Abstract
The alpha-fetoprotein (AFP) gene is highly activated in fetal liver but is dramatically repressed shortly after birth. The mechanisms that underlie AFP transcriptional repression in postpartum liver are not well understood. AFP enhancer, repressor region, and promoter are implicated to be involved in AFP postnatal repression, but the major transcriptional repressor remains undefined. We previously identified a zinc finger protein gene ZBTB20. To determine its physiological functions in vivo, we have generated hepatocyte-specific ZBTB20 knockout mice by the Cre/loxP approach and demonstrated here that ZBTB20 ablation in liver led to dramatic derepression of the AFP gene in entire liver throughout adult life, although the hepatocytes were normally under nonproliferating status. Furthermore, we found that ZBTB20 was a transcriptional repressor capable of specifically inhibiting AFP promoter-driven transcriptional activity. Liver chromatin immunoprecipitation and mobility shift assays showed that ZBTB20 bound to AFP promoter directly. ZBTB20 was developmentally activated in liver after birth and inversely correlated with its AFP gene expression, suggesting that activated ZBTB20 expression in liver mediated AFP gene repression. Our data point to ZBTB20 as a key regulator governing AFP gene transcription and postulate a new model for the postnatal gene repression of AFP in liver.
Keywords: gene regulation, transcription factor, transcriptional repressor
Alpha-fetoprotein (AFP) is the major serum protein in the developing mammalian fetus produced at high levels by the fetal liver and visceral endoderm of the yolk sac and at low levels by fetal gut and kidney (1). AFP is required for female fertility during embryonic development by protecting the developing female brain from prenatal exposure to estrogen (2, 3). Shortly after birth, the AFP gene is dramatically repressed to extremely low levels, which in liver represents a nearly 10,000-fold reduction of transcription (4). However, in adult liver, AFP can be reactivated in hepatocyte proliferation (e.g., liver regeneration and hepatocellular carcinogenesis) (1). Due to its high tissue specificity and tight temporal regulation of expression, AFP serves as an ideal model to investigate the developmental control of transcription (5, 6).
AFP transcription is governed by five distinct regulatory regions: a 250-bp tissue-specific promoter, a 600-bp repressor region directly upstream of the promoter (7), and three enhancers located 2.5, 5.0, and 6.5 kb, respectively, upstream of the AFP promoter and named enhancer I (EI), EII, and EIII (8). Postnatal repression of AFP transcription may involve combinatorial action of distinct mechanisms. The promoter region harbors multiple binding sites for liver-enriched and ubiquitous transcription factors. A region at −120 from transcription start site (+1) can be recognized by hepatocyte nuclear factor 1 (HNF-1), nuclear factor 1 (NF-1), and CAAT/enhancer binding protein (C/EBP) and is crucial for the promoter activity (9–11). A point mutation (G to A) at −119 of AFP promoter is associated with naturally occurring Hereditary Persistence of AFP (HPAFP), which is predicted to improve HNF-1 binding and decrease NF-1 binding to the mutant sequence (12). The repressor region (−838 to −250) is required for AFP postnatal repression in pericentral hepatocytes but not essential for complete AFP repression in the intermediate zone and periportal hepatocytes (7, 13). A site in the repressor region can be recognized by p53 family members, which leads to chromatin remodeling and AFP repression (14, 15). The three AFP enhancers are essential for AFP transcription in vivo and continue to be active in the adult liver in a position-dependent manner (1). Transgenic studies show that EIII may be involved in AFP repression in all hepatocytes except those encircling the central vein (16). So far, the physiological mechanism of AFP repression in postpartum liver is not well understood, including the involved trans-acting repressors and corresponding cis-acting elements.
Transcription repressors play important roles in tissue-specific and temporal gene expression (17–19). ZBTB20, also named DPZF (20), HOF (21), and ZNF288, belongs to BTB/POZ (broad complex tramtrack bric-a-brac/poxvirus and zinc finger) zinc finger family. It has two isoforms due to the alternative translation initiation, both containing an intact N-terminal BTB domain and a C-terminal zinc finger domain (21). Very recently, ZBTB20 was found to participate in neurogenesis in a transgenic model (22). However, its physiological functions are not established. In the present study, we generated liver-specific ZBTB20 knockout mice (LZB20KO) and found that ZBTB20 was required for AFP postnatal repression in liver. Our results demonstrate that ZBTB20 is a key transcriptional repressor of the AFP gene in a dominant fashion.
Results
Generation of Liver-Specific ZBTB20 Knockout Mice.
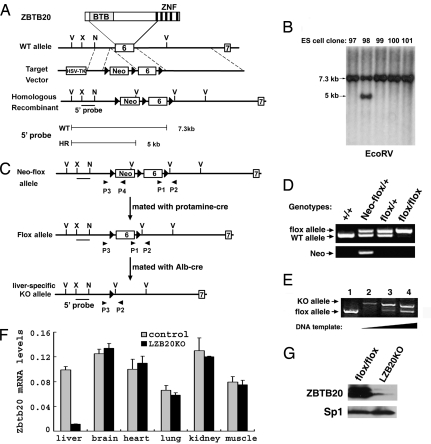
Initially, we identified the BTB zinc finger protein ZBTB20 (also known as DPZF) in human dendritic cells (20). To specifically investigate its functional role in liver, we created liver-specific ZBTB20 knockout mice (LZB20KO). First, we developed mice with loxP sites flanking exon 6 of the ZBTB20 gene (21), which encodes 540 aa of ZBTB20 protein spanning from the complete POZ domain to the first zinc finger domain (Fig. 1A). Correct initial targeting was confirmed by Southern blot analysis of genomic DNA from ES cell clones with a 5′ probe (Fig. 1B), which transmitted ZBTB20neo-flox allele to the germ line. The neo cassette was removed in the male germ line by backcrossing to protamine promoter-Cre expressing transgenic mice (23), which led to the creation of the ZBTB20flox allele (Fig. 1 C and D). The ZBTB20flox allele was bred to homozygosity, which did not affect the viability or fertility of these mice, suggesting that the flanking loxP sites did not significantly alter the expression from the targeted locus.
Fig. 1.
Generation of LZB20KO mice. (A) Schematic demonstration for the gene targeting of exon 6 of the ZBTB20 gene. The diagram of ZBTB20 protein is shown at the top. The probe 5′ is used for Southern blot analysis, which detects a 7.3-kb band for the wild-type (WT) allele and a 5-kb band for the homologously recombined (HR) Neo-flox allele in the EcoRV-digested genomic DNA. The loxP sites are represented as filled triangles. V, EcoRV; X, XhoI; N, NcoI; Neo, neomycin resistance cassette. (B) Southern blot analysis of ES cell clones after EcoRV digestion and hybridization with the 5′ probe. Clone number is indicated above each lane. (C) Schematic demonstration for the removal of Neo and tissue-specific deletion of the floxed exon 6 in liver by Cre/loxP-mediated recombination. ZBTB20neo-flox mice were mated with protamine-Cre mice to remove the Neo cassette and produce mice with the ZBTB20flox allele. Repeated mating of ZBTB20flox mice and ALB-Cre transgenic mice generated LZB20KO (ZBTB20flox/flox/Alb-cre) mice. Primers P1 and P2 were used for PCR analysis to confirm the presence of exon 6 and its downstream loxP site, and P3 and P4 were used to detect the Neo cassette. (D) PCR analysis of tail genomic DNA distinguishing the ZBTB20flox allele from the ZBTB20neo-flox and wild-type alleles. PCR with the primers P1 and P2 gave a 0.8-kb band for the floxed allele and a 0.7-kb band for the WT allele (upper row). PCR with primers P3 and P4 produced a 1.0-kb band for the ZBTB20neo-flox allele and no specific band from the ZBTB20flox allele (lower row). Genotypes are indicated above each lane. (E) PCR analysis for Cre-mediated ZBTB20 deletion in liver. Genomic DNA from control (ZBTB20flox/flox, lane 1) or LZB20KO liver (lane 2–4) was subjected to PCR amplification with primers P1, P2, and P3. Deletion of the floxed exon 6 produced a 1.2-kb band amplified by primers P3 and P2, whereas the 0.8-kb band indicates the presence of floxed exon 6. Efficient deletion of the ZBTB20 target exon in liver was shown by PCR products from increasing DNA template inputs (lane 2–4). (F) By real-time PCR, ZBTB20 mRNA expression was shown in control (ZBTB20flox/flox) and LZB20KO mice. (G) Western blot analysis for ZBTB20 protein expression in the adult livers from control (ZBTB20flox/flox) and LZB20KO mice. The nuclear lysate from liver was probed with anti-ZBTB20 (upper row) and anti-Sp1 antibodies (lower row).
LZB20KO mice were generated by repeated mating of ZBTB20flox/+ mice with rat albumin promoter-Cre (ALB-Cre) transgenic mice (24). PCR analysis of genomic DNA showed that exon 6 was efficiently disrupted in LZB20KO liver, which gave a 1.2-kb band and that the residual signal of a 0.8-kb band reflects the un-recombined ZBTB20flox allele, most likely from nonhepatocyte cells in the liver (Fig. 1E). Efficient deletion of the ZBTB20 gene in LZB20KO liver was confirmed at the mRNA level by real-time RT-PCR and at the protein level by immunoblot analysis with ZBTB20-specific antibody (Fig. 1 F and G). By contrast, no deletion was detected in the brain, muscle, heart, kidney, or lung from LZB20KO mice. There was no effect on ZBTB20 expression in ZBTB20flox/flox littermates (data not shown).
LZB20KO (ZBTB20flox/flox/ALB-Cre) and control mice (ZBTB20flox/flox and ALB-Cre) were born normally and survived equally into adulthood with no significant difference in development. Both male and female LZB20KO mice were fertile and produced normal-sized litters. The livers from LZB20KO mice were normal in appearance and size, and histological examination revealed no abnormality in their architecture [supporting information (SI) Fig. S1], suggesting that ZBTB20 was dispensable for liver development.
Dysregulation of AFP in Liver.
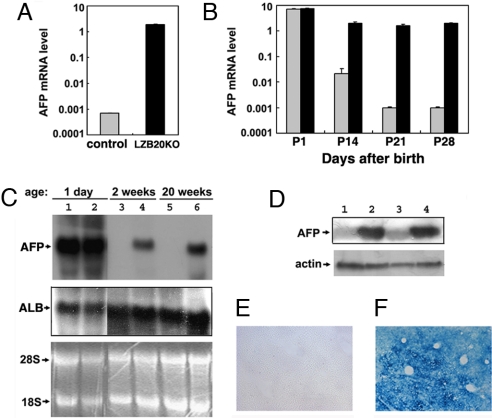
To evaluate the differentiation status of hepatocytes, we analyzed the expression pattern of albumin and AFP. Real-time RT-PCR analysis showed that adult LZB20KO liver expressed very high levels of AFP mRNA, ≈3,000-fold higher than the basal levels of control littermates (Fig. 2A). To distinguish between the failure of shutoff and reactivation after birth that led to the dysregulated expression of AFP in adult LZB20KO liver, we explored the ontogeny of AFP gene expression during the postnatal development of ZBTB20-deficient liver. Consistent with a previous report (1, 25), AFP mRNA was activated gradually in fetal liver (Fig. S2), declined >10,000-fold to basal levels in wild-type and ZBTB20flox/flox liver within four weeks after birth. Whereas, AFP levels in LZB20KO liver were comparable at birth but decreased <5-fold in the first four weeks of age, which was followed by the high and constitutive expression throughout adult life (Fig. 2 A and B and data not shown). Northern blot and western blot analysis also confirmed the abundant expression of AFP in adult LZB20KO liver (Fig. 2 C and D). This indicated that aberrant expression of AFP in the postnatal LZB20KO liver was due to the failure of transcriptional shutoff. On the other hand, LZB20KO livers from either young (2-week-old) or adult (20-week-old) mice expressed comparable levels of ALB mRNA (Fig. 2C). These data suggest that AFP postnatal dysregulation in LZB20KO liver was gene-specific, supporting the current view on the autonomous regulation of AFP and ALB gene expression (1).
Fig. 2.
Dysregulated AFP expression in LZB20KO liver. (A) AFP mRNA expression in liver from 4-month-old control (gray bar) and LZB20KO (black bar) mice was shown by real-time RT-PCR. n = 3 experiments. (B) Real-time RT-PCR analysis for AFP mRNA expression in liver from control (gray bar) and LZB20KO (black bar) mice during postnatal days 1–28. Each group at individual time points included at least 5–8 mice. (C) Northern blot analysis for AFP and ALB mRNA expression in the livers from 1-day-, 2-week-, and 20-week-old LZB20KO mice (lanes 2, 4, and 6) and their control littermates (lanes 1, 3, and 5). The RNA loading control was demonstrated by EtBr staining of the gel. (D) Western blot analysis of AFP expression in whole liver lysates from 3-week-old control (lanes 1 and 3) and LZB20KO (lanes 2 and 4) mice. The loading control was demonstrated by reprobing with anti-actin antibody. (E and F) In situ β-galactosidase staining showed LacZ expression throughout the liver from 3-week-old ZBTB20flox/flox/Alb-cre/AFP−/− compound knockout mice (F) rather than the same-aged ZBTB20flox/flox/AFP−/− knockouts (E), which reflected the AFP gene transcriptional activity. Error bar represents SD.
We further examined the zonal expression pattern of AFP in the LZB20KO liver. To this end, we took advantage of the AFP mutant mice harboring the LacZ reporter gene in the AFP locus under the control of AFP P1 promoter (2), which allowed us to assess the transcriptional activity of the AFP gene by X-Gal staining. Consistent with nearly complete repression of AFP gene transcription, β-galactosidase was undetectable in the adult liver from AFP single mutant mice (Fig. 2E). However, robust β-galactosidase activity was detected throughout the adult livers from ZBTB20flox/flox/Alb-Cre/AFP−/− compound mice with the pericentral region tending to show stronger activity (Fig. 2F), indicating that AFP dysregulation occurred throughout the adult LZB20KO liver.
Proliferation Status of the Hepatocytes.
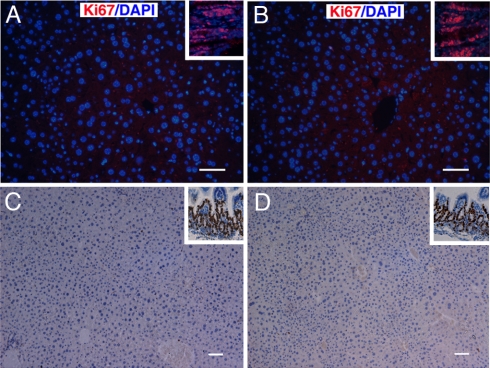
AFP expression in liver often reflects hepatocyte proliferation. So, we examined the in situ cell proliferation status of LZB20KO liver. Immunostaining of Ki67, a cell proliferation marker, revealed that Ki67-positive cells were hardly detectable in the livers either from LZB20KO or control adult mice (2-month-old), whereas some populations of Ki67-positive cells were persistently detected in the intestine (Fig. 3 A and B). Similarly, in vivo labeling with BrdU (an analogue of deoxythymidine) demonstrated that few BrdU-positive hepatocytes could be detected in adult liver from either LZB20KO or control mice after 24 h of labeling, whereas some cells in intestine were positive (Fig. 3 C and D). These results indicated that the hepatocytes from adult LZB20KO liver were normally nonproliferating, although they aberrantly produced AFP.
Fig. 3.
Proliferation status of hepatocytes in adult liver. (A and B) Ki67 staining analysis for the livers from ZBTB20flox/flox (A) and LZB20KO (B) mice at the age of 2 months. The inserts show positive Ki67 staining of intestines from the same animals. (C and D) BrdU staining in the livers from the 2-month-old ZBTB20flox/flox (C) and LZB20KO (D) mice after 24h of in vivo labeling. The inserts show positive BrdU staining (C and D) of intestines from the same animals. n = 4 experiments. (Scale bar, 50 μm.)
Transcriptional Repression of ZBTB20.
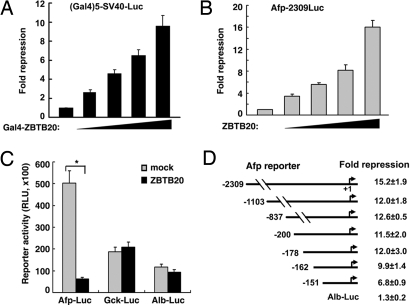
To evaluate the potential transcription-regulatory activity of ZBTB20, we established a Gal4-based transcriptional assay. A Gal4-ZBTB20 fusion vector was cotransfected in 293T cells together with the luciferase reporter pG5SV40luc containing five Gal4-binding sites. Expression of the Gal4-ZBTB20 fusion protein was verified by immunoblotting (data not shown). When tethered to DNA by Gal4, ZBTB20 exhibited marked inhibitory effects on pG5SV40luc reporter transcription in a dose-dependent manner (Fig. 4A). This result suggests that ZBTB20 act as a transcriptional repressor.
Fig. 4.
ZBTB20-mediated transcriptional repression of AFP activity. (A) 293T cells were cotransfected with different amounts of Gal4-ZBTB20 fusion expression vector, (GAL)5-sv40-Luc reporter, and pCMV-β-Gal. Results are expressed as fold repression of luciferase normalized to the internal control. (B–E) HepG2 cells were cotransfected with ZBTB20 expression plasmids, different luciferase reporters, and the internal control pCMV-β-Gal. Results are expressed as fold repression or relative luciferase activity normalized to the internal control. n = 3 experiments. (B) ZBTB20 overexpression repressed AFP-2309Luc reporter (−2309 to +27) activity in HepG2 cells in a dose-dependent manner. n = 4 experiments. (C) ZBTB20 overexpression repressed AFP-2309Luc reporter activity in HepG2 cells but showed no effects on Gck or Alb reporter activity. Gray bar = mock control of empty vector; black bar = ZBTB20 expression vector. n = 3 experiments. (D) ZBTB20 overexpression repressed the activity of the AFP reporter driven by the AFP promoter −2309/+27 or 5′ truncations at −1103, −837, −200, −178, −162, or −151. n = 5 experiments. ∗, P < 0.01; Error bar represents SD.
Next, we determined whether ZBTB20 could repress AFP-driven transcriptional activity by using a transiently transfected reporter assay. When transiently overexpressed in human hepatoma HepG2 cells, ZBTB20 exerted up to a 16-fold repression of the reporter AFP-2309Luc, driven by −2309 to +27 of the AFP gene, in a dose-dependent manner (Fig. 4B). However, it had no significant effects on ALB or glucokinase (GCK)-driven reporters (Fig. 4C), suggesting that ZBTB20-mediated transcriptional repression was gene-specific. To localize the potential region in the AFP gene mediating transcriptional repression by ZBTB20, we made serial 5′ deletions of the AFP gene in the AFP reporter construct and analyzed the effects of ZBTB20 on their activity (Fig. 4D). Serial deletion of the fragment from −2309 to −163 of the AFP gene did not significantly alter the repressive effects of ZBTB20 on the resultant AFP reporters, suggesting that the ZBTB20-responsive region might be located in the −162 to +27 region of the AFP promoter. The effort to analyze the effects of ZBTB20 by further 5′ deletions of the AFP gene was hampered by their weak reporter activity. These data suggest that the minimal AFP promoter (−162/+27) contains a cis-acting element mediating ZBTB20 repressive activity.
Binding of ZBTB20 to AFP Promoter.
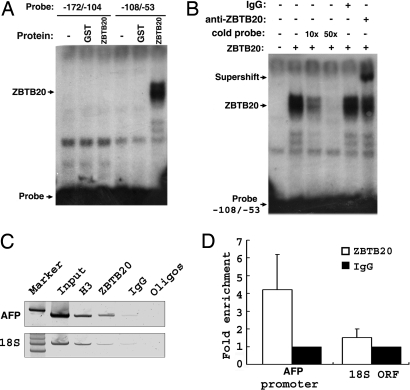
To determine whether ZBTB20 binds directly to DNA in the AFP promoter, we performed gel mobility shift assayby using recombinant GST fusion protein of ZBTB20 zinc finger domain (531–741aa). Double-stranded oligonucleotides corresponding to −108 to −53 of mouse Afp gene bound to ZBTB20 with high specificity, rather than the fragment −172/−104 or −56/+27 (Fig. 5A and data not shown). The DNA-protein complex was supershifted by anti-ZBTB20 antibodies and was abolished by excess unlabeled DNA fragment −108/−53 as a competitor (Fig. 5B), indicating that ZBTB20 could bind to the AFP promoter directly at −108/−53 (Fig. S3).
Fig. 5.
Binding of ZBTB20 to the AFP promoter. (A) ZBTB20 bound to fragment −108/−53 of the Afp gene to form a DNA-protein complex. (B) The DNA-protein complex formed by labeled Afp −108/−53 and ZBTB20 was blocked by excessive unlabeled probe and supershifted by anti-ZBTB20 antibody. (C) ChIP analysis of normal mouse liver at the age of 2 months. Conventional PCR analysis was performed to determine the association of ZBTB20 with the promoter region of AFP in comparison to mouse 18S ORF (ORF). H3 antibody is shown as an internal control for chromatin recovery. IgG antibody is shown as a negative control. (D) Real-time PCR was performed to quantify antibody-bound DNA fragments of multiple liver ChIPs. The graph represents fold enrichment of ZBTB20 bound at AFP promoter region compared with 18S ORF. All values generated by real-time PCR analysis were normalized to control IgG precipitations. Each bar represents the average result from two independent ChIP experiments and six real-time PCR analyses.
To determine whether endogenous ZBTB20 interacts with the AFP promoter in AFP postnatal repression, we performed a chromatin immunoprecipitation (ChIP) assay in adult liver, where ZBTB20 is highly expressed and AFP is fully repressed. We found that ZBTB20 was bound at the AFP promoter in normal adult liver (Fig. 5 C and D). With histone H3 as a positive control for chromatin recovery, no ZBTB20 binding was detected at a control distal genomic locus, the 18S ORF, indicating that association of ZBTB20 to the AFP promoter was gene-specific.
Developmental Expression of ZBTB20 and Postnatal Repression of AFP Transcription in Liver.
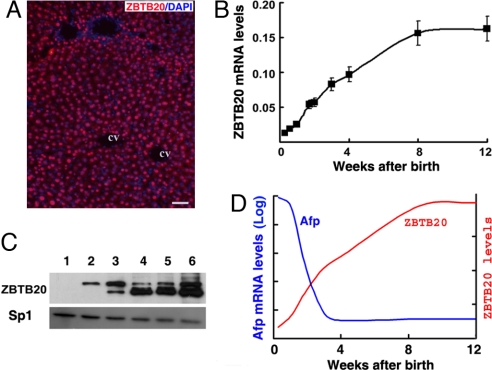
Last, we analyzed the expression pattern of ZBTB20 in liver development. Immunostaining showed that ZBTB20 was a nuclear protein nearly uniformly detectable in most hepatocytes from adult WT liver (Fig. 6A), whereas no ZBTB20 staining was detected in the hepatocytes from LZB20KO mice (Fig. S4). RT-PCR analysis demonstrated that ZBTB20 mRNA was poor in fetal liver (data not shown) and developmentally activated in postpartum liver with a 4-fold increase at the age of two weeks, a 12-fold increase at the age of 8 weeks, and after that reaching a plateau (Fig. 6B). Western blot analysis revealed that ZBTB20 protein was undetectable in the nuclear extracts from the neonatal liver and elevated significantly during postnatal development (Fig. 6C). We detected two distinct isoforms of ZBTB20 in liver, which are likely produced by alternative utilization of the start codon in translation (21). In the early stage of postnatal development, the long one was dominant in liver. However, the short one became dominant after four weeks of age.
Fig. 6.
Developmental activation of ZBTB20 leads to postnatal AFP repression in liver. (A) Immunohistochemical staining of ZBTB20 in the adult liver from normal C57BL/6 mice. ZBTB20 was stained with anti-ZBTB20 antibodies and visualized by Alexa Fluor 594 (red). Nuclei were counterstained with DAPI (blue). CV, central vein. (Scale bar, 50 μm.) (B) Real-time RT-PCR analysis for ZBTB20 mRNA levels in the developing postpartum livers from C57BL/6 mice at different ages. (C) Immunoblot of liver nuclear extracts from C57BL/6 mice at different weeks of age with anti-ZBTB20 (upper row) and anti-Sp1 (lower row) antibodies. n = 2 experiments. (D) The postulated model for AFP repression in postpartum liver. The ZBTB20 gene, which is developmentally activated in postpartum liver with the expression level (red line) increasing gradually, binds to the AFP promoter and leads to the dramatic repression of AFP gene transcription in liver (blue line).
Taken together, we postulate a model for postnatal repression of AFP gene transcription in liver. In this model, ZBTB20 is developmentally up-regulated in postpartum liver and acts as a dominant transcriptional repressor by interacting with the AFP promoter to repress AFP gene transcription to basal levels (Fig. 6D). Ablation of ZBTB20 in postpartum liver leads to derepressed aberrant expression of AFP gene independent of hepatocyte proliferation.
Discussion
Our results establish zinc finger protein ZBTB20 as a key repressor governing AFP gene transcription in postpartum liver. First, expression of AFP and ZBTB20 is developmentally regulated and exhibits an inverse correlation with each other in postpartum liver, where ZBTB20 is gradually activated while AFP is dramatically repressed to extremely low levels. Moreover, our unpublished data show that ZBTB20 expression was decreased in chemically induced rodent hepatoma, where AFP was reactivated. Second, hepatocyte-specific disruption of ZBTB20 gene in mice results in dramatic derepression of AFP in the entire liver throughout adult life with expression levels almost close to fetal liver. Third, forced expression of ZBTB20 represses AFP-driven transcriptional activity in vitro. Last, ZBTB20 binds to the AFP promoter both in vitro and in liver, which mediates postnatal repression of AFP transcription.
AFP gene transcription is regulated by positively and negatively acting factors that bind to specific elements in different regions of the AFP gene. Hepatocyte-enriched transcriptional activators, such as HNF1, HNF3, and C/EBP, are thought to play critical roles in AFP activation in a tissue-specific manner. On the contrary, transcriptional repressors involved in AFP gene regulation are poorly defined. There is a DNA-binding site located at −135 of the AFP promoter for transcriptional repressor COUP-TF (26, 27), but its physiological significance in regulation of AFP transcription is still unknown. Overexpression of c-Jun in hepatoma cells inhibits AFP promoter activity in a DNA binding-independent manner (28). Also, p53 mediates AFP repression by competing with HNF3 to bind DNA in the repressor region of the AFP gene (−838 to −250). The p53-null mice display a delay in AFP postnatal repression in liver with eventual repression at four months of age (15). The Zhx2 gene, which encodes a zinc finger and homeobox protein, regulates AFP postnatal repression in liver, and its mutation in BALB/cJ mice leads to 5–20-fold higher adult serum AFP levels (29). Enhancer III participates in AFP postnatal repression (16), but the involved transacting repressor remains undefined. Our finding that ZBTB20 interacts with the AFP promoter and represses AFP gene transcription in a dominant fashion sheds light on the physiological nature of AFP gene regulation.
The most likely scenario is that ZBTB20 interferes with transcriptional stimulation by activators or recruits corepressors to inhibit transcription and/or remodel chromatin structure. Transgenic studies show EIII may be responsible for postnatal repression of AFP transcription (16). It will be worthwhile to examine whether ZBTB20 is involved in EIII-mediated repression or whether there is synergy between these distinct repression mechanisms. We note that there is also a several-fold decrease of AFP mRNA in adult ZBTB20-null liver compared with fetal liver, suggesting that other mechanisms also play important roles in AFP postnatal repression. It is possible that postnatal repression in ZBTB20-null liver is mediated by other identified (e.g., p53, Zhx2, and COUP-TF) or unidentified transcriptional repressors. Less likely, it may result from the reduction of transcriptional activators. We are further investigating the potential repression mechanisms of ZBTB20 and the mechanisms of AFP repression in ZBTB20-deficient liver. We speculate that multiple factors and multiple mechanisms may be involved in complete repression of AFP gene in postpartum liver.
The relation between liver cell proliferation and AFP gene transcription has been noted but not fully addressed yet. Postnatal AFP repression in liver is coincident with the cessation of hepatocyte cell division, and AFP reactivation is often related to the hepatocyte proliferation (e.g., regeneration and carcinogenesis) (1). It is thought that there may be a mechanistic link behind this temporal correlation, which is further supported by in vitro studies of AFP reactivation (30). However, the adult LZB20KO liver has a normal architecture and remains in a normally nonproliferating status, despite their aberrant expression of AFP. This raises the possibility that AFP expression is independent of cell proliferation in the absence of ZBTB20. Furthermore, although not essential in the absence of ZBTB20, aberrant proliferation may facilitate loss of ZBTB20 interaction with chromatin to promote AFP derepression during HCC or liver regeneration. Nevertheless, it should be interesting to determine whether AFP reactivation in HCC reflects the decreased levels or activity of ZBTB20. Our unpublished data demonstrated that ZBTB20 expression was down-regulated in diethylnitrosamine-induced mouse HCC, where AFP was markedly reactivated. Whether the AFP derepression in ZBTB20-deficient hepatocytes implies the potentially enhanced susceptibility of hepatocellular carcinogenesis is under examination.
Taken together, our identification of ZBTB20 as a dominant repressor of AFP gene transcription provides insight into AFP gene regulation, which will help to unravel the biochemical basis of AFP reactivation in carcinogenesis and facilitate earlier diagnosis of a disease that is generally untreatable by the time of its discovery.
Materials and Methods
Generation of LZB20KO Mice.
The targeted ES cells carrying the ZBTB20neo-flox allele were injected into C57BL/6 blastocysts to produce chimeric mice, which transmitted the ZBTB20neo-flox allele to the progeny. The neomycin resistance cassette was removed by breeding with protamine promoter-cre transgenic mice. Liver-specific ZBTB20 knockout mice were generated by crossing ZBTB20flox mice with ALB-Cre transgenic mice. See SI Methods for detail. LZB20KO mice were repeatedly crossed to AFP-deficient mice (AFPtm1/bmm, C57BL/6 background), which harbor the LacZ gene in the AFP mutant allele driven by the AFP promoter (2). Animal experiments were done following institutional guidelines.
Expression Plasmids.
Full-length mouse ZBTB20 cDNA encoding 741 aa was cloned from liver by RT-PCR and inserted into the expression vectors pcDNA3 (Invitrogen) and pCMV-FA (Stratagene). In the latter, it was in-frame fused with Gal4-DBD (1–147 aa) at the N terminus. The fragment containing five Gal4 binding sites was subcloned upstream of the SV40 promoter in the pGL2-promoter vector (Promega). The mouse AFP promoter with different lengths of 5′ flanking sequence was removed from the plasmid pUC19-AFP (18) and inserted into the pGL3-basic vector (Promega). The mouse GCK promoter (−980 ≈ +16) and ALB promoter (−790 ≈ +23) were cloned by PCR and inserted into the pGL3-basic vector.
Analysis of RNA.
Total RNA was extracted from the TRIzol (Invitrogen) homogenates. Total RNA (20 μg per lane) were loaded on a 1.2% formaldehyde agarose gel and transferred to a nylon membrane. The membranes were hybridized with 32P-labeled mouse AFP or ALB cDNA probes. Real-time RT-PCR was performed in a two-step reaction. First strand cDNA was synthesized with a SuperScript III RT-PCR kit (Invitrogen), and the second step was performed in a fluorescent temperature cycler (Mastercycler ep realplex, Eppendorf) with SYBR green and specific primers for each of the genes. Every plate included the 36B4 gene as an internal control. Primer sequences are available on request. Results were analyzed with the Student's unpaired t test.
Western Blot Analysis.
Liver nuclear extracts were prepared by sucrose gradient centrifugation of homogenates and dissolved in buffer containing 20 mM Hepes (pH 7.9), 1.5 mM MgCl2, 410 mM KCl, 0.2 mM EDTA, 25% glycerol, 0.5% Nonidet P-40, 0.5 mM DTT, 0.5 mM PMSF, 4 μg/ml leupeptin, 4 μg/ml aprotinin, and 4 μg/ml pepstatin. We resolved lysates (100 μg of protein per lane) with SDS/PAGE and immunoblotted membranes with anti-ZBTB20 antibodies, rabbit anti-AFP serum (2), or rabbit anti-Sp1 antibodies (Santa Cruz Biotechnologies).
BrdU Labeling and Immunohistochemistry.
BrdU (50 mg/kg) was injected i.p. into mice, and tissues were harvested 24h later, fixed overnight in 4% paraformaldehyde, embedded in paraffin, cut into 6-μm sections, immunostained with the BrdU detection kit (BD Bioscience), and developed by DAB staining. Immunostaining of rat anti-mouse Ki67 antibody (Dako, clone TEC-3) was visualized by Alexa Fluor 594 (Molecular Probe) and counterstained with DAPI. For X-Gal staining, mice were perfused with 4% paraformaldehyde, and the livers were further immersed in the fixative at 4 °C for 4 h before cryosectioning. The 15-μm sections were stained at 37 °C for 24 h with X-gal (5-bromo-4-chloro-3- indolyl-β-d-galactosidase) staining solution containing 1 mg/ml X-gal, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, and 2 mM MgCl2.
Reporter Assay.
Human hepatoma cells (HepG2) were transiently transfected in 24-well plates with plasmids by using Effectene (Qiagen). Forty-eight hours after transfection, cells were disrupted with passive lysis buffer (Promega) and subjected to a luminescent assay in a luminometer (MiniLumat LB9506, Berthold GmbH). SV40-Renilla luciferase plasmid or pCMV-β-Gal was used as an internal control to normalize the luciferase activity, and the β-galactosidase activity was measured by using a luminescent detection kit (Clontech). The fold repression of transcription was calculated relative to transcription of the reporters in the presence of the relevant empty expression vector and normalized to the internal control. Expression of all of the transfected proteins was confirmed by immunoblotting with the appropriate antibodies.
Chromatin Immunoprecipitation Assay.
Liver tissue was isolated from normal adult mice and flash-frozen in liquid nitrogen. ChIP assays of liver tissue were performed as described (15), with minimal modifications. The fragmented, precleared chromatin lysate was incubated overnight with specific antibodies for ChIP: histone H3 (Abcam), ZBTB20, and normal rabbit IgG (Upstate). To analyze specific, antibody- and protein-bound DNA, conventional PCR and quantitative real-time PCR were performed. Conventional PCR primers were generated to detect the AFP core promoter region (−82 ≈ +94) as described (15). Real-time PCR was conducted in a 7500 FAST ABI instrument.
Gel Mobility Shift Assay.
The double-stranded oligomers from the AFP promoter were labeled with [32P]ATP by T4 polynucleotide kinase, purified with a G25 DNA purification column, and used for the mobility shift assay. The 20-μl binding reaction contained 10 mM Hepes (pH 7.6), 50 mM NaCl, 5 mM MgCl2, 10 mM EDTA, 1 mM DTT, 5% glycerol, 1 μg of poly (dI-dC) (Amersham), 50,000 cpm probe, and 4 ng of GST-fusion protein. After incubation at room temperature for 15 min, the reactions were separated on a 5% PAGE gel containing 0.5× TBE. Excessive unlabeled oligomers were used as competitors, and 2 μg of anti-ZBTB20 antibodies were added in the binding reaction for the supershift assay.
Supplementary Material
Acknowledgments.
We thank B. Spear, H. Nakabayashi, and J. Dannan for the plasmids and X. Ma and G. Gu for technical assistance. This work was supported by National Natural Science Foundation of China Grants 90608026 (to W.Z.) and 30772478 (to Z.X.), National “973” Program of China Grant 2006CB503910 (to W.Z.), National “863” Program of China Grant 2007AA02Z173 (to W.Z.), and fellowships from the Ministry of Education of China (to W.Z.), Shanghai Municipal Science and Technology Commission, and Education Commission (to W.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800647105/DCSupplemental.
References
- 1.Tilghman SM. The structure and regulation of the alpha-fetoprotein and albumin genes. Oxf Surv Eukaryot Genes. 1985;2:160–206. [PubMed] [Google Scholar]
- 2.Gabant P, et al. Alpha-fetoprotein, the major fetal serum protein, is not essential for embryonic development but is required for female fertility. Proc Natl Acad Sci USA. 2002;99:12865–12870. doi: 10.1073/pnas.202215399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakker J, et al. Alpha-fetoprotein protects the developing female mouse brain from masculinization and defeminization by estrogens. Nat Neurosci. 2006;9:220–226. doi: 10.1038/nn1624. [DOI] [PubMed] [Google Scholar]
- 4.Belayew A, Tilghman SM. Genetic analysis of alpha-fetoprotein synthesis in mice. Mol Cell Biol. 1982;2:1427–1435. doi: 10.1128/mcb.2.11.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, Egan JO, Chiu JF. Regulation and activities of alpha-fetoprotein. Crit Rev Eukaryot Gene Expr. 1997;7:11–41. doi: 10.1615/critreveukargeneexpr.v7.i1-2.20. [DOI] [PubMed] [Google Scholar]
- 6.Spear BT. Alpha-fetoprotein gene regulation: Lessons from transgenic mice. Semin Cancer Biol. 1999;9:109–116. doi: 10.1006/scbi.1998.0087. [DOI] [PubMed] [Google Scholar]
- 7.Vacher J, Tilghman SM. Dominant negative regulation of the mouse alpha-fetoprotein gene in adult liver. Science. 1990;250:1732–1735. doi: 10.1126/science.1702902. [DOI] [PubMed] [Google Scholar]
- 8.Godbout R, Ingram R, Tilghman SM. Multiple regulatory elements in the intergenic region between the alpha-fetoprotein and albumin genes. Mol Cell Biol. 1986;6:477–487. doi: 10.1128/mcb.6.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feuerman MH, Godbout R, Ingram RS, Tilghman SM. Tissue-specific transcription of the mouse alpha-fetoprotein gene promoter is dependent on HNF-1. Mol Cell Biol. 1989;9:4204–4212. doi: 10.1128/mcb.9.10.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang DE, Ge X, Rabek JP, Papaconstantinou J. Functional analysis of the trans-acting factor binding sites of the mouse alpha-fetoprotein proximal promoter by site-directed mutagenesis. J Biol Chem. 1991;266:21179–21185. [PubMed] [Google Scholar]
- 11.Bois-Joyeux B, Danan JL. Members of the CAAT/enhancer-binding protein, hepatocyte nuclear factor-1 and nuclear factor-1 families can differentially modulate the activities of the rat alpha-fetoprotein promoter and enhancer. Biochem J. 1994;301(Pt 1):49–55. doi: 10.1042/bj3010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McVey JH, et al. A G–>A substitution in an HNF I binding site in the human alpha-fetoprotein gene is associated with hereditary persistence of alpha-fetoprotein (HPAFP) Hum Mol Genet. 1993;2:379–384. doi: 10.1093/hmg/2.4.379. [DOI] [PubMed] [Google Scholar]
- 13.Emerson JA, Vacher J, Cirillo LA, Tilghman SM, Tyner AL. The zonal expression of alpha-fetoprotein transgenes in the livers of adult mice. Dev Dyn. 1992;195:55–66. doi: 10.1002/aja.1001950106. [DOI] [PubMed] [Google Scholar]
- 14.Lee KC, Crowe AJ, Barton MC. p53-mediated repression of alpha-fetoprotein gene expression by specific DNA binding. Mol Cell Biol. 1999;19:1279–1288. doi: 10.1128/mcb.19.2.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen TT, Cho K, Stratton SA, Barton MC. Transcription factor interactions and chromatin modifications associated with p53-mediated, developmental repression of the alpha-fetoprotein gene. Mol Cell Biol. 2005;25:2147–2157. doi: 10.1128/MCB.25.6.2147-2157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peyton DK, Ramesh T, Spear BT. Position-dependent activity of alpha -fetoprotein enhancer element III in the adult liver is due to negative regulation. Proc Natl Acad Sci USA. 2000;97:10890–10894. doi: 10.1073/pnas.200290397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson AD. The price of repression. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- 18.Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 19.Gray S, Levine M. Transcriptional repression in development. Curr Opin Cell Biol. 1996;8:358–364. doi: 10.1016/s0955-0674(96)80010-x. [DOI] [PubMed] [Google Scholar]
- 20.Zhang W, et al. Identification and characterization of DPZF, a novel human BTB/POZ zinc finger protein sharing homology to BCL-6. Biochem Biophys Res Commun. 2001;282:1067–1073. doi: 10.1006/bbrc.2001.4689. [DOI] [PubMed] [Google Scholar]
- 21.Mitchelmore C, et al. Characterization of two novel nuclear BTB/POZ domain zinc finger isoforms. Association with differentiation of hippocampal neurons, cerebellar granule cells, and macroglia. J Biol Chem. 2002;277:7598–7609. doi: 10.1074/jbc.M110023200. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen JV, Nielsen FH, Ismail R, Noraberg J, Jensen NA. Hippocampus-like corticoneurogenesis induced by two isoforms of the BTB-zinc finger gene Zbtb20 in mice. Development. 2007;134:1133–1140. doi: 10.1242/dev.000265. [DOI] [PubMed] [Google Scholar]
- 23.O'Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Postic C, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 25.Tilghman SM, Belayew A. Transcriptional control of the murine albumin/alpha-fetoprotein locus during development. Proc Natl Acad Sci USA. 1982;79:5254–5257. doi: 10.1073/pnas.79.17.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Chiu JF. Transactivation and repression of the alpha-fetoprotein gene promoter by retinoid X receptor and chicken ovalbumin upstream promoter transcription factor. Nucleic Acids Res. 1994;22:1079–1086. doi: 10.1093/nar/22.6.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leng X, Cooney AJ, Tsai SY, Tsai MJ. Molecular mechanisms of COUP-TF-mediated transcriptional repression: Evidence for transrepression and active repression. Mol Cell Biol. 1996;16:2332–2340. doi: 10.1128/mcb.16.5.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bois-Joyeux B, et al. The c-jun proto-oncogene down-regulates the rat alpha-fetoprotein promoter in HepG2 hepatoma cells without binding to DNA. J Biol Chem. 1995;270:10204–10211. doi: 10.1074/jbc.270.17.10204. [DOI] [PubMed] [Google Scholar]
- 29.Perincheri S, Dingle RW, Peterson ML, Spear BT. Hereditary persistence of alpha-fetoprotein and H19 expression in liver of BALB/cJ mice is due to a retrovirus insertion in the Zhx2 gene. Proc Natl Acad Sci USA. 2005;102:396–401. doi: 10.1073/pnas.0408555102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crowe AJ, Piechan JL, Sang L, Barton MC. S-Phase progression mediates activation of a silenced gene in synthetic nuclei. Mol Cell Biol. 2000;20:4169–4180. doi: 10.1128/mcb.20.11.4169-4180.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.