Abstract
Glycerophosphocholine (GPC) is an abundant osmoprotective renal medullary organic osmolyte. We previously found that its synthesis from phosphatidylcholine is catalyzed by tonicity-regulated activity of the phospholipase B, neuropathy target esterase. We also found that its degradation is catalyzed by glycerophosphocholine phosphodiesterase (GPC-PDE) activity and that elevating osmolality from 300 to 500 mosmol/kg by adding NaCl or urea, inhibits GPC-PDE activity, which contributes to the resultant increase of GPC. In the present studies we identify GDPD5 (glycerophosphodiester phosphodiesterase domain containing 5) as a GPC-PDE that is rapidly inhibited by high NaCl or urea. Recombinant GDPD5 colocalizes with neuropathy target esterase in the perinuclear region of HEK293 cells, and immuno-precipitated recombinant GDPD5 degrades GPC in vitro. The in vitro activity is reduced when the cells from which the GDPD5 is immuno-precipitated have been exposed to high NaCl or urea. In addition, high NaCl decreases mRNA abundance of GDPD5 via an increase of its degradation rate, although high urea does not. At 300 mosmol/kg siRNA knockdown of GDPD5 increases GPC in mouse inner medullary collecting duct-3 cells, and over expression of recombinant GDPD5 increases cellular GPC-PDE activity, accompanied by decreased GPC. We conclude that GDPD5 is a GPC-PDE that contributes to osmotic regulation of cellular GPC.
Keywords: high urea, hypertonicity, RNA stability
Interstitial NaCl and urea normally are very high in the renal inner medulla, associated with their role in concentrating the urine. High NaCl and urea can damage cells, necessitating osmoprotective responses, as reviewed in ref. 1. The cellular perturbations include cytoskeletal rearrangement, cell cycle delay, DNA breaks, oxidation of DNA and proteins, and apoptosis. Many damaging effects of high NaCl occur because it is hypertonic. It causes osmotic efflux of water from cells, which reduces their volume and elevates the concentration of all intracellular components. As a result, macromolecular crowding increases, which greatly elevates the activity of cellular macromolecules. In addition, intracellular ionic strength increases, which also affects most biochemical processes. Cells generally respond to the osmotic shrinkage by regulatory volume increase in which transporters are activated that mediate rapid uptake of inorganic ions, followed by an osmotic influx of water that restores cell volume, albeit at the expense of continued elevation of intracellular ionic strength. Over a period of hours, however, compatible organic osmolytes accumulate and the ionic strength gradually decreases toward that in the normotonic state, with cell volume being maintained. The benefit is that compatible organic osmolytes perturb less than do inorganic ions because they stabilize proteins by interacting favorably with the protein backbone. The principal compatible organic osmolytes in renal medullary cells are sorbitol, glycine betaine (betaine), myo-inositol, and glycerophosphocholine (GPC).
GPC is synthesized from phosphatidylcholine (PC) (2–4), catalyzed by neuropathy target esterase (NTE) (5, 6) and broken down into choline and α-glycerophosphate, catalyzed by glycerophosphocholine phosphodiesterase (GPC-PDE) (2, 3, 7–10). High NaCl increases GPC synthesis and reduces its degradation. High NaCl increases NTE activity by elevating its transcription, and thereby its mRNA and protein (5). GPC is special in that it is the only organic osmolyte also accumulated by renal cells in response to high urea and hypertonicity (11, 12). High urea elevates GPC solely by inhibiting GPC-PDE activity, which reduces the rate at which it is degraded (9). Although high NaCl and high urea both inhibit GPC-PDE, their effects differ in some respects. Thus, inhibition by high urea is sustained for over a week in Madin–Darby canine kidney (MDCK) cells, but inhibition by high NaCl disappears progressively between the second and seventh day of stress, and by that time high NaCl is sustaining high GPC solely by increasing its synthesis (9).
Although the phospholipase B responsible for tonicity-dependent regulation of GPC synthesis has been identified as NTE (5), the molecular identity of the GPC-PDE responsible for its osmoregulated degradation has remained unidentified. Glycerophosphoryl diester phosphodiesterases (GP-PDE, EC 3.1.4.46) hydrolyze glycerophosphodiesters including GPC. The GP-PDE family has homologues in both prokaryotes and eukaryotes. The purpose of the present studies was to determine whether any of the several homologues that exist in Mus musculus might contribute to osmoregulation of GPC-PDE in renal cells.
Results
Glycerophosphoryl Diester Phosphodiesterase Homologs.
Bacteria express a family of glycerophosphoryl diester phosphodiesterases (Pfam accession no. PF03009) that catalyze hydrolysis of a broad range of glycerophosphodiesters like glycerophosphocholine, glycerophosphoethanolamine, and glycerophosphoglycerol. As a start toward molecular identification of the osmoregulated GPC-PDE in renal cells, we searched for mouse homologues of this family, finding at that time (2003) five candidate genes that contain a similar catalytic site, namely BC024955 (GDPD5), NM 023608 (GDPD2), AK122510 (mKIAA1434), NM 025638 (GDPD1), and AK003726. Using RT-PCR, we found that all five are expressed in mouse inner medullary collecting duct (mIMCD)3 cells, and that expression of one of them, GDPD5 (glycerophosphodiester phosphodiesterase domain containing 5), is reduced when osmolality is increased from 300 to 700 mosmol/kg by adding NaCl (200 mosmol/kg) and urea (200 mosmol/kg) in combination (Table 1). With this as a clue, we examined the osmotic regulation of GDPD5 in more detail and tested to find out whether it is a GPC-PDE.
Table 1.
Effect of high NaCl and urea in combination on expression of mouse genes homologous to bacterial glycerophosphoryl diester phosphodiesterases
| Gene | mRNA ratio (700/300 mosmol/kg) |
|---|---|
| BC024955 (GDPD5) | 0.53 |
| NM_023608 (GDPD2) | 1.66 |
| AK122510 (mKIAA1434) | 0.91 |
| NM_025638 (GDPD1) | 0.93 |
| AK003726 (unknown) | 0.87 |
mRNA levels in mIMCD3 cells were measured by RT-PCR. Cells were either maintained at 300 mosmol/kg or increased to 700 mosmol/kg by addition of NaCl (200 mosm/kg) and urea (200 mosm/kg) for 24 h. Results are expressed by the ratio of mRNA abundance at 700 to that at 300 mosmol/kg (n = 3).
Regulation of GDPD5 mRNA Expression by Hypertonicity in Vivo and in Cell Culture.
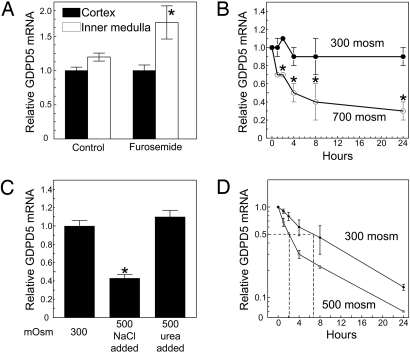
Knowing that exposure of mIMCD3 cells to high NaCl and urea decreases GDPD5 mRNA abundance, and that interstitial levels of NaCl and urea are normally high in the renal medulla associated with intracellular accumulation of GPC (13), we checked to see if decreasing renal medullary osmolality might increase GDPD5 mRNA expression. The level of GDPD5 mRNA is similar in the inner medulla and cortex of mouse kidney (Fig. 1A). Treatment with furosemide for 4 h, which reduces medulla interstitial osmolality, increases GDPD5 mRNA abundance in the inner medulla without changing the level in the cortex.
Fig. 1.
GDPD5 mRNA abundance in vivo and in mIMDC3 cells. (A) Effect of furosemide on GDPD5 mRNA abundance in mouse renal cortex and medulla. Kidneys were removed for analysis 4 h after i.p. injection of 1.5 mg of furosemide per 20 g of body weight. (B) Effect of hyperosmolality in mIMCD3 cells relative to the initial value at 300 mosmol/kg. (C) Effect on mIMCD3 cells of elevating NaCl or urea for 24 h, relative to the initial value at 300 mosmol/kg. (D) Effect on GDPD5 mRNA stability mIMCD3 cells of increasing osmolality from 300 to 500 mosmol/kg by adding NaCl. Five μg/ml actinomycin D was added 2 h after increasing NaCl. mRNA abundance was measured by real-time RT-PCR. (Mean ± SEM, n = 3, *, P < 0.05).
Next, we checked the time course of the decrease of GDPD5 mRNA after increased osmolality in cell culture. When mIMCD3 cells are exposed to high NaCl and urea in combination, GDPD5 mRNA abundance decreases by 30% within 2 h and by 70% within 24 h (Fig. 1B). To determine the separate effects of NaCl and urea we exposed mIMCD3 cells to elevated levels of either alone (Fig. 1C). Raising osmolality from 300 to 500 mosmol/kg by adding NaCl decreases GDPD5 mRNA abundance in mIMCD3 cells but urea does not. To test the role of mRNA stability in down regulation of GDPD5 mRNA, we added actinomycin D to stop transcription 2 h after increasing NaCl and then measured the rate at which TonEBP/OREBP mRNA decreased (Fig. 1D). At 300 mosmol/kg, the half-life of GDPD5 mRNA is ∼6 h. In contrast, after osmolality is increased to 500 mosmol/kg by adding NaCl, GDPD5 mRNA half-life decreases to 2 h. We conclude that high NaCl destabilizes GDPD5 mRNA. Given that the half-life of GDPD5 mRNA is 6 h at 300 mosmol/kg, if we assume that transcription of GDPD5 mRNA is not affected by high NaCl, a more than twofold increase in its degradation is sufficient to reduce its level by 50%, as observed Fig. 1B. We conclude that destabilization of GDPD5 mRNA accounts for the decrease in its abundance when NaCl is increased.
Osmotic Regulation of GPC and GPC-PDE Activity in mIMCD3 Cells.
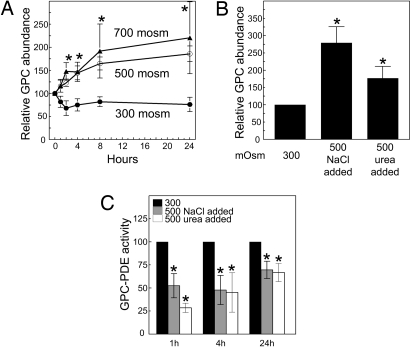
We found that high NaCl and urea, separately and in combination, increase GPC in MDCK cells and inhibit GPC-PDE activity (8, 9). Because we wanted to test the role of GDPD5 in mouse cells, we began by reexamining these findings in mIMCD3 cells. When the osmolality bathing mIMCD3 cells is elevated from 300 to 500 mosm/kg by adding NaCl, or to 700 mosm/kg by adding NaCl and urea in combination, GPC increases rapidly and remains elevated for at least 24 h (Fig. 2A). Both high NaCl alone and high urea alone increase GPC abundance (Fig. 2B). Furthermore, both alone inhibit GPC-PDE activity, beginning within one hour (Fig. 2C), confirming the previous results in MDCK cells.
Fig. 2.
Effect of hyperosmolality on GPC abundance and GPC-PDE activity in mIMCD3 cells. (A) Effects of increasing osmolality on GPC abundance (percentage of the initial value of 17.6 ± 0.6 nmoles/mg protein at 300 mosmol/kg). (B) Effects on GPC abundance of increasing NaCl or urea for 24 h. GPC abundance is expressed as percentage of the initial value at 300 mosmol/kg. (C) Effect on GPC-PDE activity (expressed as percentage of the initial value at 300 mosmol/kg) of increasing NaCl or urea. (Mean ± SEM, n = 3, *, P < 0.05).
Subcellular Localization of GDPD5.
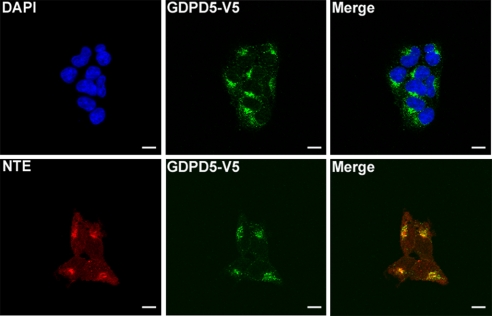
Recombinant GDPD5 was reported to localize to the perinuclear region of COS7 cells (14). In agreement, recombinant GDPD5-V5 is perinuclear when transfected into HEK293 cells (Fig. 3). Localization is the same in mIMCD3 cells (data not shown). GDPD5 possesses seven putative transmembrane domains, consistent with membrane localization, in this case most likely to the endoplasmic reticulum (ER) (14). To test this we compared the location of GDPD5-V5 and NTE, which is known to be an ER protein (15) and which catalyzes osmotically induced increase of synthesis of GPC from PC (5). GDPD5-V5 colocalizes with NTE in the ER of HEK293 cells (see Fig. 3). This observation raises the possibility that these enzymes may be colocalized to jointly regulate PC homeostasis. Nevertheless, we do not observe coimmuno-precipitation of NTE with recombinant GDPD5 (data not shown).
Fig. 3.
Subcellular localization of GDPD5. HEK293 cells were transfected with GDPD5-V5 at 300-mosmol/kg medium for 48 h, then fixed to visualize GDPD5-V5. Top panels: GDPD5-V5 (green) is localized in the perinuclear region. Nuclei are stained with DAPI (blue). Bottom panels: colocalization between GDPD5-V5 (green) and NTE (red) in the perinuclear region. (Scale bar = 20 μm.)
GDPD5 Regulates GPC Abundance and GPC-PDE Activity in mIMCD3 Cells.
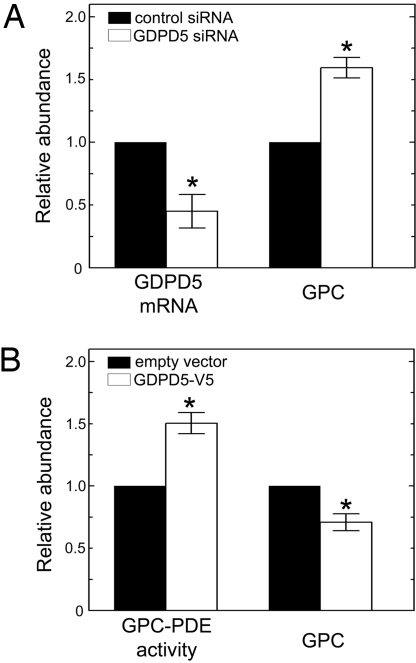
We transiently transfected a specific siRNA to knock down GDPD5 and a scrambled siRNA as control (Fig. 4A). Knocking down its expression by 55% in mIMCD3 cells at 300 mosmol/kg increases GPC by 60%. Conversely, overexpressing recombinant GDPD5-V5 increases GPC-PDE activity by 50%, while decreasing GPC by 30% (Fig. 4B). We conclude that expression of GDPD5 increases GPC-PDE activity and reduces cellular GPC abundance.
Fig. 4.
Effects of GDPD5 expression in mIMCD3 cells. (A) GDPD5 knock down with a specific siRNA at 300 mosmol/kg. The cells were transfected for 48 h with either control siRNA or GDPD5-specific siRNA. Left bars: Relative GDPD5 mRNA abundance. Right bars: Relative GPC abundance. (B) The cells were transfected with either empty vector or GDPD5-V5 construct at 300 mosmol/kg for 24 h. Left bars: Relative GPC abundance. Right bars: Relative GPC-PDE activity normalized by the total protein per dish of cells. (Mean ± SEM, n = 3, *, P < 0.05).
GDPD5 Is a GPC-PDE.
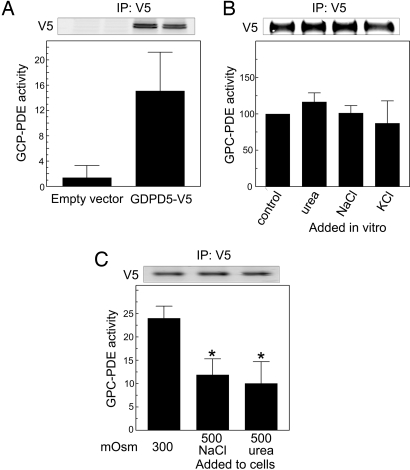
We transfected recombinant GDPD5-V5 into HEK293 cells at 300 mosmol/kg, then immuno-precipitated it and assayed the immuno-precipitate in vitro for GPC-PDE activity (Fig. 5A). The immuno-precipitate has significant GPC-PDE activity, whereas control immuno-precipitate from cells transfected with empty vector does not. We conclude that GDPD5 is a GPC-PDE.
Fig. 5.
GPC-PDE activity of GDPD5 immuno-precipitated from HEK293 cells and assayed in vitro. HEK293 cells were transfected for 24 h at 300 mosmol/kg with empty vector-V5 or GDPD5-V5. GPC-PDE activity was measured in immuno-precipitates from cell extracts and is normalized to the amount of extracted protein used for the immuno-precipitation. GDPD5-V5 expression was measured in the immuno-precipitates. Top panels are representative Western blots showing equivalent expression of GDPD5-V5 protein. (A) GPC-PDE activity is present in immuno-precipitates of GDPD5-V5, but not of empty vector-V5. (B) GDPD5 was immuno-precipitated from cells at 300 mosmol/kg, and its GPC-PDE activity measured in vitro, using the default buffer or buffers to which 200 mosmol/kg of NaCl, KCl, or urea was added. (C) After transfection, cells were incubated for 1 h either at 300 mosmol/kg or at 500 mosmol/kg (200-mosmol/kg NaCl or urea added). Note that GPC-PDE activity measured under identical conditions in vitro varies with the osmolality to which the cells had been exposed. (Mean ± SEM, n = 3, *, P < 0.05).
High NaCl or High Urea Alone Each Inhibits the GPC-PDE Activity of GDPD5.
High NaCl or high urea alone each inhibits GPC-PDE activity in MDCK (9) and mIMCD3 cells (see Fig. 2C). To investigate the mechanism involved, we tested the effects of high levels of salts and urea on the activity of recombinant GDPD5 by immuno-precipitating GDPD5-V5 from HEK293 cells at 300 mosmol/kg and directly adding the solutes to the GPC-PDE assay in vitro. Adding 200 mosmol/kg of NaCl, KCl, or urea to the assay buffer does not change GPC-PDE activity (Fig. 5B). Thus, we find no evidence that salts or urea directly inhibit GDPD5 activity. We also measured the GPC-PDE activity of recombinant GDPD5-V5 immuno-precipitated from HEK293 cells that had been exposed to an increase in osmolality from 300 to 500 mosmol/kg for 1 h by adding either NaCl or urea (Fig. 5C). GPC-PDE activity is reduced by >60% in immuno-precipitates from cells exposed to either high NaCl or high urea. It is important to note that, while the osmotic stress was applied to the cells, the subsequent assays of GPC-PDE activity were all performed in vitro under identical conditions. We conclude that exposure of cells to high NaCl or urea inhibits the activity of GDPD5 in a way that persists even after the enzyme is separated from the cells, perhaps by a posttranslational modification of the enzyme.
Discussion
GDPD5 Is an Osmoregulated Mammalian GPC-PDE.
The glycerophosphodiester phosphodiesterase motif (Pfam accession no. PF03009) was identified in a family of bacterial genes. GlpQ and UgpQ in Escherichia coli hydrolyze deacylated phospholipid glycerophosphodiesters, including glycerophosphocholine (16). In yeast, Gde1p is a GPC-PDE that is involved in retrieving choline from GPC as a substrate for the Kennedy pathway (17, 18). In mammals, several GP-PDEs have recently been identified. MIR16 (GDE1) interacts with RGS16 (regulator of G protein 16) and hydrolyzes glycerophosphoinositol, but not GPC (19). The substrates of the others were not reported. GDE3 is involved in differentiation and cytoskeletal regulation (20). GDE6 is mostly expressed in spermatocytes, where it plays a role in male germ cell differentiation (14). GDPD5 (GDE2), which we now identify as a GPC-PDE, is widely expressed, including in lung, heart, brain, kidney, and testis (14). It is a serpentine membrane protein containing seven putative transmembrane domains (14). It localizes to the perinuclear region in COS7 (14) and HEK293 cells (see Fig. 3). GDPD5 is necessary for spinal motor neuron differentiation (21) and retinoid-induced neuronal outgrowth (22). We find that GDPD5 also is an osmoregulated GPC-PDE that contributes to regulation of GPC in mIMCD3 cells. At this point, we do not know whether the other GP-PDEs also hydrolyze GPC or whether they, in addition to GDPD5, are involved in osmotic regulation of GPC.
Osmotic Regulation of GDPD5.
We find that either high NaCl or high urea decreases GDPD5 enzymatic activity (see Figs. 2 C and 5 B), but we do not yet know the mechanisms of its regulation by NaCl and urea. There is a difference, however, between the effects of the two solutes. High NaCl, but not high urea, reduces GDPD5 mRNA (see Fig. 1C). High NaCl reduces the half-life of GDPD5 mRNA (see Fig. 1D) enough to account for the rapid decrease in mRNA without any change in transcription. MRNAs, whose stability is regulated, often contain adenine and uracil (AU) rich elements (AREs) in their 3′-untranslated regions. However, we cannot identify AREs in the GDPD5 mRNA and we do not know how its stability is regulated. The level of GDPD5 mRNA apparently is also osmotically regulated in the renal medulla in vivo. Furosemide, which lowers medullary NaCl, increases GDPD5 mRNA, suggesting that the mRNA is normally lowered by the high level of NaCl in the medulla.
Elevating either NaCl or urea inhibits GPC-PDE activity in MDCK (9) and mIMCD3 (see Fig. 2C) cells. We find no evidence that the mechanism is direct inhibition of GDPD5 activity because adding NaCl, KCl, or urea to the assay buffer does not change GPC-PDE activity of GDPD5 (see Fig. 5B). Nevertheless, it is striking that when cells are exposed to high NaCl or urea, the resultant decrease in GDPD5 enzymatic activity (see Fig. 2C) is maintained in recombinant GDPD5 immuno-precipitated from the cells and assayed under fixed conditions in vitro (see Fig. 5C). One possibility is that hyperosmolality causes a posttranslational modification in GDPD5. GDPD5 contains many potential sites for phosphorylation (Motif Scan and Minimotif Miner). Several of them are possible targets for kinases whose activity is modulated by high NaCl or urea, including Erk (23), ATM (24), and PKA (25). Nevertheless, migration of recombinant GDPD5 in SDS-PAGE is not noticeably retarded by elevation of extracellular NaCl or urea, as it might be by increased phosphorylation. Other possibilities include that high NaCl or urea might alter association of some other protein with GDPD5 or modify the associated protein.
Physiological Roles of NTE and GPC-PDEs.
In Saccharomyces cerevisiae NTE catalyses synthesis of GPC from phosphatidylcholine, and YPL110c (Gde1p) degrades GPC to choline, providing substrate for renewed synthesis of phosphatidylcholine (18). Such cooperative activity of NTE and GPC-PDE is important for phosphatidylcholine homeostasis mammalians (26) and yeast (27). In the present study, we demonstrate that GDPD5 is a GPC-PDE, making it a plausible candidate for a mammalian GPC-PDE involved in the membrane phosphatidylcholine homeostasis. In support of this idea, GDPD5 and NTE are involved in outgrowth of neuronal cells (22, 28), a process that depends on membrane phosphatidylcholine renewal. Further investigation is required to confirm that GDPD5 actually has such a role in renewal of membrane phosphatidylcholine. Furthermore, GDPD5, being expressed in the brain (14), could be involved in the human central nervous system disorders in which altered levels of GPC have been reported (29). In addition to these other possible roles of GDPD5, our present studies demonstrate that it contributes to osmoregulation of GPC in the renal medulla. We show that GDPD5 is a GPC-PDE that contributes to accumulation of GPC in cells exposed to osmotic stress. We found that NTE contributes to the osmoregulated accumulation of GPC (5), so NTE and GDPD5 apparently act synergistically to adapt cells to stress of the normally high NaCl and urea in the renal medulla.
Materials and Methods
Cell Culture.
HEK293 cells were grown in Eagle's minimal essential medium (American Type Culture Collection) supplemented with 10% FBS (HyClone Laboratories). Mouse inner medullary collecting duct (mIMCD3) cells (30) were grown in 45% low-glucose DMEM (Irvine Scientific) plus 45% Coon's improved medium F-12 (BioSource) plus 10% FBS. Osmolality of the control medium was 300 mosmol/kg. All cells were cultured at 37°C in 5% CO2 and were used while subconfluent.
Animal Experiments.
Two- to 3-month-old male black six-strain mice (Taconic Farms) were injected intraperitonially with 1.5 mg/20 g furosemide or the same volume of saline solution (controls). Kidneys were removed 4 h later and inner medullas and cortexes were dissected for RNA analysis. Experiments were approved by the National Heart, Blood and Lung Institute Animal Care and Use Committee (Animal Study Proposal H-0130).
Plasmids and siRNA.
Mouse GDPD5 cDNA was cloned by PCR with specific primers (sense 5′-CACCATGGTGAGACACCAG-3′and anti-sense 5′-ATGCCCACTCTGCTCTGTG-3′) and inserted into pcDNA6.2/GW/D-TOPO (Expression Kit, Invitrogen). The sequence of the GDPD5 cDNA was confirmed. Double-stranded small interfering RNA (Dicer substrate RNAi duplex) against GDPD5 and nonspecific control were designed following the Integrated DNA Technologies instructions and recommendations (31). The siRNAs were synthesized by Integrated DNA Technologies. The sequences for GDPD5 siRNA were sense 5′-UGACUACGAUGAGUUCAACUGGUAC-3′ and antisense 5′-GUACCAGUUGAACUCAUCGUAGUCAUU-3′; and for nonspecific control were sense 5′AACUCCCUCCCUGGGUCAGGUUCAUU-3′ and antisense, 5′-UGAACCUGACCCAGGGGAGGGAGTT-3′. The siRNAs and plasmids were transiently transfected for 48 h total using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions and as described in ref. 5.
RNA Isolation and cDNA Preparation.
Total RNA was isolated from mIMCD3 cells using QiaShredder columns, followed by Qiagen RNeasy columns (Qiagen), according to the manufacturer's directions. RNA was treated with DNase for 15 min at room temperature (Qiagen) while bound to the RNeasy column. Total RNA concentration was measured by spectrophotometry, and the RNA was run on agarose gels to assess its quality. Then, 2 μg of total RNA were reverse transcribed with random hexamers, using TaqMan reverse transcription reagent kits (Applied Biosystems), following the manufacturer's recommendations.
Real-Time PCR.
Real-time PCR specific primers were designed for mouse GDPD5 mRNA using Primer Express software. The forward primer was 5′-GAAGTGACA TCACCGTCTCC-3′, the reverse primer was 5′-CAGGTGAGTGTGAGGCTGAA-3′. Multiplex PCR was performed with an ABI Prism 7900 sequence detection system, using a Quantitect SYBR Green PCR kit (Qiagen). Triplicates of each sample at each of two levels of added cDNA (8 and 80 ng) were included in each PCR run. We analyzed the results using ABI Prism 7900 system software.
Analysis of mRNA Stability.
Osmolality bathing subconfluent mIMCD3 cells was increased from 300 to 500 mosmol/kg for 2 h by adding NaCl or was kept at 300. Then, 5-μg/ml actinomycin D was added and cells were harvested at indicated times for measurement of mRNA abundance by real-time RT-PCR.
Immuno-Precipitation.
HEK293 cells transiently expressing recombinant GDPD5-V5 were grown in 300-mosmol/kg medium. Fresh medium at 300 or 500 mosmol/kg (NaCl or urea added) was substituted for the designated times. Then, cells were trypsinized and pelleted by centrifugation. Subsequent steps were at 4°C. The pellet from one 10-cm dish was extracted for 5 min with 1 ml of lysis buffer containing 50-mM Tris·HCl, pH 8.0, 150-mM NaCl, 1% Triton X-100, protease (Complete Mini, Roche Applied Science), and phosphatase inhibitor cocktails (Phosphatase Inhibitor Cocktails 1 and 2, Sigma) and centrifuged (15,000 × g, 10 min). Samples were precleared with 1-mg Dynabeads (Invitrogen) and 2-μg rabbit IgG biotin-conjugated (Santa Cruz Biotechnology) for 1 h. Precleared supernatants were incubated overnight with 4-μg rabbit anti-V5 biotin-conjugated (Immunology Consultants Laboratory) and 1-mg Dynabeads (Invitrogen). As a negative control, IgG-biotin conjugate (Santa Cruz Biotechnology) was substituted for anti-V5. Beads-GDPD5V5 were resuspended in GPC-PDE assay Buffer (see GPC-PDE assay) or incubated 5 min at 95°C in Laemmli buffer for Western blot analysis.
Western-Blotting.
Western blot analysis was performed as described in ref. 5. Briefly, total protein concentration was measured in cell extracts by bicinchoninic acid assay (Pierce) and proteins were separated on 4 to 12% gradient acrylamide-Tris-glycine gels and transferred electrophoretically to nitrocellulose membranes Invitrogen). Membranes were blocked with blocking buffer (LI-COR Biosciences,) plus Tween (0.1%), then incubated overnight at 4°C with the mouse anti-V5 monoclonal antibody (Invitrogen), followed by Alexa Fluor 680 goat anti-rabbit secondary antibody (Invitrogen) for 1 h at room temperature. Blots were visualized and quantitated by using a LI-COR Odyssey Infrared Imager (LI-COR Biosciences).
Confocal Immunofluorescence Microscopy.
HEK293 cells transfected with recombinant GDPD5-V5 grown on 2 wells Lab-Tek II glass Slide (Nalge Nunc International) were fixed for 20 min (all procedures at room temperature) with 3% paraformaldehyde in PBS, washed with PBS, permeabilized for 5 min with 0.1% Triton X-100 in PBS, washed with PBS, and incubated for 1 h with blocking buffer (3% BSA in PBS). Cells were washed with PBS and incubated with a mouse anti-V5 and a rabbit anti-NTE overnight, washed with PBS, and incubated for 1 h with Alexa Fluor 488 goat anti-mouse IgG and Alexa Fluor 633 goat anti-rabbit IgG (both from Invitrogen). After washing with PBS, nuclei were stained with 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI, Invitrogen). Cells were mounted in SlowFade Gold antifade reagent (Invitrogen) and inspected with a confocal fluorescence microscope (LSM 510; Carl Zeiss).
Measurement of GPC.
Cell GPC was measure by chemiluminescence as described in refs. 5 and 9. Briefly, perchloric acid (PCA, 7%) was added to attached cells for 10 min on ice after removal of the experimental medium and the PCA extract was kept at −80°C until analyzed. 0.5-N NaOH was added to the cell remnants for protein determination. Fifty microliters of the PCA extract were diluted 1:1 with 7% PCA in duplicate. One aliquot was heated at 95°C for 1 h to hydrolyze GPC to choline. The other aliquot was kept on ice for 1 h to prevent hydrolysis. Then, 36 μl of 2M K2CO3 were added to each aliquot, followed by centrifugation at 16,000 × g for 15 min. Twenty microliters of each supernatant were diluted to 1 ml with 67-mM glycine buffer, pH 8.6. One hundred microliters of this dilution plus 300 μl of glycine buffer were analyzed for choline in a luminometer (model 2010; Monolight) by rapidly adding 100 μl of reagent containing 67-mM glycine buffer, pH 8.6, 20-μg horseradish peroxidase, 0.4-U choline oxidase, and 6-nmoles Luminol.
GPC-PDE activity was assayed as described in ref. 32. Briefly, subconfluent mIMCD3 were exposed to medium at 300 or 500 mosmol/kg (NaCl or urea added) for the specified time. The cells were then rinsed twice with sterile ice-cold 0.25-M sucrose. The cells were scraped into 1.0 ml of 0.25 M sucrose and then homogenized with a Polytron (Kinematica). Deoxycholic acid (DOCA) was added to the homogenate to a final concentration of 0.39%, and the mixture was assayed immediately, using as substrate GPC (Sigma) from which traces of choline were first removed with a cation-exchange resin (AG 5OW-X12, Bio-Rad). Sixty microliters of glycine buffer pH 9.0 (200 mM glycine-NaOH, Sigma Chemical) plus 40 μl of 50-mM GPC were added to 100 μl of the homogenate-DOCA mixture. Eighty microliters of this solution were immediately mixed with 8 μl of 35% PCA for determination of choline in the homogenate at time 0. Another 80 μl of this solution were incubated at 37°C for 60 min, at which time the reaction was stopped by addition of 8 μl of PCA. Assay mixtures were centrifuged and supernatants were neutralized with 2M K2CO3. After centrifugation, the supernatant was diluted 25- to 50-fold with 67-mM glycine buffer pH 8.6 and analyzed for choline by chemiluminescence, as described above. GPC-PDE assays were performed by using the same method on the beads (above) containing immuno-precipitated GDPD5-V5. After the enzyme assay, GDPD5-V5 was removed from the beads, using Laemmli buffer and quantified by Western analysis, as above.
Statistical Analysis.
Data were compared by ANOVA, followed by Student-Newman-Keuls post test. Normalized data were log-transformed before ANOVA. Results are expressed as means ± SE (n = number of independent experiments). Differences were considered significant for P < 0.05.
Acknowledgments.
This work was supported by the Intramural Research Program of the National Institutes of Health, National Heart Lung and Blood Institute.
Footnotes
The authors declare no conflict of interest.
References
- 1.Burg MB, Ferraris JD, Dmitireva NI. Cellular response to hyperosmotic stresses. Physiol. Rev. 2007;87:1441–1474. doi: 10.1152/physrev.00056.2006. [DOI] [PubMed] [Google Scholar]
- 2.Bauernschmitt HG, Kinne RK. Metabolism of the ‘organic osmolyte’ glycerophosphorylcholine in isolated rat inner medullary collecting duct cells. I. Pathways for synthesis and degradation. Biochim Biophys Acta. 1993;1148:331–341. doi: 10.1016/0005-2736(93)90147-r. [DOI] [PubMed] [Google Scholar]
- 3.Bauernschmitt HG, Kinne RK. Metabolism of the ‘organic osmolyte’ glycerophosphorylcholine in isolated rat inner medullary collecting duct cells. II. Regulation by extracellular osmolality. Biochim Biophys Acta. 1993;1150:25–34. doi: 10.1016/0005-2736(93)90117-i. [DOI] [PubMed] [Google Scholar]
- 4.Kwon ED, Jung KY, Edsall LC, Kim HY, Garcia-Perez A, Burg MB. Osmotic regulation of synthesis of glycerophosphocholine from phosphatidylcholine in MDCK cells. Am J Physiol. 1995;268:C402–C412. doi: 10.1152/ajpcell.1995.268.2.C402. [DOI] [PubMed] [Google Scholar]
- 5.Gallazzini M, Ferraris JD, Kunin M, Morris RG, Burg MB. Neuropathy target esterase catalyzes osmoprotective renal synthesis of glycerophosphocholine in response to high NaCl. Proc Natl Acad Sci USA. 2006;103:15260–15265. doi: 10.1073/pnas.0607133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaccheo O, Dinsdale D, Meacock PA, Glynn P. Neuropathy target esterase and its yeast homologue degrade phosphatidylcholine to glycerophosphocholine in living cells. J Biol Chem. 2004;279:24024–24033. doi: 10.1074/jbc.M400830200. [DOI] [PubMed] [Google Scholar]
- 7.Ullrich KJ, Pehling G. Über das Vorkommen von Phosphorverbindungen in verschiedenen Nierenabschnitten und Ánderungen ihrer Konzentration in Abhängigkeit vom Diuresezustand. Pflügers Archiv Eur J Physiol. 1956;262:551–561. doi: 10.1007/BF00362117. [DOI] [PubMed] [Google Scholar]
- 8.Zablocki K, Miller SP, Garcia-Perez A, Burg MB. Accumulation of glycerophosphocholine (GPC) by renal cells: osmotic regulation of GPC: choline phosphodiesterase. Proc Natl Acad Sci USA. 88:7820–7824. doi: 10.1073/pnas.88.17.7820. and correction (1991) 88(21):9907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon ED, Zablocki K, Jung KY, Peters EM, Garcia-Perez A, Burg MB. Osmoregulation of GPC: choline phosphodiesterase in MDCK cells: different effects of urea and NaCl. Am J Physiol. 1995;269:C35–C41. doi: 10.1152/ajpcell.1995.269.1.C35. [DOI] [PubMed] [Google Scholar]
- 10.Nakanishi T, Turner RJ, Burg MB. Osmoregulation of betaine transport in mammalian renal medullary cells. Am J Physiol. 1990;258:F1061–F1067. doi: 10.1152/ajprenal.1990.258.4.F1061. [DOI] [PubMed] [Google Scholar]
- 11.Nakanishi T, Burg MB. Osmoregulation of glycerophosphorylcholine content of mammalian renal cells. Am J Physiol. 1989;257:C795–C801. doi: 10.1152/ajpcell.1989.257.4.C795. [DOI] [PubMed] [Google Scholar]
- 12.Peterson DP, Murphy KM, Ursino R, Streeter K, Yancey PH. Effects of dietary protein and salt on rat renal osmolytes: covariation in urea and GPC contents. Am J Physiol. 1992;263:F594–F600. doi: 10.1152/ajprenal.1992.263.4.F594. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Perez A, Burg MB. Renal medullary organic osmolytes. Physiol Rev. 1991;71:1081–1115. doi: 10.1152/physrev.1991.71.4.1081. [DOI] [PubMed] [Google Scholar]
- 14.Nogusa Y, Fujioka Y, Komatsu R, Kato N, Yanaka N. Isolation and characterization of two serpentine membrane proteins containing glycerophosphodiester phosphodiesterase, GDE2 and GDE6. Gene. 2004;337:173–179. doi: 10.1016/j.gene.2004.04.026. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Dinsdale D, Glynn P. Protein domains, catalytic activity, and subcellular distribution of neuropathy target esterase in Mammalian cells. J Biol Chem. 2003;278:8820–8825. doi: 10.1074/jbc.M210743200. [DOI] [PubMed] [Google Scholar]
- 16.Tommassen J, Eiglmeier K, Cole ST, Overduin P, Larson TJ, Boos W. Characterization of two genes, glpQ and ugpQ, encoding glycerophosphoryl diester phosphodiesterases of Escherichia coli. Mol Gen Genet. 1991;226:321–327. doi: 10.1007/BF00273621. [DOI] [PubMed] [Google Scholar]
- 17.Fisher E, Almaguer C, Holic R, Griac P, Patton-Vogt J. Glycerophosphocholine-dependent growth requires Gde1p (YPL110c) and Git1p in Saccharomyces cerevisiae. J Biol Chem. 2005;280:36110–36117. doi: 10.1074/jbc.M507051200. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez-Murray JP, McMaster CR. Glycerophosphocholine catabolism as a new route for choline formation for phosphatidylcholine synthesis by the Kennedy pathway. J Biol Chem. 2005;280:38290–38296. doi: 10.1074/jbc.M507700200. [DOI] [PubMed] [Google Scholar]
- 19.Zheng B, Chen D, Farquhar MG. MIR16, a putative membrane glycerophosphodiester phosphodiesterase, interacts with RGS16. Proc Natl Acad Sci USA. 2000;97:3999–4004. doi: 10.1073/pnas.97.8.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yanaka N, et al. Novel membrane protein containing glycerophosphodiester phosphodiesterase motif is transiently expressed during osteoblast differentiation. J Biol Chem. 2003;278:43595–43602. doi: 10.1074/jbc.M302867200. [DOI] [PubMed] [Google Scholar]
- 21.Rao M, Sockanathan S. Transmembrane protein GDE2 induces motor neuron differentiation in vivo. Science. 2005;309:2212–2215. doi: 10.1126/science.1117156. [DOI] [PubMed] [Google Scholar]
- 22.Yanaka N, Nogusa Y, Fujioka Y, Yamashita Y, Kato N. Involvement of membrane protein GDE2 in retinoic acid-induced neurite formation in Neuro2A cells. FEBS Lett. 2007;581:712–718. doi: 10.1016/j.febslet.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 23.Yang XY, Zhang Z, Cohen DM. ERK activation by urea in the renal inner medullary mIMCD3 cell line. Am J Physiol. 1999;277:F176–F185. doi: 10.1152/ajprenal.1999.277.2.F176. [DOI] [PubMed] [Google Scholar]
- 24.Irarrazabal CE, Liu JC, Burg MB, Ferraris JD. ATM, a DNA damage-inducible kinase, contributes to activation by high NaCl of the transcription factor TonEBP/OREBP. Proc Natl Acad Sci USA. 2004;101:8809–8814. doi: 10.1073/pnas.0403062101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferraris JD, Persaud P, Williams CK, Chen Y, Burg MB. cAMP-independent role of PKA in tonicity-induced transactivation of tonicity-responsive enhancer/ osmotic response element-binding protein. Proc Natl Acad Sci USA. 2002;99:16800–16805. doi: 10.1073/pnas.222659799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baburina I, Jackowski S. Cellular responses to excess phospholipid. J Biol Chem. 1999;274:9400–9408. doi: 10.1074/jbc.274.14.9400. [DOI] [PubMed] [Google Scholar]
- 27.Dowd SR, Bier ME, Patton-Vogt JL. Turnover of phosphatidylcholine in Saccharomyces cerevisiae. The role of the CDP-choline pathway. J Biol Chem. 2001;276:3756–3763. doi: 10.1074/jbc.M003694200. [DOI] [PubMed] [Google Scholar]
- 28.Akassoglou K, Malester B, Xu J, Tessarollo L, Rosenbluth J, Chao MV. Brain-specific deletion of neuropathy target esterase/swisscheese results in neurodegeneration. Proc Natl Acad Sci USA. 2004;101:5075–5080. doi: 10.1073/pnas.0401030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walter A, et al. Glycerophosphocholine is elevated in cerebrospinal fluid of Alzheimer patients. Neurobiol Aging. 2004;25:1299–1303. doi: 10.1016/j.neurobiolaging.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 30.Rauchman MI, Nigam SK, Delpire E, Gullans SR. An osmotically tolerant inner medullary collecting duct cell line from an SV40 transgenic mouse. Am J Physiol. 1993;265:F416–F424. doi: 10.1152/ajprenal.1993.265.3.F416. [DOI] [PubMed] [Google Scholar]
- 31.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 32.Kwon ED, Zablocki K, Peters EM, Jung KY, Garcia-Perez A, Burg MB. Betaine and inositol reduce MDCK cell glycerophosphocholine by stimulating its degradation. Am J Physiol. 1996;270:C200–C207. doi: 10.1152/ajpcell.1996.270.1.C200. [DOI] [PubMed] [Google Scholar]