti-CYP1B1 scFv-QCM sensor sensitivity and specificity in the presence of contaminants were ascertained by adding proteins [rabbit IgG, FBS, CYP1A1 (a eukaryotic P450 similar but not identical to CYP1B1), BMV-3 (a P450 from the bacterium Bacillus megatarium)], lipids, detergents, or fractionated microsomes to the anti-CYP1B1 scFv QCM assay (Figure 4). Almost little or no nonspecific adsorption was detected when CYP1A1 (curve 1), P450 BMV3 (curve 2), microsomes devoid of CYP1B1 (curve 3), rabbit IgG and FBS (curve 4), or lipids and detergents (curve 5) were added to the anti-CYP1B1 QCM sensor. Less than 10 Hz signal was observed showing nonsignificant cross-activity of our CYP1B1 QCM immunosensors for CYP1A1 and P450 BMV-3 (compared to ~200 Hz decrease when 64.4 nM CYP1B1 was added to the same system).
Figure 4.
The scFv B-66 QCM sensors exposed to 60 nM CYP1A1 (curve 1); 30 nM P450 BMV-3 (curve 2); 2.4 µg/mL microsome without CYP1B1 (curve 3); 14.4 µg/mL FBS and 130 nM rabbit IgG (curve 4); 0.01% cholate, 0.02% Triton, and 6.43 × 10−5 M lipid (1,2-dilauroyl-sn-glycero-3-phosphocholine) (curve 5). Curve 6: Negative control scFv (designated 210E) immobilized onto QCM surface exposed to 60 nM CYP1B1.
A recombinant antibody 210E scFv-cys (binds specifically to rabbit IgG antigen14) modified QCM surface was used additionally as a negative control to examine the specificity of the sensor. Negligible frequency change was observed by the addition of CYP1B1 to the negative control of 210E scFv modified sensor surface (curve 6). These results indicated that the scFv-based QCM electrodes specifically detected CYP1B1 in the presence of contaminants.
Binding Affinity
In order to obtain accurate affinity constant for the binding between CYP1B1 and scFvs, we need to analyze the energy losses introduced to the oscillatory system due to (1) a viscoelastic porous structure that is strained during oscillation, (2) trapped liquid that moves between or in and out of pores due to the deformation of the film, or (3) the load from the bulk liquid, which increases the strain of film. Measurements were made using a QCM impedance analyzer (Agilent 4395A network impedance analyzer). The QCM impedance analyzer allows for the simultaneous measurements of the frequency and the energy dissipation during antibody and antigen-binding processes. By obtaining the damping resistance through fitting the Butworth-van Dyek circuit, we can verify the validity of Sauerbrey equation and the dissipative properties if the modified layer shows viscoelastic properties. In the course of all experiments, the damping resistance fluctuated within a range of |ΔRq|/Rq ≤ 1.0% (Table 1). This result proved that the attached biofilms behaved as a rigidly attached mass and the Sauerbrey equation is valid in our systems.
Table 1.
Changes in Damping Resistances for QCM Experiments
| Figure | |ΔRq|/Rq (%) | Figure | |ΔRq|/Rq (%) |
|---|---|---|---|
| 2a | 0.6 | 4(1) | 0.6 |
| 3A | 1.0 | 4(2) | 0.2 |
| 3B | 0.04 | 4(3) | 0.2 |
| 6b | 0.3 | 4(4) | 0.3 |
| 7 | 0.7 | 4(5) | 0.1 |
| 4(6) | 0.2 |
The CYP1B1/scFv binding association constant (Ka), binding amount at the nanogram level (ΔM), can be obtained from the time relationship of frequency decrease at various CYP1B1 antigen concentrations.
The CYP1B1/scFv binding interaction is described in eq 1.
| (1) |
Based on a Langmuir adsorption isotherm,21 the association (Ka) and dissociation (Kd) constants for CYP1B1/scFv binding can be described by eq 2. In eq 2, ΔMmax is the maximum amount that
| (2) |
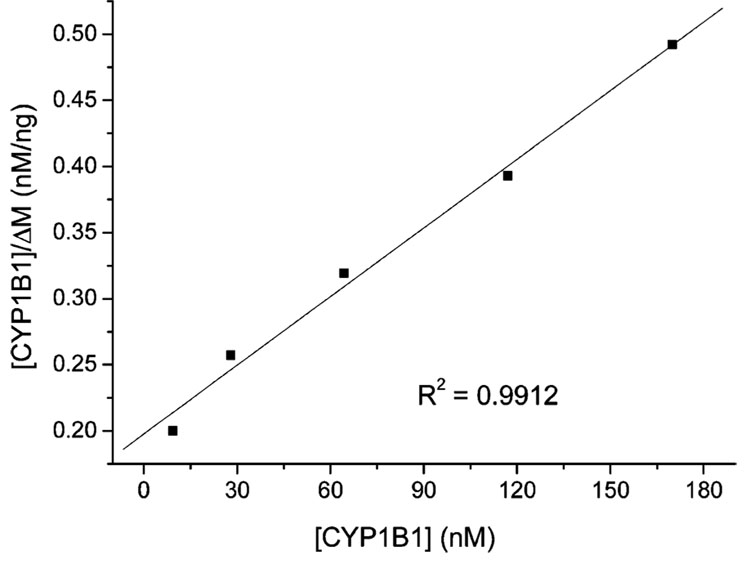
CYP1B1 can be bound and ΔMmax is the characteristics of the sensor surface, which is equivalent to the binding of CYP1B1 when all the scFvs are converted to the CYP1B1-scFv complex. ΔM is the measured CYP1B1 binding amount at equilibrium, which is a function of the CYP1B1 concentration and will not change with the time, and [CYP1B1] is the original concentration of CYP1B1. Since ΔM is unrelated to the previous addition of CYP1B1,19 based on the data obtained from Figure 2a, we can obtain Figure 5, a representative plot of [CYP1B1]/ΔM versus [CYP1B1]. According to eq 2, the ratio of the slope to the intercept gave the association constant (Ka). Dissociation constant (Kd) can be calculated as 1/Ka. Different anti-CYP1B1 scFv immobilized onto biosensor surfaces gave similar Kd as summarized in Table 2.
Figure 5.
[CYP1B1]/ΔM vs [CYP1B1] of CYP1B1 binding with scFv B-66.
Table 2.
Dissociate Constants (Kd) for scFv–CYP1B1 Interactions
| scFv QCM | scFv D-23 | scFv B-66 | scFv L-21 |
|---|---|---|---|
| Kd (10−7 M) (n = 3) | 1.59 ± 0.91 | 1.52 ± 0.34 | 1.52 ± 0.68 |
ScFv-QCM Biosensor Quantification of CYP1B1 in Microsome and Cell Lysate Samples
ELISA and CO-difference spectrometry were attempted in our laboratories to analyze CYP1B1 in normal and cancer cell lysates. However, our study showed that CO-difference spectrometry was not sensitive enough and required a large amount of samples to obtain a suitable spectrum to quantify CYP1B1. Two antibodies specific for different sites on the same antigen can be used in an ELISA to detect and quantify and antigen in a sample. Typically, one antibody is used to capture the antigen out of solution while a second antibody, labeled with a reporter molecule such as an enzyme, is used to detect the captured antibody bound to the first antibody. It can be difficult to find antibody pairs that can be used to capture and detect an antigen in an ELISA. On the other hand, scFv-based QCM biosensors use only one unlabeled scFv to detect and quantify antigens in samples that contain irrelevant molecules or contaminants. Being label free, it exempt from the time and cost demanding labeling step, which consequently eliminates any possible interference to the “true” binding process due to the presence of the labels. Furthermore, they showed excellent sensitivity and specificity in standard conditions. The little nonspecific adsorption observed for the scFv-immobilized sensor surface might be the combination of high density of correct oriented scFv on the surface and effective use of the BSA blocking reagents. As a result, scFv-QCM biosensors were applied to quantify CYP1B1 in cell samples such as breast cancer microsomes, Hela cells, HCC, and normal cell lysates.
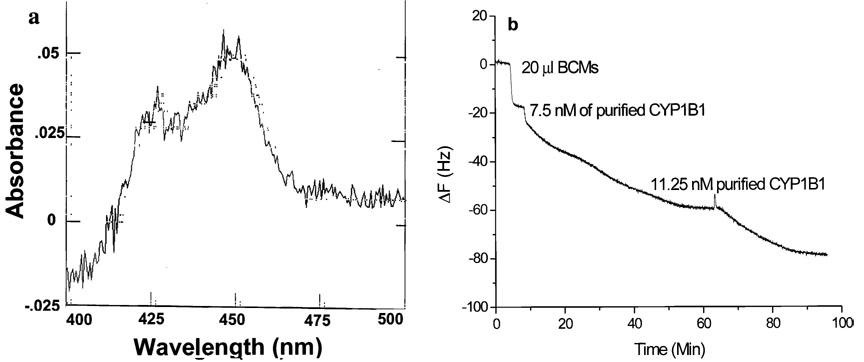
An example of the scFv B-66 QCM response curves obtained after addition of the breast cancer microsome sample is presented in Figure 6b. A small volume of diluted T47D human breast cancer microsomes (BCMs) was added to the scFv B-66-modified QCM sensor, followed by the successive addition of standard CYP1B1; the final concentration of CYP1B1 in BCMs was calculated to be 7 × 10−7 M according to the QCM analysis. To validate this result, we analyzed the same batch of samples using CO-difference spectrometry (Figure 6a) and the CYP1B1 concentration was determined to be 1.1 × 10−6 M. The estimated amount of CYP1B1, calculated through the scFv-QCM biosensor, was relatively lower than the value obtained by CO-difference spectrometry. This can be explained by the presence of other P450 subfamilies, such as CYP1A1 in the sample, which absorb light at same wavelength, thereby increasing the total estimated total amount as estimated by CO-difference spectrometry. In contrast, anti-CYP1B1 scFv-based QCM analysis measures only CYP1B1 and provides a more precise concentration.
Figure 6.
(a) CO-difference spectrum of diluted T47D human BCMs; (b) scFv B-66 QCM sensor response to the addition 20 µL of BCMs (1:4 dilution by PBS-T containing 0.5% Triton X-100), 7.5 nM of purified CYP1B1, and 11.25 nM purified CYP1B1.
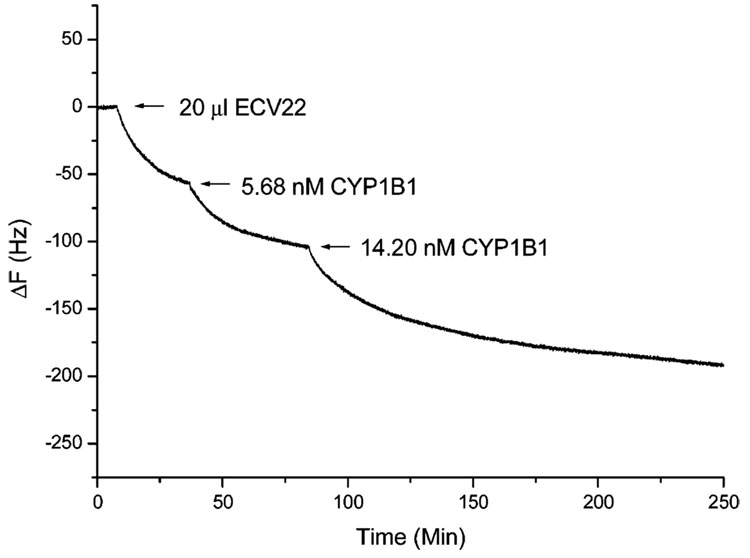
The scFv-QCM sensors were also used for the direct detection of CYP1B1 in cancer cell lysates (Hela, HCC, 4T1) and in normal cell lysates (ECV22, Cos7, Huvev, mouse liver) via the method described above. Figure 7 is a representative dose–response curve for analysis of CYP1b1 concentration in ECV22 cell lysate sample. The final concentrations of CYP1B1 are listed in Table 3.
Figure 7.
scFv D-23 QCM sensor response to the addition 20 µL of ECV22, 5.68 nM of purified CYP1B1, and 14.20 nM purified CYP1B1. Similar procedures were used to analyze all other samples, and results are presented in Table 3.
Table 3.
CYP1B1 Concentrations in Cell Lysates Measured by scFv-QCM Sensorsa
| cancer cell lysates |
normal cell lysates |
|||
|---|---|---|---|---|
| scFv-QCM | Hela | HCC | 4T1 | ECV22 |
| B66-QCM | 118.8 µg/mL | 6.9 µg/mL | 74.8 µg/mL | 10.3 µg/mL |
| (6.3%) | (4.3%) | (4.8%) | (0.67%) | |
| D-23 QCM | 105.6 µg/mL | 6.2 µg/mL | 60.5 µg/mL | 14.8 µg/mL |
| (5.6%) | (3.9%) | (3.9%) | (0.97%) | |
| L-21 QCM | 127.0 µg/mL | 3.5 µg/mL | 50.6 µg/mL | 12.3 µg/mL |
| (6.8%) | (2.2%) | (3.3%) | (0.80%) | |
| total protein concns | 1.88 mg/mL | 0.16 mg/mL | 1.55 mg/mL | 1.53 mg/mL |
| average | 117.1 ± 10.8 µg/mL | 5.9 ± 1.8 µg/mL | 60.0 ± 12.2 µg/mL | 12.5 ± 2.2 µg/mL |
| results | (6.2 ± 0.6%) | (3.5 ± 1.1%) | (4.0 ± 0.8%) | (0.81 ± 0.15%) |
The first number in each block represents the concentration of CYP1B1 (µg/mL), while the second number (in parentheses) represents the percentage of CYP1B1 relative to the total protein concentration in the sample.
The anti-CYP1B1 scFvs B-66, D-23, and L-21 used to determine CYP1B1 concentration in QCM assays gave similar results. The percentage of CYP1B1 in cancer cell lysates (e.g., Hela, 6.2%) was higher than in normal cell lysates (e.g., ECV22, 0.81%). When normal (Huvec) cell lysate samples, which expressed very little CYP1B1, were added to the scFv-QCM sensors, no response was observed. This result showed little CYP1B1 expression in normal mammary epithelial cells but a high level of expression in the cancer cells, which was consistent with our early studies in which the scFvs were used in immunohistochemistry to detect CYP1B1 in normal and breast cancer human samples. The immunohistochemical study showed that large numbers of tumor cells were stained with scFv I-20 while scFv I-20 did not react with normal human breast tissues (results not shown). Our current sensor analysis suggested that the anti-CYP1B1 scFv QCM assay could be used to specifically detect CYP1B1 in cell samples.
CONCLUSION
Our results demonstrated that an anti-CYP1B1 scFv-based QCM assay could be used to detect and quantify CYP1B1 in cell samples. Results of this study suggest that CYP1B1 enzymes are overexpressed in some tumor cell lysates (Hela, HCC, and 4T1). Normal cell lysates (ECV22, Cos7, and mouse liver) also express CYP1B1, but at a much lower level. The presence of specific P450 enzymes in individual tumors may have diagnostic and therapeutic applications. The anti-CYP1B1 scFv-based QCM assay can potentially be used to determine the stage during tumor development when P450s are upregulated and the identification of the endogenous function(s) of P450 in tumor cells. To answer these questions will require the development of appropriate systems for the systematic study of P450 expression, regulation, and function in tumor cells by using sensor systems, DNA microarray, and proteomic technologies. We described here the first scFv-QCM biosensor for CYP1B1 detection in model solutions and cell samples. Compared to traditional methods like ELISA and CO-difference spectrometry, scFv-QCM offers several advantages including higher selectivity and specificity and therefore shows promise as a possible way to clarify questions regarding cancer diagnostics, monitoring, and treatment. The method can be used for analysis and characterization of other cytochrome P450 enzymes.
ACKNOWLEDGMENT
This research was supported by NIH (1R21EB000672-01, 4R33EB000672-02, 5P30 CA68485-07, 5P30 ES00267-36), Oakland University Research Excellent Fund, and faculty start-up funds. X.Z. thanks Dr. Arthur Bull for his helpful discussions during this study and Mr. Yijun Tan and Dr. Tatiana Delaney’s contribution in preparing the manuscript. Zhihong Shen and Heping Yan contributed equally to this work.
References
- 1.Guengerich FP. Chem. Res. Toxicol. 2001;14:611–650. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- 2.Fishbain DA, Fishbain D, Lewis J, Cutler RB, Cole B, Rosomoff HL, Rosomoff RS. Pain Med. 2004;5:81–93. doi: 10.1111/j.1526-4637.2004.04007.x. [DOI] [PubMed] [Google Scholar]
- 3.Yager JD, Davidson NE. N. Engl. J. Med. 2006;354:270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 4.Murray GI, Taylor MC, Burke MD, Melvin WT. Br. J. Cancer. 1998;77:1040–1044. doi: 10.1038/bjc.1998.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maecker B, Sherr DH, Vonderheide RH, von Bergwelt-Baildon MS, Hirano N, Anderson KS, Xia ZN, Butler MO, Wucherpfennig KW, O’Hara C, Cole G, Kwak SS, Ramstedt U, Tomlinson AJ, Chicz RM, Nadler LM, Schultze JL. Blood. 2003;102:3287–3294. doi: 10.1182/blood-2003-05-1374. [DOI] [PubMed] [Google Scholar]
- 6.McFadyen MM, Graeme I. Future Oncol. 2005;1:259–263. doi: 10.1517/14796694.1.2.259. [DOI] [PubMed] [Google Scholar]
- 7.Omura T, Sato R. J. Biol. Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 8.Phillips IR, Shephard EA, editors. Cytochrome P450 protocols. Totowa, NJ: Humana Press; 2006. [Google Scholar]
- 9.Murray GI, Taylor MC, McFadyen MCE, McKay JA, Greenlee WF, Burke MD, Melvin WT. Cancer Res. 1997;57:3026–3031. [PubMed] [Google Scholar]
- 10.McFadyen MCE, Breeman S, Payne S, Stirk C, Miller ID, Melvin WT, Murray GI. J. Histochem. Cytochem. 1999;47:1457–1464. doi: 10.1177/002215549904701111. [DOI] [PubMed] [Google Scholar]
- 11.Sauerbrey GZ. Phys. 1959;155:206–222. [Google Scholar]
- 12.Sota H, Yoshimine H, Whittier RF, Gotoh M, Shinohara Y, Hasegawa Y, Okahata Y. Anal. Chem. 2002;74:3592–3598. doi: 10.1021/ac025526b. [DOI] [PubMed] [Google Scholar]
- 13.Shen ZH, Mernaugh RL, Yan HP, Yu L, Zhang Y, Zeng XQ. Anal. Chem. 2005;77:6834–6842. doi: 10.1021/ac0507690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen Z, Stryker GA, Mernaugh RL, Yu L, Yan HP, Zeng XQ. Anal. Chem. 2005;77:797–805. doi: 10.1021/ac048655w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hennig EE, Mernaugh R, Edl J, Cao P, Cover TL. Infect. Immun. 2004;72:3429–3435. doi: 10.1128/IAI.72.6.3429-3435.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spink DC, Spink BC, Cao JQ, Gierthy JF, Hayes CL, Li Y, Sutter TR. J. Steroid Biochem. Mol. Biol. 1997;62:223–232. doi: 10.1016/s0960-0760(97)00024-1. [DOI] [PubMed] [Google Scholar]
- 17.Hanna IH, Dawling S, Roodi N, Guengerich FP, Parl FF. Cancer Res. 2000;60:3440–3444. [PubMed] [Google Scholar]
- 18.Clark B, Waterman M. J. Biol. Chem. 1991;266:5898–5904. [PubMed] [Google Scholar]
- 19.Tang Y, Mernaugh R, Zeng X. Anal. Chem. 2006;78:1841–1848. doi: 10.1021/ac051868g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Luo S, Tang Y, Yu L, Hou K-Y, Cheng J-P, Zeng X, Wang PG. Anal. Chem. 2006;78:2001–2008. doi: 10.1021/ac051919+. [DOI] [PubMed] [Google Scholar]
- 21.Hengerer A, Decker J, Prohaska E, Hauck S, Kösslinger C, Wolf H. Biosens. Bioelectron. 1999;14:139–144. doi: 10.1016/s0956-5663(98)00111-0. [DOI] [PubMed] [Google Scholar]