Abstract
Inward-rectifier K+ (Kir) channels play many important biological roles and are emerging as important therapeutic targets. Subtype-specific inhibitors would be useful tools for studying the channels' physiological functions. Unfortunately, available K+ channel inhibitors generally lack the necessary specificity for their reliable use as pharmacological tools to dissect the various kinds of K+ channel currents in situ. The highly conserved nature of the inhibitor targets accounts for the great difficulty in finding inhibitors specific for a given class of K+ channels or, worse, individual subtypes within a class. Here, by modifying a toxin from the honey bee venom, we have successfully engineered an inhibitor that blocks Kir1 with high (1 nM) affinity and high (>250-fold) selectivity over many commonly studied Kir subtypes. This success not only yields a highly desirable tool but, perhaps more importantly, demonstrates the practical feasibility of engineering subtype-specific K+ channel inhibitors.
Keywords: block, inhibition, K channel, tertiapin, toxin
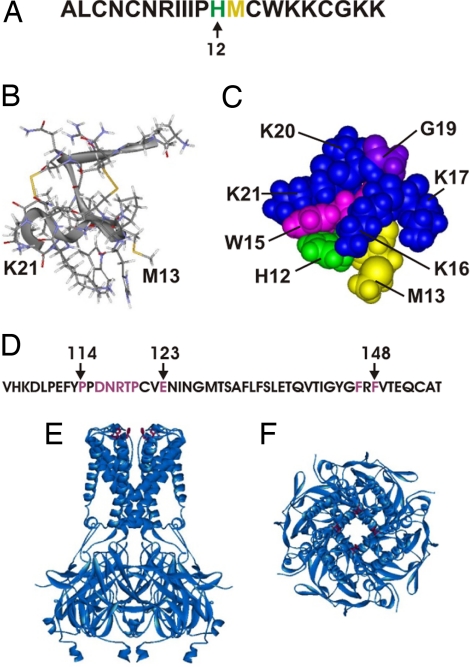
Tertiapin (TPN), a toxin present in honey bee venom (Fig. 1A), inhibits certain subtypes of inward-rectifier K+ channels (1). Kinetic studies indicate that the toxin inhibits the channels with 1:1 stoichiometry (2, 3). It does so by binding to the external vestibule of the K+-conduction pore that is formed by the linker between the first and second transmembrane (M1–M2) segments. This latter inference is derived from the following observations. First, transferring the linker of a TPN-sensitive channel to a nonsensitive channel confers TPN sensitivity on the recipient channel (4). Second, alanine (and other) mutation at many residues (114–123, 146, and 148 of Kir1.1) within the linker (Fig. 1D) significantly affects toxin binding to the channel (2, 3).
Fig. 1.
Sequences and structures of TPN and Kir. (A) Primary sequence of TPN with residues 12 (green) and 13 (yellow). (B) Ribbon-and-stick representation of the NMR structure of TPN (5). (C) CPK representation of the channel interaction surface of TPN. (D) Primary sequence of the M1–M2 linker of Kir1.1, where the residues whose alanine mutation affects TPN binding >1.5 kcal/mol (2) are colored purple. (E and F) Side (E) and extracellular (F) views of a ribbon representation of the crystal structure of a chimera, which is formed by joining the transmembrane pore of a bacterial Kir with the cytoplasmic pore of eukaryotic Kir3.1 (28). The four equivalent residues that correspond to F148 of Kir1.1 are maroon.
TPN is a highly compact protein of 21 residues. Its C-terminal portion (histidine 12 to glycine 19) adopts an α-helical structure, whereas the N-terminal half acquires extended conformations (5) (Fig. 1B). Two pairs of disulfide bonds help to hold the two parts together. Alanine-scanning mutagenesis shows that TPN′s channel-interacting surface is mainly formed by its α-helix (2) (Fig. 1C), whereas classic pore-blocking scorpion toxins bind to the pore of a voltage- or Ca2+-activated K+ (Kv or Kca) channel mainly via certain β-sheets (6–9). A mutant cycle study shows that residue 13 of TPN (Fig. 1 A–C) interacts with F148 of Kir1.1 (10) (Fig. 1 D–F), a channel residue located near the external opening of the pore.
There are many small protein-based toxins that inhibit Kv or Kca channels; each toxin usually inhibits several Kv subtypes with different affinities. However, they generally do not have the necessary specificity to be used reliably as pharmacological tools to dissect the various kinds of K+ channel currents in situ (11). This is also the case for TPN. For example, TPN inhibits homotetrameric Kir1.1 (12) with a Kd of ≈1 nM and heterotetrameric Kir3.1/3.4 (13–15) with ≈10-fold lower affinity (1). The affinity of Kir3.1/3.4 for TPN seems to reside in the Kir3.4, not Kir3.1, subunit, because the mutant homomeric Kir3.4, not Kir3.1, channels are sensitive to tertiapin (4).
The cardiac Kir3.1/3.4 mediates vagal control of pacemaker activity, whereas the renal Kir1 mediates salt reabsorption. Genetic defects in Kir1 lead to type II Bartter's syndrome with characteristic volume depletion and hypotension (16). The fact that mutations in Kir1 cause hypotension makes this channel a plausible novel target in the treatment of hypertension. Unfortunately, TPN′s expected cardiac side-effects would thwart attempts to test the concept of this novel anti-hypertensive approach.
Results
To develop a TPN derivative highly (>100-fold) selective for Kir1 we had carried out extensive studies that included systematic mutagenesis (2, 4), mutant cycle analysis, and examination of semirandom peptide libraries. Through these studies, we found that the H12K mutant preferentially inhibits Kir1 over Kir3.1/3.4 with additional 8-fold higher selectivity than the wild type. We thus first focused on the role of residue 12 (Fig. 1 A and C) in determining TPN′s channel selectivity. The overall amino acid sequences of members within a given Kir subtype (e.g., Kir3.1 versus Kir3.4) are significantly different, except the Kir1 subtype. The Kir1.1, Kir1.2 and Kir1.3 are alternative splicing variants that differ only in the length of their N terminus (12, 17–19) and all three should thus interact with TPN in the same way. For continuity, we used Kir1.1 to carry out the study.
Effects of Mutations of TPN on Channel Selectivity.
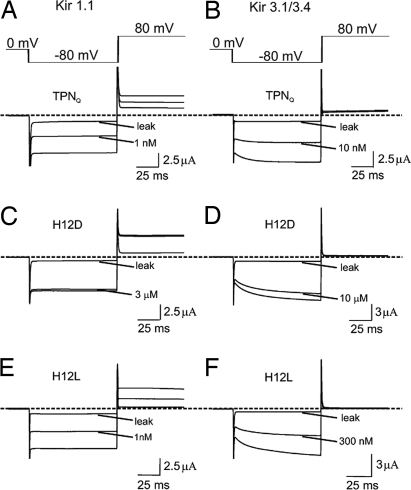
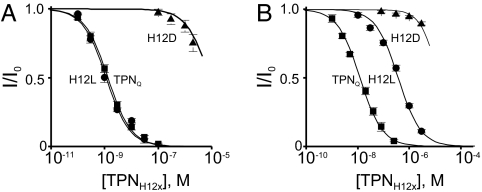
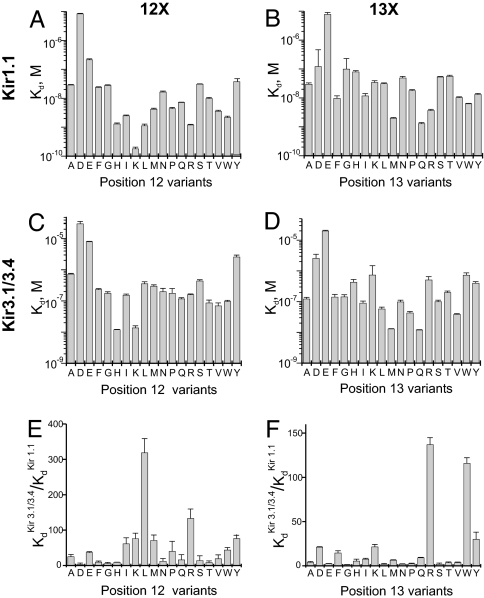
We first examined the selectivity of TPN variants in which H12 was replaced with each of the remaining natural residues except cysteine, omitted so as not to interfere with the formation of two essential disulfide bridges. The replacement was done on the background of the M13Q mutation, which prevents the spontaneous oxidation of TPN that lowers its affinity (10) (the derivative with the M13Q mutation is denoted as TPNQ). Fig. 2 shows current traces of Kir1.1 and Kir3.1/3.4 channels, recorded in the presence of TPNQ or of its derivatives H12D and H12L. Inhibition constants (Kd values) were obtained from plots of the fraction of current remaining, as illustrated for the inhibition of Kir1.1 (Fig. 3A) and Kir3.1/3.4 (Fig. 3B) by TPNQ and its H12D and H12L derivatives. Kd values of all 19 variants for Kir1.1 and Kir3.1/3.4 are graphed in Fig. 4 A and C, respectively. The channel selectivity of individual variants, defined as the ratio of Kd between the two channels tested, varies greatly, ranging from only 4-fold for the H12D derivative to as high as 320-fold for the H12L derivative (Fig. 4E). Recall that TPN itself (as well as TPNQ (10) has a 10-fold higher affinity for Kir1.1 than for Kir3.1/3.4. Thus, the H12L mutation increases the selectivity by an additional 30-fold. The H12L variant has a slightly higher affinity (Kd = 1.1 nM) for Kir1.1 than TPNQ itself (Kd = 1.3 nM) but a 30-fold lower affinity for Kir3.1/3.4 (Kd = 361 nM). Therefore, the increased Kir1.1 selectivity of the H12L variant derives almost completely from the 30-fold reduction in its affinity for Kir3.1/3.4, a desired outcome. We will refer to this H12L variant of TPNQ as TPNLQ.
Fig. 2.
Inhibition of Kir1.1 and Kir3.1/3.4 by TPN derivatives. Currents of Kir1.1 (A, C, and E) and Kir3.1/3.4 (B, D, and F) elicited with the voltage protocol shown, in the absence or presence of TPNQ or it derivatives. All leak currents were obtained after adding TPNQ at concentrations >100 times the Kd value. Dashed lines indicate the zero current levels.
Fig. 3.
Analysis of Kir1.1 and Kir31./3.4 inhibition by TPN derivatives. Fraction of Kir1.1 (A) or Kir3.1/3.4 (B) current remaining (mean ± SEM., n = 5) plotted against the toxin concentration. The curves through the data are fits of the equation I/Io = Kd/(Kd + [toxin]).
Fig. 4.
Kd values of 37 TPN variants for Kir1.1 and Kir3.1/3.4, and their selectivity for Kir1.1 over Kir3.1/3.4. (A–D) Kd values (mean ± SEM., n = 5) for Kir1.1 (A and B) and for Kir3.1/3.4 (C and D), where position 12 (A and C) or position 13 (B and D) of TPN was occupied by each possible natural residue except cysteine. The position 12 series was examined with Q in position 13, and the position 13 series with H in position 12. (E and F) Ratios of Kd values of each variant for Kir3.1/3.4 versus Kir1.1.
We next examined whether mutations at the neighboring residue 13 also affect channel selectivity, which has been shown to interact with a channel residue that surrounds the external opening of the ion conduction pore (10). Most of the 18 substitutions lower the affinity of TPN for both channels (Fig. 4 B and D), and most exhibit little or no channel selectivity. However, the 13R and 13W variants exhibit 140- and 116-fold selectivity for Kir1.1 over Kir3.1/3.4 (Fig. 4F), which, again, results mainly from reduced affinity for Kir3.1/3.4.
Nonadditivity of Mutations of TPN′s Residues 12 and 13.
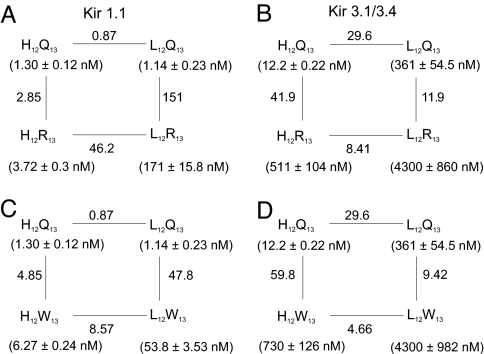
In the bound state, the residues at positions 12 and 13 are located near the external opening of the pore. How these two residues are packed in this spatially constrained region may be a critical factor underlying the observed channel selectivity. If so, it is unlikely that they would act independently. To test this, we compared the effects of single mutations at position 12 or 13 with those of the corresponding double mutations. A leucine residue at position 12 produces the highest selectivity (Fig. 4E), whereas at position 13 an arginine or a tryptophan residue produces comparably higher selectivity (Fig. 4F). Below, we first discuss the results with the H12L and Q13R pair (Fig. 5 A and B).
Fig. 5.
Comparison of single and double mutations at residues 12 and 13. In the four boxes, residue 12 of TPN is tested against residue 13 of TPN in Kir1.1 (A and C) or in Kir3.1/3.4 (B and D), where histidine is compared with leucine at position 12 (A–D) and glutamine with either arginine (A and B) or tryptophan (C and D) at position 13. Each of the four corners of a mutant cycle box represents a tested residue pair in single letter codes. Kd values for each tested pair are shown in parentheses. Numbers next to the lines are ratios of the adjacent Kd values.
Fig. 5B shows that in the case of Kir3.1/3.4, the H12L mutation with Q in position 13 increases the Kd by 30-fold (upper passage), but only 8-fold with R in position 13 (lower passage). Similarly, the Q13R mutation increases the Kd by 42-fold when H is in position 12 (left passage), but only 12-fold with L in position 12 (right passage). Thus, not surprisingly, the effect of a substitution at one residue depends on the nature of the adjacent residue. This energetically nonadditive behavior of mutations at the two positions indicates that they do not act independently but rather interactively. Consequently, the observed Kd of the double mutant (4.3 μM) is 0.28 of what is expected if the two mutations acted independently. The opposite effect occurs for Kir1.1 where the observed Kd of the double mutant (171 nM) is 53-fold what is expected (3.2 nM) if the two mutations acted independently (Fig. 5A). Consequently, the double-mutant toxin (H12L and Q13R) is only 25-fold selective between the two channels, which is less selective than either single-mutant toxin. For similar reasons (Fig. 5 C and D), the H12L and Q13W double mutant toxin is only 60-fold selective.
Because the presented studies involve only residues at two positions, we did not formerly invoke the concept of thermodynamic mutant cycle analysis (8, 20) to rationalize the data. However, for those who are now more accustomed to thinking in that way, we point out that the values of the coupling coefficient Ω (8) for the mutant cycle box in Fig. 5 A–D, are 53, 0.28, 9.9, and 0.16, respectively.
Channel Selectivity of TPNLQ.
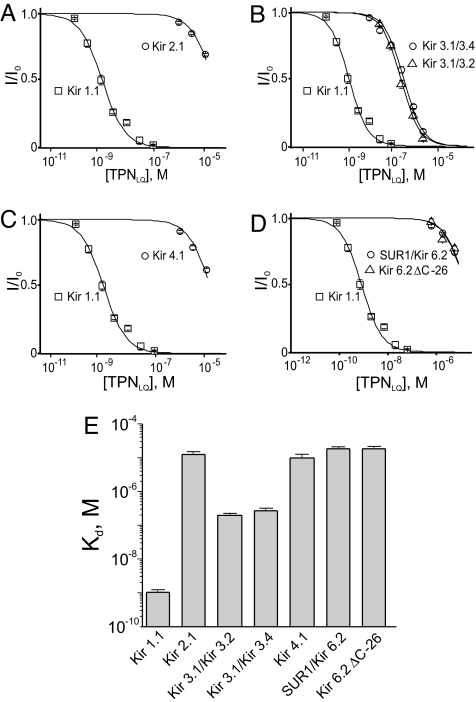
Among all derivatives examined, TPNLQ is most selective for Kir1.1 over Kir3.1/3.4. We further tested it against the four other commonly studied Kir subtypes Kir2.1 (21), Kir3.1/3.2 (13, 14, 22), Kir4.1 (23), and Kir6.2 (24) [coexpressed with a sulfonylurea receptor (SUR)] or Kir6.2-ΔC26 (25) (a constitutively active Kir6.2 mutant lacking the 26 C-terminal residues). For each of these channels, and for the control channels, the fraction of current not blocked is plotted against the TPNLQ concentration in Fig. 6 A–D. Fifty percent inhibition of Kir 2.1, Kir4.1 or Kir6.2 requires >10 μM TPNLQ. The Kd values of TPNLQ for each of the six homo- and heteromeric channels are summarized in Fig. 6E. It is noteworthy that TPNLQ inhibits neural Kir3.1/.3.2 with a Kd of 274 nM, a value comparable to the Kd of 361 nM for Kir3.1/3.4. Thus, among these six commonly studied Kir channels, TPNLQ exhibits >250-fold selectivity for Kir1.1.
Fig. 6.
Comparison of Kir subtype inhibition by TPNLQ. Currents of Kir2.1 (A), Kir 3.1/3.2 (B), Kir 4.1 (C), and Kir6.2/SUR1A or Kir6.2-ΔC26 (D), relative to the unblocked value, are plotted against the TPNLQ concentration. For easy comparison, the data for Kir1.1 are also plotted in A–D, and those for Kir3.1/3.4 are plotted in B. All data are presented as mean ± SEM (n = 5), and the curves through the data are fits of the equation I/Io = Kd/(Kd + [toxin]). The Kd values obtained from the fits are shown in E.
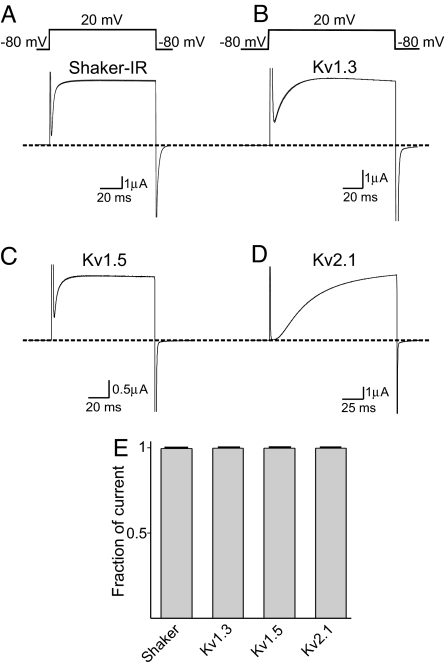
Additionally, we tested TPNLQ against several Kv channels. Fig. 7 A–D shows the current traces of four Kv channels, recorded in the absence or presence of 5 μM TPNLQ. At this high concentration, TPNLQ has no inhibitory effects on these channels, as the current traces before and after the toxin application are superimposed. The percentage of currents of all channels that remained after the application of 5 μM TPNLQ is summarized in Fig. 7E.
Fig. 7.
Effects of TPNLQ on Kv channels. (A–D) Current traces of four Kv channels in the absence or presence of 5 μM TPNLQ elicited with the shown voltage protocol. (E) Fraction of currents (mean ± SEM, n = 3–9) that remained after 5-min exposure to 5 μM TPNLQ.
Discussion
TPN is, thus far, the only reported nanomolar-affinity inhibitor for any Kir channels. It inhibits Kir1 and Kir3 channels with modest selectivity. Practically, an inhibitor with >100-fold selectivity is required to achieve selective inhibition of a given channel current, because at a concentration of 10 times Kd, it would inhibit the targeted current ≥90% but other currents ≤10%. In the present study, we successfully developed a TPN derivative, TPNLQ, that inhibits Kir1 with not only high affinity (Kd = 1 nM) but also high specificity (>250-fold) among six commonly studied Kir (and some Kv) channels tested here. This was achieved by decreasing the affinity of TPN for (heteromeric) Kir 3 but retaining its nanomolar affinity for Kir1. Also importantly, TPNLQ is devoid of two previously identified adverse features. Residue H12 in wild-type TPN renders its channel affinity highly sensitive to pH (26), whereas M13 undergoes a spontaneous oxidation that significantly lowers the toxin's affinity (10). TPNLQ does not have these undesirable features as H12 is replaced by nontitratable leucine and M13 by nonoxidizable glutamine.
TPN inhibits a Kir channel by occluding its ion conduction pore from the extracellular side. The TPN′s channel interaction surface is formed by the residues in its C-terminal half (H12-K21) (Fig. 1C). A mutant-cycle study from our laboratory showed that residue 13 of TPN (Fig. 1 A–C) is interacting with residue F148 in Kir1.1 (10) (Fig. 1 D–F), a residue that corresponds to the residue that forms the high-affinity tetraethylammonium site in Kv channels. Four F148 residues, one from each subunit, surround the external opening of the narrow K+ selectivity filter (27, 28). Thus, in the bound state, much of the toxin's channel-interacting surface must adhere to the channel's vestibule (which involves residues 114–123) in such a manner that residue 12 and its neighboring residue 13 lie near the F148 of Kir1.1 (2, 4, 10), four of which surround the external opening of the narrow K+ pore. Given these spatial constraints and because the effects of the H12L and M13R (or M13W) mutations are not additive (i.e., not independent) in increasing selectivity, how residues 12 and 13 of TPN sterically fit into that spatially constrained region appears to be a crucial determinant of TPN′s selectivity among Kir subtypes.
Although wild-type TPN (or TPNQ) could be used to distinguish and isolate Kir1 and some Kir3 channels from other Kir subtypes, TPNLQ developed here can further distinguish between homomeric Kir1 and heteromeric Kir3.1/3.2 or Kir3.1/3.4. More important conceptually is our demonstration of the feasibility of engineering high-affinity and subtype-specific K+ channel inhibitors.
Materials and Methods
Synthesis of cRNAs and Their Expression in Xenopus oocytes.
The cDNA of Kir1.1 was subcloned in the pSPORT vector, and those of Kv1.3 and Kv1.5 were subcloned in the pSP64 vector. The cDNAs of Kir2.1, Kir3.1, Kir3.2, Kir3.4, Kir4.1 Kir6.2, Shaker(-IR), Kv2.1, SUR1, and the M2 receptor were all subcloned in the pGEM-HE vector. All mutant cDNAs were obtained through PCR-based mutagenesis and confirmed by DNA sequencing; cRNAs were synthesized by using RNA polymerase from the corresponding linearized cDNAs. Channel currents were recorded from whole oocytes (previously injected with cRNA encoding relevant channels), using a two-electrode voltage clamp amplifier (Warner OC-725C). The recorded signal was filtered at 1 kHz and sampled at 5 kHz, using an analog-to-digital converter (DigiData 1322A) interfaced with a personal computer. The pClamp8 software was used to control the amplifier and acquire the data. The resistance of electrodes filled with 3 M KCl was ≈0.2 MΩ. The bath solution for recording Kir currents contained 100 mM K+ (Cl− plus OH−), 0.3 mM CaCl2; 1 mM MgCl2, and 10 mM HEPES (pH 7.6 adjusted with KOH). The bath solution for recording Kv currents contained the same ingredients, except that K+ and Na+ were 20 mM and 80 mM, respectively. Kir3.1/3.2 and Kir3.1/3.4 [coexpressed with mascarinic type 2 (M2) receptors] were activated with 1 or 150 μM acetylcholine, and Kir6.2 (coexpressed with SUR1) with 3 mM sodium azide. All TPN derivatives were prepared as described in ref. 1.
Acknowledgments.
We thank K. Ho (Washington University School of Medicine, St. Louis) and S. Hebert (Yale University, New Haven, CT) for Kir1.1 cDNA, L. Jan (University of California, San Francisco) for Kir2.1 and Kir3.1 cDNAs, J. Yang (Columbia University, New York) for Kir2.1 cDNA subcloned in the pGEM-HESS vector, H. Lester (California Institute of Technology, Pasadena, CA) for Kir3.2 cDNA, D. Clapham (Children's Hospital, Boston) for Kir3.4 cDNA, R. Buono (University of Pennsylvania) and T. Ferraro (Baylor College of Medicine, Houston) for Kir4.1 cDNA, J. Bryan (Harvard University, Cambridge, MA) for Kir6.2 and SUR1 cDNAs, E. G. Peralta (National Institute of Neurological Disorders and Stroke, Bethesda, MD) for HM2 cDNA, K. Swartz (University of Pennsylvania) for Shaker-IR cDNA in the pGEM-HESS vector, C. Deutsch (Vanderbilt University, Nashville, TN) for Kv1.3 cDNA, K. Murray (University of Texas Southwestern Medical Center, Dallas) for Kv1.5 cDNA, R. Joho (University of Pennsylvania) for Kv 2.1 cDNA, and P. De Weer for critical review of our manuscript. This work was supported by National Institutes of Health Grant GM61929 and a pilot grant from University of Pennsylvania Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Jin W, Lu Z. A novel high-affinity inhibitor for inward-rectifier K+ channels. Biochemistry. 1998;37:13291–13299. doi: 10.1021/bi981178p. [DOI] [PubMed] [Google Scholar]
- 2.Jin W, Klem AM, Lewis JH, Lu Z. Mechanisms of inward-rectifier K+ channel inhibition by tertiapin-Q. Biochemistry. 1999;38:14294–14301. doi: 10.1021/bi991206j. [DOI] [PubMed] [Google Scholar]
- 3.Felix JP, et al. Characterization of Kir1.1 channels with the use of a radiolabeled derivative of tertiapin. Biochemistry. 2006;45:10129–10139. doi: 10.1021/bi060509s. [DOI] [PubMed] [Google Scholar]
- 4.Ramu Y, Klem AM, Lu Z. Short variable sequence acquired in evolution enables selective inhibition of various inward-rectifier K+ channels. Biochemistry. 2004;43:10701–10709. doi: 10.1021/bi049125x. [DOI] [PubMed] [Google Scholar]
- 5.Xu X, Nelson JW. Solution structure of tertiapin determined using nuclear magnetic resonance and distance geometry. Proteins. 1993;17:124–137. doi: 10.1002/prot.340170203. [DOI] [PubMed] [Google Scholar]
- 6.Stampe P, Kolmakova-Partensky L, Miller C. Intimations of K+ channel structure from a complete functional map of the molecular surface of charybdotoxin. Biochemistry. 1994;33:443–450. doi: 10.1021/bi00168a008. [DOI] [PubMed] [Google Scholar]
- 7.Goldstein SA, Pheasant DJ, Miller C. The charybdotoxin receptor of a Shaker K+ channel: Peptide and channel residues mediating molecular recognition. Neuron. 1994;12:1377–1388. doi: 10.1016/0896-6273(94)90452-9. [DOI] [PubMed] [Google Scholar]
- 8.Hidalgo P, MacKinnon R. Revealing the architecture of a K+ channel pore through mutant cycles with a peptide inhibitor. Science. 1995;268:307–310. doi: 10.1126/science.7716527. [DOI] [PubMed] [Google Scholar]
- 9.Ranganathan R, Lewis JH, MacKinnon R. Spatial localization of the K+ channel selectivity filter by mutant cycle-based structure analysis. Neuron. 1996;16:131–139. doi: 10.1016/s0896-6273(00)80030-6. [DOI] [PubMed] [Google Scholar]
- 10.Jin W, Lu Z. Synthesis of a stable form of tertiapin: A high-affinity inhibitor for inward-rectifier K+ channels. Biochemistry. 1999;38:14286–14293. doi: 10.1021/bi991205r. [DOI] [PubMed] [Google Scholar]
- 11.Miller C. The charybdotoxin family of K+ channel-blocking peptides. Neuron. 1995;15:5–10. doi: 10.1016/0896-6273(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 12.Ho K, et al. Cloning and expression of an inwardly rectifying ATP-regulated potassium channel. Nature. 1993;362:31–38. doi: 10.1038/362031a0. [DOI] [PubMed] [Google Scholar]
- 13.Kubo Y, Reuveny E, Slesinger PA, Jan YN, Jan LY. Primary structure and functional expression of a rat G-protein-coupled muscarinic potassium channel. Nature. 1993;364:802–806. doi: 10.1038/364802a0. [DOI] [PubMed] [Google Scholar]
- 14.Dascal N, et al. Expression of an atrial G-protein-activated potassium channel in Xenopus oocytes. Proc Natl Acad Sci USA. 1993;90:6596–6600. doi: 10.1073/pnas.90.14.6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krapivinsky G, et al. The G-protein-gated atrial K+ channel IKACh is a heteromultimer of two inwardly rectifying K+ channel proteins. Nature. 1995;374:135–141. doi: 10.1038/374135a0. [DOI] [PubMed] [Google Scholar]
- 16.Simon DB, et al. Genetic heterogeneity of Bartter's syndrome revealed by mutations in the K+ channel, ROMK. Nat Genet. 1996;14:152–156. doi: 10.1038/ng1096-152. [DOI] [PubMed] [Google Scholar]
- 17.Zhou H, Tate SS, Palmer LG. Primary structure and functional properties of an epithelial K channel. Am J Physiol. 1994;266:C809–C824. doi: 10.1152/ajpcell.1994.266.3.C809. [DOI] [PubMed] [Google Scholar]
- 18.Boim MA, et al. ROMK inwardly rectifying ATP-sensitive K+ channel. II. Cloning and distribution of alternative forms. Am J Physiol. 1995;268:F1132–F1140. doi: 10.1152/ajprenal.1995.268.6.F1132. [DOI] [PubMed] [Google Scholar]
- 19.Lee WS, Hebert SC. ROMK inwardly rectifying ATP-sensitive K+ channel. I. Expression in rat distal nephron segments. Am J Physiol. 1995;268:F1124–F1131. doi: 10.1152/ajprenal.1995.268.6.F1124. [DOI] [PubMed] [Google Scholar]
- 20.Horovitz A, Fersht AR. Strategy for analysing the cooperativity of intramolecular interactions in peptides and proteins. J Mol Biol. 1990;214:613–617. doi: 10.1016/0022-2836(90)90275-Q. [DOI] [PubMed] [Google Scholar]
- 21.Kubo Y, Baldwin TJ, Jan YN, Jan LY. Primary structure and functional expression of a mouse inward rectifier potassium channel. Nature. 1993;362:127–133. doi: 10.1038/362127a0. [DOI] [PubMed] [Google Scholar]
- 22.Lesage F, et al. Cloning provides evidence for a family of inward rectifier and G-protein coupled K+ channels in the brain. FEBS Lett. 1994;353:37–42. doi: 10.1016/0014-5793(94)01007-2. [DOI] [PubMed] [Google Scholar]
- 23.Bond CT, et al. Cloning and expression of a family of inward rectifier potassium channels. Receptors Channels. 1994;2:183–191. [PubMed] [Google Scholar]
- 24.Inagaki N, et al. Reconstitution of IKATP: An inward rectifier subunit plus the sulfonylurea receptor. Science. 1995;270:1166–1170. doi: 10.1126/science.270.5239.1166. [DOI] [PubMed] [Google Scholar]
- 25.Tucker SJ, Gribble FM, Zhao C, Trapp S, Ashcroft FM. Truncation of Kir6.2 produces ATP-sensitive K+ channels in the absence of the sulphonylurea receptor. Nature. 1997;387:179–183. doi: 10.1038/387179a0. [DOI] [PubMed] [Google Scholar]
- 26.Ramu Y, Klem AM, Lu Z. Titration of tertiapin-Q inhibition of ROMK1 channels by extracellular protons. Biochemistry. 2001;40:3601–3605. doi: 10.1021/bi002584n. [DOI] [PubMed] [Google Scholar]
- 27.Kuo A, et al. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 2003;300:1922–1926. doi: 10.1126/science.1085028. [DOI] [PubMed] [Google Scholar]
- 28.Nishida M, Cadene M, Chait BT, MacKinnon R. Crystal structure of a Kir3.1-prokaryotic Kir channel chimera. EMBO J. 2007;26:4005–4015. doi: 10.1038/sj.emboj.7601828. [DOI] [PMC free article] [PubMed] [Google Scholar]