Abstract
Increased α-synuclein gene (SNCA) dosage due to locus multiplication causes autosomal dominant Parkinson's disease (PD). Variation in SNCA expression may be critical in common, genetically complex PD but the underlying regulatory mechanism is unknown. We show that SNCA and the heme metabolism genes ALAS2, FECH, and BLVRB form a block of tightly correlated gene expression in 113 samples of human blood, where SNCA naturally abounds (validated P = 1.6 × 10−11, 1.8 × 10−10, and 6.6 × 10−5). Genetic complementation analysis revealed that these four genes are co-induced by the transcription factor GATA-1. GATA-1 specifically occupies a conserved region within SNCA intron-1 and directly induces a 6.9-fold increase in α-synuclein. Endogenous GATA-2 is highly expressed in substantia nigra vulnerable to PD, occupies intron-1, and modulates SNCA expression in dopaminergic cells. This critical link between GATA factors and SNCA may enable therapies designed to lower α-synuclein production.
Keywords: α-synuclein dosage, GATA-1, GATA-2, gene expression, microarray
Dosage of α-synuclein appears to be central to the pathogenesis of both rare familial and common sporadic forms of human Parkinson's disease (PD) (1–3). Inclusions of α-synuclein (4, 5), together with loss of dopamine neurons and elevated iron levels in the substantia nigra (6) are pathologic hallmarks of the disease. In patients with PD due to a duplication or triplication of the SNCA locus, copies of functionally normal SNCA message and protein in brain and blood are increased by 50–100% (3, 7). Although small, over years, this increase is sufficient to bring death to a majority of vulnerable dopamine neurons. Even in sporadic PD, ≈3% of individuals carry a SNCA promoter variant, which confers susceptibility to PD possibly by increasing SNCA expression (2, 8). Toxic effects of wild-type SNCA overexpression are seen in human dopaminergic cells (9) and model organisms (reviewed in ref. 4).
The transcriptional mechanisms regulating the cellular concentration of SNCA copies may thus hold a key for understanding PD pathobiology and for developing therapeutics designed to keep α-synuclein levels within normal range. Whereas cis-acting variation (such as copy number variations and promoter polymorphisms) may explain up to 25–35% of interindividual differences in gene expression (10), heritable gene expression differences from trans-acting mechanisms appear to be quantitatively more important (10).
Although PD symptoms reflect preferential neuronal death, molecular changes in dopamine metabolism and other biologic processes are detected in blood cells (references in ref. 11). SNCA has been initially characterized as “expressed only in nervous system tissue, not in … muscle, liver, spleen, heart, or kidney” (12), although select reports have detected α-synuclein in plasma (13) and platelets (14). We observed surprisingly high levels of SNCA in human red blood cells. This dramatic and tissue-specific expression of SNCA in hematopoietic cells and neuronal cells suggested to us that these two cell types may share a common mechanism activating SNCA transcription.
Results
SNCA mRNA and Protein Are Highly Abundant in Human and Mouse Erythroid Cells.
Relative SNCA mRNA abundance was determined by comparing the SNCA mRNA abundance in each target tissue to the calibrator Universal Human Reference RNA (Fig. 1A). SNCA mRNA abundance was high in whole blood from human donors (relative abundance 10 {range 6.6–15.3}) with very high levels in packed red blood cells (33.8 {25.2–45.5}). SNCA mRNA abundance in peripheral blood mononuclear white cells (PBMC) (0.9 {0.7–1.3}), as well as in two brain regions vulnerable to PD pathology, human frontal cortex and substantia nigra, was comparably low (3.7 {3.3–4.1} and 2.6 {1.7–3.9}, respectively). mRNA extracted from packed red blood cells is derived from reticulocytes, immature red blood cells originating from transferrin receptor (CD71)-positive early erythroid cells in the bone marrow (15). Consistently, SNCA mRNA levels were exceedingly high in immunopurified CD71+ cells from human bone marrow (relative abundance of 9.2 {8.8–9.6}). High SNCA mRNA levels in erythroid cells were confirmed when GAPDH instead of the ribosomal gene RPL13 was used to control for input RNA (data not shown).
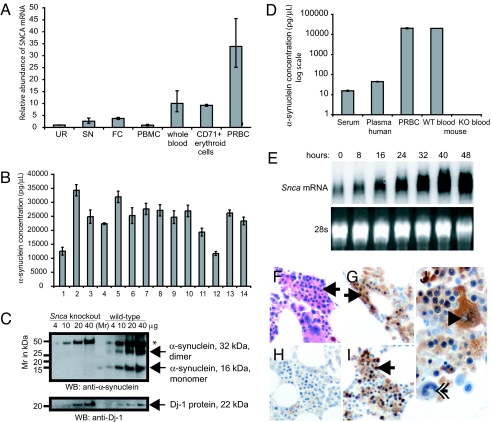
Fig. 1.
SNCA mRNA and protein is abundantly expressed in human and mouse erythroid cells. (A) SNCA mRNA was quantified by quantitative PCR using the ribosomal gene RPL13 as reference and Human Universal Reference RNA (UR) as calibrator. Relative SNCA mRNA abundance was high in whole blood of human donors without neurologic disease (relative abundance 10 {range 6.6–15.3}) and in immunopurified CD71+ erythroid cells (9.2 {8.8–9.6}), and also very high in packed red blood cells (PRBC) (33.8 {25.2–45.5}) after removal of plasma and buffy coat containing white blood cells and platelets. Relative SNCA mRNA abundance in peripheral blood mononuclear white cells (PBMC) (0.9 {0.7–1.3}), as well as in two brain regions vulnerable to PD pathology, human frontal cortex (FC) and substantia nigra (SN), was low (3.7 {3.3–4.1} and 2.6 {1.7–3.9}, respectively). (B–D) Detailed characterization of α-synuclein protein abundance revealed high levels in cell lysates of whole blood from 14 healthy humans by sandwich ELISA (B) and mice by Western blot analysis (C). (C) Wild-type, full-length murine α-synuclein was found in lysates of whole blood of wild-type mice (right) in the form of monomers (16 kDa) and dimers (32 kDa) and was absent in Snca knockout mice (left). Western blot analysis with anti-Dj-1 antibodies is shown as loading control. *, abundant, nonspecific, ≈50-kDa band due to cross-reactivity of the secondary anti-mouse antibody with mouse antigen. (D) α-Synuclein was particularly abundant in the cellular blood compartments, PRBC, and whole blood by sandwich ELISA (16). α-Synuclein concentrations measured 15.0 ± 0.9 pg/μl in fresh serum, 45.0 ± 1.4 pg/μl in fresh plasma, and 24.16 ± 1.7 ng/μl in whole blood lysates and PRBC. (E) SNCA mRNA was strongly and progressively expressed in a model system of terminal erythroid differentiation by Northern blot analysis. Transformed erythroblasts were harvested after 0, 8, 16, 24, 32, 40, and 48 h. (F–J) Erythroblasts (arrows) in human bone marrow smears (F) (H&E stain) show strong α-synuclein-immunoreactivity with monoclonal antibody Syn-1 (G) and rabbit-based, affinity-purified hSA-2 (I). (H) In the absence of primary antibody, no α-synuclein-immunoreactivity is detected. (J) α-Synuclein-immunoreactivity is also detected in megakaryocytes by Syn-1 (arrowhead) but not in myeloid cells (double arrow).
Characterization of α-synuclein protein abundance (Fig. 1B) revealed that the relative concentration of detectable α-synuclein as a constituent of total protein in whole human blood was 0.012 ± 0.003% by sensitive sandwich ELISA (16). α-Synuclein was specifically detected in mouse and human whole blood by Western blot analysis (Fig. 1C) and ELISA (Fig. 1D). α-Synuclein was particularly abundant in cellular blood compartments, packed red blood cells, and whole blood (Fig. 1D). Importantly, progressive expression of Snca was detected in a model system of terminal erythroid differentiation, erythroblasts of Friend virus-infected mice (Fig. 1E). α-Synuclein immunoreactivity was also found in erythroblasts in human bone marrow (Fig. 1 F–J). Collectively, these data unequivocally demonstrate that SNCA is abundantly expressed during different steps of erythropoiesis.
Expression of SNCA and Three Heme Metabolism Genes Is Tightly Correlated.
We hypothesized that transcripts whose levels are tightly correlated represent a transcriptionally controlled expression block. To test this hypothesis we correlated the expression of the query gene SNCA with the expression of the 14,500 genes assayed by 22,283 probe sets in an established dataset from blood specimens of 22 control individuals [Fig. 2 A and B and supporting information (SI) Methods] (11). The expression of 35 genes was tightly correlated with SNCA expression (Spearman's rank correlation coefficient ≥0.81; see SI Methods, Fig. S1, and Tables S1 and S2). Importantly, three of the coexpressed genes, 5-aminolevulinate synthase 2 (ALAS2; R = 0.80–0.85; P = 4.4 × 10−7; Fig. 2 A and B), ferrochelatase (FECH, R = 0.84–0.91, P ≥ 4.7 × 10−7), and biliverdin reductase B (BLVRB; R = 0.74–0.89, P = 1.6 × 10−6), encode critical steps in heme metabolism. These correlations were significant after conservative Bonferroni-correction for multiple testing of 22,283 probe sets with P < 0.05. Beyond searching for genes specifically correlated with SNCA expression, we generally examined the frequency of strong correlations in expression (R ≥ 0.81) for all possible combinations of any two probe sets. Only 0.0036% of all unique combinations met this threshold (data not shown).
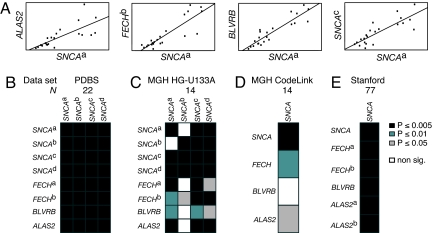
Fig. 2.
Expression of SNCA and heme metabolism genes ALAS2, FECH, and BLVRB is tightly and significantly correlated in human blood in four datasets. (A) Scatterplots of SNCAa and ALAS2, FECHb, and BLVRB expression are shown, respectively. SNCA expression measured by two distinct SNCA probes is plotted for comparison (rightmost plot). (B–E) Heatmaps visualize the P-value of the pairwise Spearman rank correlation between expression of SNCA (columns) and expression of SNCA, ALAS2, FECH, and BLVRB (rows) in four datasets. Correlations are shown as black (P ≤ 0.005), dark gray (P ≤ 0.01), or light gray cells (P ≤ 0.05). Nonsignificant correlations are represented as white cells. Probe-level correlations are shown for four distinct SNCA probes in B and C and two FECH probes in B, C, and E. (B) P-values of the correlations of SNCA and ALAS2, FECH, and BLVRB expression in the discovery set. These remain significant after Bonferroni correction. (C–E) Coexpression of SNCA with BLVRB was robustly replicated in two and coexpression of SNCA with ALAS2 and FECH in three validation studies comprising two independent populations of 14 and 77 control individuals and three different array platforms (C, Affymetrix; D, CodeLink; E, cDNA array). Correlations with P ≤ 0.05 (black and gray cells) are significant in the validation sets. Probe SNCAb is inefficient. See the main text and SI Methods for details.
Coexpression of SNCA and Three Heme Metabolism Genes in Two Independent Populations Assayed on Three Platforms.
If the coexpression of ALAS2, FECH, and BLVRB with SNCA is a robust and biologically relevant finding, it should be a universal signature in human blood. We therefore precisely validated the coexpression in three validation datasets comprising a total of 89 healthy individuals without multiple testing of other probe sets.
The first validation set (17) (Fig. 2C) of 14 healthy volunteers probed by a total of eight probes for SNCA, ALAS2, FECH, and BLVRB (four probes for SNCA, two for FECH) on HG-U133A Affymetrix oligonucleotide microarrays. This validation study confirmed that expression levels of ALAS2, FECH, and BLVRB were highly and significantly correlated with SNCA (P = 0.001, 0.0004, and 0.005 for select ALAS2, FECH, and BLVRB probes, respectively; Fig. S1 and Table S3).
To examine whether this block of coexpressed genes is independent of the microarray platform, we analyzed a human blood gene expression dataset derived from the same subjects but assayed by four oligonucleotide probes for SNCA, ALAS2, FECH, and BLVRB spotted on GE Healthcare CodeLink Uniset 20K arrays. In this second validation set (17) (Fig. 2D), a high and significant correlation for ALAS2 and FECH, with SNCA expression levels was replicated (P = 0.02 and 0.009, respectively; Fig. S1 and Table S3) thus confirming the correlation on a different platform. On the CodeLink arrays, the correlation between SNCA and BLVRB signals did not reach significance, likely due to an inefficient BLVRB probe on the CodeLink Uniset 20K arrays (P = 0.07).
Next, we analyzed a third, independent dataset (Fig. 2E) of blood samples from a large and diverse group of healthy humans (18). In this dataset, 77 RNA samples of cellular whole blood from 75 individuals were analyzed by cDNA microarrays spotting six cDNA probes, representing the four target genes (two probes for FECH and two probes for ALAS2). Consistent with our previous results, there was a strong and significant correlation between SNCA and ALAS2, FECH, and BLVRB expression levels (P = 1.6 × 10−11, 1.8 × 10−10, and 6.6 × 10−5 for ALAS2, FECHb, and BLVRB, respectively; Fig. S1 and Table S3).
Collectively, these four independent studies showed that variation in FECH, ALAS2, BLVRB, and SNCA expression is tightly and significantly linked—independent of sample collection, study population, and array platform.
Hematopoietic Transcription Factor GATA-1 Activates Snca Transcription in Erythroid Precursor Cells.
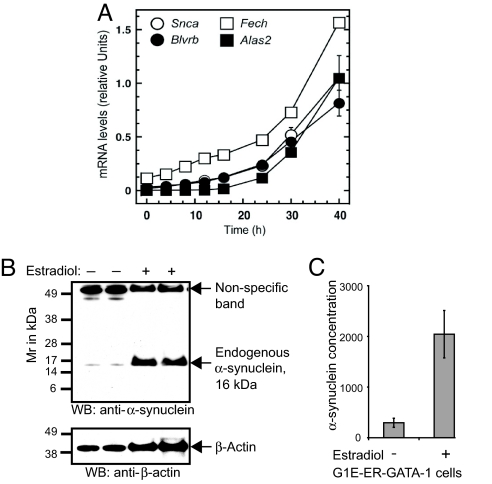
It is plausible that the SNCA, ALAS2, FECH, and BLVRB coexpression block is coordinately transcribed. The transcription factors regulating SNCA and BLVRB are unknown. However, insights gained over the past decades about the transcriptional regulation of ALAS2 and FECH offered a critical clue. ALAS2 expression is activated by the hematopoietic transcription factor GATA-1 (19, 20), and the FECH gene contains GATA-1 binding sites (21). Thus, GATA-1 represented a prime candidate for a trans-acting factor coordinating this expression block. To test this, we conducted genetic complementation analysis in GATA-1-null erythroid precursor cells (G1E-ER-GATA-1) stably expressing an estrogen receptor ligand binding domain fused to GATA-1 (ER-GATA-1). In this system, β-estradiol-mediated activation of ER-GATA-1 induces a gene expression program that recapitulates a normal window of erythropoiesis (22). This is a powerful system for identifying GATA-1 target genes (22, 23). G1E-ER-GATA-1 cells were treated with 1 μM β-estradiol for up to 40 h. The relative levels of murine Snca, Alas2, Fech, and BlvrB mRNA were normalized to Gapdh mRNA and quantified by real-time PCR (Fig. 3A). Snca, Alas2, Fech, and BlvrB messages were co-induced by GATA-1. GATA-1 induced a 62-fold increase in Snca mRNA copy numbers at 40 h compared with uninduced cells. GATA-1 induced a 27-fold increase in BlvrB, a 14-fold increase in Fech, and a 6,687-fold increase in Alas2 mRNA levels. Consistent with a highly specific regulation of these genes by GATA-1, transcription of BlvrA, the paralog of BlvrB, and Sncb (encoding β-synuclein) the paralog of Snca, were not activated (data not shown). Western blot analysis indicated that induction of Snca mRNA by GATA-1 was accompanied by an increase in α-synuclein protein (Fig. 3B). Quantification by ELISA indicated that GATA-1 induced a 6.9-fold increase in α-synuclein concentration relative to uninduced cells (Fig. 3C; mean ± standard deviation, 2,042.7 ± 467.5 and 295.6 ± 90 pg/μl, respectively).
Fig. 3.
The hematopoietic transcription factor GATA-1 activates Snca transcription in GIE-ER-GATA-1 cells. (A) Expression of murine Snca and the heme metabolism genes Alas2, Fech, and BlvrB are co-induced by conditionally active GATA-1 (ER-GATA-1). Relative levels of Snca, Alas2, Fech, and BlvrB mRNA are quantified by real-time PCR at 2–40 h postinduction of ER-GATA-1. The mRNA levels are normalized by Gapdh mRNA and expressed as relative expression. (B) ER-GATA-1 activation in G1E-ER-GATA-1 cells induces endogenous α-synuclein protein, as detected by Western blot analysis (Upper). (Lower) Western blot with anti-actin after stripping and reprobing. (C) α-Synuclein concentration (pg/μl) is increased 6.9-fold in lysates of estradiol-induced compared with uninduced G1E-ER-GATA-1 cells when quantified by sandwich ELISA (hSA-2/Syn1-B).
GATA-1 Specifically Occupies a Highly Restricted Region Within Intron-1 of Snca.
To investigate whether GATA-1 directly activates Snca transcription, we examined the distribution of conserved GATA motifs at and surrounding the murine Snca locus (Fig. 4A). Ten conserved GATA motifs exist in the Snca locus (Fig. 4B). Each of these motifs would bind GATA-1 with high affinity in vitro, based on the established DNA binding specificity of GATA-1 (24, 25). However, high affinity GATA motifs are abundantly distributed throughout genomes, and the mere presence of a conserved GATA motif does not imply functional significance (26). Functional insights can be derived from evaluating evolutionary conservation of GATA motifs (Fig. 4), but even a conserved motif does not equate to a functional motif (23, 26–30).
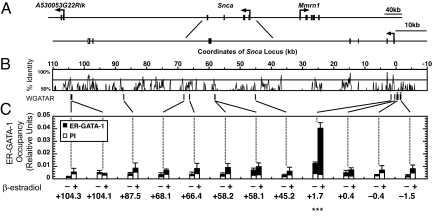
Fig. 4.
GATA-1 occupies a highly restricted region within intron-1 of Snca. (A) The organization of the murine Snca locus with respect to neighboring genes on chromosome 6 is shown at the top. (B) VISTA plot (49) of a ≈100-kb region of the Snca locus showing percentage identity of the human and mouse sequences. Ten GATA motifs in the Snca locus are evolutionary conserved between mice and humans (WGATAR, indicated by vertical lines). Coordinates are based on the predicted Snca transcription start site, which was designated as 1. Although the mouse 2.0-kb Snca promoter region does not contain conserved GATA sites, it contains three and four nonconserved GATA motifs at −0.4 and −1.5 kb of the mouse Snca promoter region, respectively (indicated by vertical lines with *). (C) Analysis of the 10 conserved and the two nonconserved GATA motifs by ChlP revealed that ER-GATA-1 occupied a single, highly restricted region within intron-1 of Snca (indicted by ***). The bar graphs depict relative ER-GATA-1 occupancy in G1E-ER-GATA-1 cells at each of the 12 sites (mean ± standard error, at least three independent experiments) in untreated and 1 μM β-estradiol-treated (24 h) G1E-ER-GATA-1 cells measured by ChIP analysis. No GATA-1 occupancy was detected at the other nine conserved and the two nonconserved sites. Preimmune serum (PI) was used as a control.
The Snca gene consists of six exons (Fig. 4A) (31) with the translation start codon ATG encoded by exon-2. Intron-1 is 1,097 bp in size (31). To determine whether the GATA motifs in Snca were occupied by GATA-1, we conducted quantitative chromatin immunoprecipitation analysis (ChIP) in G1E-ER-GATA-1 cells. Analysis of 10 regions spanning all of the predicted and evolutionary conserved GATA motifs revealed that ER-GATA-1 occupied a single, highly restricted region within intron-1 of Snca (Fig. 4C). Although the mouse 2.0-kb Snca promoter region does not contain conserved GATA sites, it contains three and four nonconserved GATA motifs at −0.4 kb and −1.5 kb, respectively (Fig. 4C). No significant occupancy was detected at these sites. Thus, GATA-1 selects exquisitely among the 10 conserved GATA motifs within the Snca locus, which strongly suggests that GATA-1 activates Snca transcription via interaction with the intron-1 site.
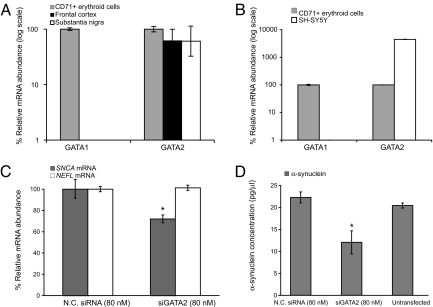
GATA-2 Is Abundantly Expressed in Dopamine Cells and Brain Regions Affected by PD, Occupies Intron-1 of Snca, and Regulates Expression of Endogenous Neuronal α-Synuclein.
The GATA family members GATA-1 and GATA-2 have overlapping activities in the control of embryonic erythropoiesis (33). In the absence of GATA-1, GATA-2 is up-regulated during erythropoiesis but does not promote erythropoiesis (34). Whereas GATA-1 is not expressed in neurons, GATA-2 is critical in neuronal development, particularly in cell fate specification of catecholaminergic sympathetic neurons (35, 36). In knockout mice, neurogenesis is severely impaired (37). In Caenorhabditis elegans, the GATA homolog elt-1 has a key role in regulating mature differentiated neurons in the locomotor circuit (38). To determine whether GATA-2 may be transcribed in human substantia nigra and cortex, two brain regions preferentially affected by PD, we performed quantitative PCR in human postmortem brain (Fig. 5A). GATA2 mRNA levels were high in these regions (61% and 62%, respectively, of its abundance in the calibrator CD71+ erythroblasts). This was confirmed at the level of GATA-2 protein expression (data not shown). In dopaminergic SH-SY5Y neuroblastoma cells, GATA2 mRNA abundance was exceedingly high (Fig. 5B; 4,500% of the abundance in the calibrator). As expected, GATA-1 was highly expressed in erythroid cells but undetectable in brain and neuroblastoma cells (Fig. 5 A and B).
Fig. 5.
Silencing of endogenous neuronal GATA-2 represses the expression of SNCA mRNA and α-synuclein protein in dopaminergic cells. (A) GATA2 mRNA is highly expressed in postmortem substantia nigra and superior frontal cortex (total n = 9; 61% and 62%, respectively, of its abundance in the calibrator CD71+ erythroblasts). (B) In dopaminergic SH-SY5Y neuroblastoma cells, GATA2 mRNA abundance was exceedingly high (4,500% of the abundance in the calibrator). GATA-1 was highly expressed in erythroid cells but undetectable in human brain and neuroblastoma cells (A and B). Note the log scales. (C and D) Silencing of neuronal GATA-2 induced a decrease in neuronal expression of both SNCA mRNA and α-synuclein protein. (C) Silencing of neuronal GATA-2 induced a 28% reduction in relative SNCA mRNA abundance compared with cells transfected with negative control siRNA (P = 0.008). Transcript levels of the neuronal control gene neurofilament light polypeptide 68 kDa (NEFL) were unaltered. (D) Silencing of neuronal GATA-2 induced a 46% reduction in α-synuclein protein concentration compared with cells transfected with negative control siRNA by ELISA (P = 0.01; mean and standard deviation of three independent experiments).
As GATA-2 precedes GATA-1 at certain chromatin sites during hematopoiesis (26), we asked whether GATA-2 occupies the same conserved GATA-binding motif in intron-1 of the Snca gene in the absence of GATA-1. We measured endogenous GATA-2 occupancy in uninduced G1E-ER-GATA-1 cells. In the absence of GATA-1, endogenous GATA-2 specifically occupied the conserved GATA-binding motif in intron-1 of the Snca locus (Fig. S3). Analogous to our findings for GATA-1, none of the other nine conserved GATA motifs in the SNCA locus and neither of the two unconserved GATA motifs in the Snca promoter region were occupied by GATA-2.
To examine the mechanistic role of neuronal GATA-2 on SNCA expression, we knocked down endogenous GATA-2 using GATA2 small interfering RNA (siRNA) in dopaminergic SH-SY5Y neuroblastoma cells (Fig. 5 and Fig. S4). Quantitative PCR showed a dose-dependent reduction of GATA2 mRNA abundance after transfection with 1–160 nM GATA2 siRNA (Fig. S4) with maximal silencing achieved at 160 nM GATA2 siRNA. Transfection with 80 nM GATA2 siRNA reliably knocked down GATA2 mRNA abundance to 40% of the abundance in cells transfected with negative control siRNA (Fig. S4). Similar results were obtained when GAPDH instead of the ribosomal gene RPL13 was used to control for RNA loading and when the experiment was repeated at 160 nM (data not shown). Consistently, GATA-2 protein levels, determined by Western blot using a well characterized, specific anti-GATA-2 antibody were substantially reduced after transfection with 80 nM GATA2 siRNA (Fig. S4) or 160 nM GATA2 siRNA (data not shown). GATA-2 levels in cells transfected with 80 nM negative control siRNA (Fig. S4) or 160 nM negative control siRNA (data not shown) were unchanged compared with untransfected cells. Silencing of neuronal GATA-2 repressed neuronal expression of both SNCA mRNA and α-synuclein protein with 28% reduction in relative SNCA mRNA abundance (Fig. 5C; P = 0.008) and 46% reduction in α-synuclein concentration compared with cells transfected with negative control siRNA (Fig. 5D; P = 0.01). Transcript levels of the neuronal marker gene neurofilament light polypeptide 68 kDa (NEFL) were unaffected by silencing of neuronal GATA-2.
Collectively, the specific occupancy of the conserved GATA-binding motif in intron-1 of the Snca gene by GATA-2, the repression of SNCA expression resulting from silencing of GATA-2 in dopamine cells, and the preferential expression of GATA-2 in dopamine cells and postmortem substantia nigra, suggest that in dopaminergic cells relevant to PD, SNCA expression is regulated by GATA-2 via occupancy at the intron-1 site.
Discussion
We have used genome-wide expression analysis to uncover a block of transcripts nonrandomly correlated with SNCA expression. By stably linking expression of SNCA to a known GATA-1 target gene in situ and genetic complementation analysis we identified a transcription factor of SNCA, the gene central to the pathobiology of PD. This approach allows hypotheses on transcriptional regulators to be generated and tested, not in vitro or in animal models (39), but in living humans.
Integrating Blood Expression and Genetic Complementation Analysis.
The identification of bona fide transcription factors of target genes of interest has been curtailed by the limitations of in vitro transcriptional assays. Vast amounts of information on transcriptional regulation are captured in rapidly expanding human datasets, but the underlying mechanism cannot be established (39). Here, we developed a method based on simple Spearman's rank correlation that uses the patterns of gene expression in human blood to identify a candidate transcription factor whose mechanistic role is confirmed by genetic analyses in cultured cells.
Trans-Acting Mechanism Causing Variation in SNCA mRNA Copy Numbers.
The transcription factors directly controlling SNCA expression are unknown. They may provide important insight into PD pathobiology and for developing therapeutic strategies designed to lower α-synuclein production. We found a critical link between GATA factors and trans-activation of SNCA expression. Gene expression analysis across 113 human blood samples from three independent populations on three distinct assay platforms showed that variation in SNCA transcript levels was tightly correlated with variation in the known GATA-1 target gene ALAS2 (Fig. 2). One specific site—in intron-1—of 10 conserved GATA-binding motifs in SNCA was directly occupied by GATA-1 (Fig. 4). The formation of a DNA-chromatin complex containing induced ER-GATA-1 and Snca intron-1 and the responsiveness of SNCA expression to GATA-1 was established by genetic complementation analysis (Fig. 3). In the absence of GATA-1, endogenous GATA-2, naturally expressed in human dopamine-producing cells and in substantia nigra, directly and specifically occupied the same GATA binding motif in intron-1 (Fig. S3). GATA-1 induced a 62-fold increase in Snca mRNA and a 6.9-fold increase in α-synuclein (Fig. 3). Silencing of endogenous neuronal GATA-2 induced a highly significant 28% decrease in SNCA mRNA and 46% decrease in α-synuclein in dopaminergic cells (Fig. 5).
Whereas classical studies on transcriptional mechanisms focused on analyzing how transcription factors function through promoter regions of genes, it is now well appreciated that common modes of transcriptional control require transcription factor interactions with far upstream, downstream, and intronic sequences (26). Strong precedence exists that such complexes encounter the promoter region through the formation of higher-order chromatin loops (40). For example, at the Gata2 locus, GATA-2 confers an important activating function through an intron 9,500 bp downstream of the promoter (28), and chromosome conformation capture analysis indicates that this region resides in close proximity to a −77-kb far upstream regulatory element (28). Examples of GATA-1 function through introns include intron 8 of the ALAS2 gene (20), intron 7 of the Tac2 gene (41), intron 1 of the Smad7 gene (42), and intron 3 of the Wilms Tumor 1 gene (43).
Despite these uniquely important insights, our understanding of the transcriptional regulation of SNCA expression is incomplete due in part to the difficulty of demonstrating GATA-2-induced SNCA activation in mammalian brain (loss of GATA-2 leads to embryonic lethality in knockout mice (33)). In addition, other factors may modulate the transcriptional regulation of SNCA in concert with GATA-2 (44).
α-Synuclein, Erythroid Cells, and Heme Synthesis.
Our study also reveals a clue into the elusive normal biological role of α-synuclein. We demonstrated the dramatic expression of SNCA during terminal steps of erythroid differentiation (Fig. 1). In human and mouse erythroid cells abundant expression of SNCA mRNA and protein was confirmed by microarray, quantitative PCR, Western blot analysis, ELISA, and immunohistochemistry (Figs. 1 and 2). These results, together with recent reports (45, 46), clearly indicate a role for SNCA during important steps of erythropoiesis. Unexpectedly, within erythroid cells, SNCA was strongly coexpressed and coinduced with critical enzymes of heme metabolism, ALAS2, FECH, and BLVRB (Fig. 2). Heme, an iron molecule coordinated within a tetrapyrrole, has unique properties that allow it to function both as an electron carrier and oxygen transporter. Almost all iron in the adult human body is bound to heme (47). ALAS2, a previously identified GATA-1 target gene (19, 20) catalyzes a rate-limiting step in heme production. FECH (here identified as GATA-1-activated gene) catalyzes the final step of heme biosynthesis, which inserts iron into protoporphyrin IX. The transcriptional coregulation may be fine-tuned at the level of protein translation through iron-responsive elements predicted in the 5′ untranslated region of ALAS2 mRNA and SNCA mRNA (48). Collectively, these observations lead us to hypothesize that heme metabolism may be a missing link between two unreconciled, alleged culprits of PD, α-synuclein aggregation (4) and iron deposition (6).
In summary, by linking the expression of SNCA and a known GATA-1 target gene in humans and by correlating the specific occupancy of a highly conserved GATA binding motif in intron-1 with genetic complementation analysis, we established the first mechanistic link between the GATA family of transcription factors and SNCA expression. Elucidating the precise regulation of SNCA expression will be critical for understanding PD pathobiology and for devising novel therapeutics designed to lower α-synuclein burden in patients with PD.
Materials and Methods
A detailed description of biospecimens, GEO accession numbers, bioinformatics, ChIP, quantitative PCR, Northern and Western blot analysis, ELISA, immunohistochemistry, and siRNA methods can be found in SI Methods.
Supplementary Material
Acknowledgments.
We thank Drs. Dennis Selkoe, Mel Feany, and Omar El-Agnaf for insightful comments. This work was supported by Paul B. Beeson K08AG024816 from the NIA and the American Federation for Aging Research (C.R.S.), Dr. George Cotzias Award of the American Parkinson Disease Association (C.R.S.), the Michael J. Fox Foundation (C.R.S. and M.G.S.), the M. E. and M. O. Hoffman Foundation (C.R.S.), American, Lebanese, and Syrian Associated Charities (P.A.N.), National Institutes of Health Grants R01 DK68634 and DK50107 (E.H.B.), P50 NS38375 (M.G.S.), and R01 CA084214 (P.A.N.).
Footnotes
Conflict of interest statement: C.R.S., E.H.B., and M.G.S. are listed as coinventors on a United States patent application related to the development of therapeutics for Parkinson's disease.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802437105/DCSupplemental.
References
- 1.Singleton AB, et al. α-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 2.Maraganore DM, et al. Collaborative analysis of α-synuclein gene promoter variability and Parkinson disease. J Am Med Assoc. 2006;296:661–670. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- 3.Farrer M, et al. Comparison of kindreds with parkinsonism and α-synuclein genomic multiplications. Ann Neurol. 2004;55:174–179. doi: 10.1002/ana.10846. [DOI] [PubMed] [Google Scholar]
- 4.Scherzer CR, Feany MB. Yeast genetics targets lipids in Parkinson's disease. Trends Genet. 2004;20:273–277. doi: 10.1016/j.tig.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Klein C, Schlossmacher MG. The genetics of Parkinson disease: Implications for neurological care. Nat Clin Pract Neurol. 2006;2:136–146. doi: 10.1038/ncpneuro0126. [DOI] [PubMed] [Google Scholar]
- 6.Gotz ME, Double K, Gerlach M, Youdim MB, Riederer P. The relevance of iron in the pathogenesis of Parkinson's disease. Ann NY Acad Sci. 2004;1012:193–208. doi: 10.1196/annals.1306.017. [DOI] [PubMed] [Google Scholar]
- 7.Miller DW, et al. α-Synuclein in blood and brain from familial Parkinson disease with SNCA locus triplication. Neurology. 2004;62:1835–1838. doi: 10.1212/01.wnl.0000127517.33208.f4. [DOI] [PubMed] [Google Scholar]
- 8.Chiba-Falek O, Nussbaum RL. Effect of allelic variation at the NACP-Rep1 repeat upstream of the α-synuclein gene (SNCA) on transcription in a cell culture luciferase reporter system. Hum Mol Genet. 2001;10:3101–3109. doi: 10.1093/hmg/10.26.3101. [DOI] [PubMed] [Google Scholar]
- 9.Xu J, et al. Dopamine-dependent neurotoxicity of α-synuclein: A mechanism for selective neurodegeneration in Parkinson disease. Nat Med. 2002;8:600–606. doi: 10.1038/nm0602-600. [DOI] [PubMed] [Google Scholar]
- 10.Pastinen T, Hudson TJ. Cis-acting regulatory variation in the human genome. Science. 2004;306:647–650. doi: 10.1126/science.1101659. [DOI] [PubMed] [Google Scholar]
- 11.Scherzer CR, et al. Molecular markers of early Parkinson's disease based on gene expression in blood. Proc Natl Acad Sci USA. 2007;104:955–960. doi: 10.1073/pnas.0610204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maroteaux L, Campanelli JT, Scheller RH. Synuclein: A neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J Neurosci. 1988;8:2804–2815. doi: 10.1523/JNEUROSCI.08-08-02804.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Agnaf OM, et al. Detection of oligomeric forms of α-synuclein protein in human plasma as a potential biomarker for Parkinson's disease. FASEB J. 2006;20:419–425. doi: 10.1096/fj.03-1449com. [DOI] [PubMed] [Google Scholar]
- 14.Michell AW, Luheshi LM, Barker RA. Skin and platelet α-synuclein as peripheral biomarkers of Parkinson's disease. Neurosci Lett. 2005;381:294–298. doi: 10.1016/j.neulet.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 15.Shintani N, et al. Expression and extracellular release of transferrin receptors during peripheral erythroid progenitor cell differentiation in liquid culture. Blood. 1994;83:1209–1215. [PubMed] [Google Scholar]
- 16.Mollenhauer B, et al. Direct quantification of CSF α-synuclein by ELISA and first cross-sectional study in patients with neurodegeneration. Exp Neurol. 2008 doi: 10.1016/j.expneurol.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Borovecki F, et al. Genome-wide expression profiling of human blood reveals biomarkers for Huntington's disease. Proc Natl Acad Sci USA. 2005;102:11023–11028. doi: 10.1073/pnas.0504921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitney AR, et al. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci USA. 2003;100:1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Surinya KH, Cox TC, May BK. Transcriptional regulation of the human erythroid 5-aminolevulinate synthase gene. Identification of promoter elements and role of regulatory proteins. J Biol Chem. 1997;272:26585–26594. doi: 10.1074/jbc.272.42.26585. [DOI] [PubMed] [Google Scholar]
- 20.Pal S, et al. Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc Natl Acad Sci USA. 2004;101:980–985. doi: 10.1073/pnas.0307612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taketani S, Mohri T, Hioki K, Tokunaga R, Kohno H. Structure and transcriptional regulation of the mouse ferrochelatase gene. Gene. 1999;227:117–124. doi: 10.1016/s0378-1119(99)00011-6. [DOI] [PubMed] [Google Scholar]
- 22.Welch JJ, et al. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- 23.Johnson KD, et al. Friend of GATA-1-independent transcriptional repression: A novel mode of GATA-1 function. Blood. 2007;109:5230–5233. doi: 10.1182/blood-2007-02-072983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merika M, Orkin SH. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ko LJ, Engel JD. DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bresnick EH, Johnson KD, Kim SI, Im H. Establishment and regulation of chromatin domains: Mechanistic insights from studies of hemoglobin synthesis. Prog Nucleic Acid Res Mol Biol. 2006;81:435–471. doi: 10.1016/S0079-6603(06)81011-1. [DOI] [PubMed] [Google Scholar]
- 27.Johnson KD, et al. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc Natl Acad Sci USA. 2002;99:11760–11765. doi: 10.1073/pnas.192285999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grass JA, et al. Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol Cell Biol. 2006;26:7056–7067. doi: 10.1128/MCB.01033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martowicz ML, Grass JA, Boyer ME, Guend H, Bresnick EH. Dynamic GATA factor interplay at a multicomponent regulatory region of the GATA-2 locus. J Biol Chem. 2005;280:1724–1732. doi: 10.1074/jbc.M406038200. [DOI] [PubMed] [Google Scholar]
- 30.Grass JA, et al. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci USA. 2003;100:8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Touchman JW, et al. Human and mouse α-synuclein genes: Comparative genomic sequence analysis and identification of a novel gene regulatory element. Genome Res. 2001;11:78–86. doi: 10.1101/gr.165801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai FY, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 33.Fujiwara Y, Chang AN, Williams AM, Orkin SH. Functional overlap of GATA-1 and GATA-2 in primitive hematopoietic development. Blood. 2004;103:583–585. doi: 10.1182/blood-2003-08-2870. [DOI] [PubMed] [Google Scholar]
- 34.Weiss MJ, Keller G, Orkin SH. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1 embryonic stem cells. Genes Dev. 1994;8:1184–1197. doi: 10.1101/gad.8.10.1184. [DOI] [PubMed] [Google Scholar]
- 35.Tsarovina K, et al. Essential role of Gata transcription factors in sympathetic neuron development. Development. 2004;131:4775–4786. doi: 10.1242/dev.01370. [DOI] [PubMed] [Google Scholar]
- 36.Bilodeau ML, Boulineau T, Greulich JD, Hullinger RL, Andrisani OM. Differential expression of sympathoadrenal lineage-determining genes and phenotypic markers in cultured primary neural crest cells. In Vitro Cell Dev Biol Anim. 2001;37:185–192. doi: 10.1290/1071-2690(2001)037<0185:DEOSLD>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 37.Nardelli J, Thiesson D, Fujiwara Y, Tsai FY, Orkin SH. Expression and genetic interaction of transcription factors GATA-2 and GATA-3 during development of the mouse central nervous system. Dev Biol. 1999;210:305–321. doi: 10.1006/dbio.1999.9278. [DOI] [PubMed] [Google Scholar]
- 38.Smith JA, McGarr P, Gilleard JS. The Caenorhabditis elegans GATA factor elt-1 is essential for differentiation and maintenance of hypodermal seam cells and for normal locomotion. J Cell Sci. 2005;118:5709–5719. doi: 10.1242/jcs.02678. [DOI] [PubMed] [Google Scholar]
- 39.Lamb J, et al. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell. 2003;114:323–334. doi: 10.1016/s0092-8674(03)00570-1. [DOI] [PubMed] [Google Scholar]
- 40.Dekker J. Gene regulation in the third dimension. Science. 2008;319:1793–1794. doi: 10.1126/science.1152850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pal S, et al. Neurokinin-B transcription in erythroid cells: Direct activation by the hematopoietic transcription factor GATA-1. J Biol Chem. 2004;279:31348–31356. doi: 10.1074/jbc.M403475200. [DOI] [PubMed] [Google Scholar]
- 42.Benchabane H, Wrana JL. GATA- and Smad1-dependent enhancers in the Smad7 gene differentially interpret bone morphogenetic protein concentrations. Mol Cell Biol. 2003;23:6646–6661. doi: 10.1128/MCB.23.18.6646-6661.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Xing G, Fraizer GC, Saunders GF. Transactivation of an intronic hematopoietic-specific enhancer of the human Wilms' tumor 1 gene by GATA-1 and c-Myb. J Biol Chem. 1997;272:29272–29280. doi: 10.1074/jbc.272.46.29272. [DOI] [PubMed] [Google Scholar]
- 44.Chiba-Falek O, Kowalak JA, Smulson ME, Nussbaum RL. Regulation of α-synuclein expression by poly(ADP ribose) polymerase-1 (PARP-1) binding to the NACP-Rep1 polymorphic site upstream of the SNCA gene. Am J Hum Genet. 2005;76:478–492. doi: 10.1086/428655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goh SH, et al. The human reticulocyte transcriptome. Physiol Genomics. 2007;30:172–178. doi: 10.1152/physiolgenomics.00247.2006. [DOI] [PubMed] [Google Scholar]
- 46.Nakai M, et al. Expression of α-synuclein, a presynaptic protein implicated in Parkinson's disease, in erythropoietic lineage. Biochem Biophys Res Commun. 2007;358:104–110. doi: 10.1016/j.bbrc.2007.04.108. [DOI] [PubMed] [Google Scholar]
- 47.Ajioka RS, Phillips JD, Kushner JP. Biosynthesis of heme in mammals. Biochim Biophys Acta. 2006;1763:723–736. doi: 10.1016/j.bbamcr.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 48.Friedlich AL, Tanzi RE, Rogers JT. The 5′-untranslated region of Parkinson's disease α-synuclein messengerRNA contains a predicted iron responsive element. Mol Psychiatry. 2007;12:222–223. doi: 10.1038/sj.mp.4001937. [DOI] [PubMed] [Google Scholar]
- 49.Mayor C, et al. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.