Abstract
Huntington's disease is a dominant autosomal neurodegenerative disorder caused by an expansion of polyglutamines in the huntingtin (Htt) protein, whose cellular function remains controversial. To gain insight into Htt function, we purified epitope-tagged Htt and identified Argonaute as associated proteins. Colocalization studies demonstrated Htt and Ago2 to be present in P bodies, and depletion of Htt showed compromised RNA-mediated gene silencing. Mouse striatal cells expressing mutant Htt showed fewer P bodies and reduced reporter gene silencing activity compared with wild-type counterparts. These data suggest that the previously reported transcriptional deregulation in HD may be attributed in part to mutant Htt's role in post-transcriptional processes.
Keywords: microRNA, poly-glutamine, RNA interference, post-transcriptional gene silencing, neuronal RNA granule
Huntington's disease (HD) is caused by an expansion of the polymorphic stretch of uninterrupted CAG trinucleotide repeats in exon 1 of the IT15 gene, resulting in a long tract of polyglutamine (polyQ) in the N terminus of the HD protein huntingtin (Htt) (1). The human Htt protein contains 3,144 amino acids, and the polyQ stretch starts at the 18th amino acid, followed by a polyproline (polyP) sequence; the remaining portion of the protein is likely to be rich in α-helices, many of which compose HEAT repeats (2). The length of the glutamine repeats in the unaffected population varies from 6 to 35; HD is caused by expansion to 36 or more repeats (3, 4). HD is a member of a group of at least nine diseases caused by CAG repeat expansions that includes spinocerebellar ataxias (SCAs) 1–3, 6, 7, and 17, spinobulbar muscular atrophy, and dentatorubral-pallidoluysian atrophy. The protein that is subject to polyQ expansion in each disease appears to be unrelated, and despite their widespread expression in the brain and other tissues, mutation in each protein leads to distinct characteristic neurodegeneration patterns.
Htt is expressed in a variety of tissues both within and outside of the nervous system. Many proteins have been demonstrated to interact with WT Htt and/or mutant Htt, and their functions support some of the mechanisms perturbed in HD, which include transcription, signaling, trafficking/endocytosis, and metabolism/mitochondrial functions (5–10). However, Htt's precise function has proven elusive, and our poor understanding of its function remains a limiting factor to the development of successful therapeutics (11). Proteolytic processing of WT and mutant Htt has been investigated extensively and appears to play a critical role in disease pathogenesis. Caspase-6 cleavage at amino acid 586 in mouse Htt and the release of the N-terminal Htt fragment are required for neuronal dysfunction and degeneration in an HD mouse model (12), but the basis for the need for this cleavage is unclear.
We set out to search for binding proteins to elucidate the normal function of Htt. We chose a biochemical approach to purify proteins associated with WT and mutant Htt and identified Argonaute (Ago2) as a copurifying protein. Colocalization studies demonstrated the presence of Htt and Ago2 in processing (P) bodies, cytoplasmic foci that contain translationally repressed mRNAs with bound proteins. P bodies have also been implicated in small RNA-mediated gene silencing. Our data suggest that normal Htt may be a component of a P body and functions in post-transcriptional repression pathways.
Results
Argonaute 2 Copurifies with Huntingtin.
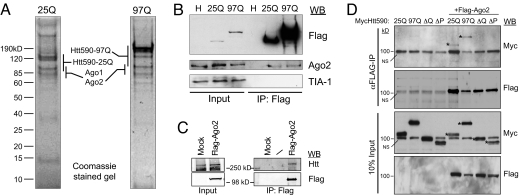
To critically assess Htt protein interactions, we have established a discovery-based Htt purification scheme aimed at identifying novel interactions and uncovering any obvious difference between proteins that copurify with WT and mutant Htt. HeLa cells, which express endogenous Htt, were used to establish stable cell lines that express Flag-tagged Htt N-terminal 590 aa with 25 or 97 glutamines (Flag-Htt590–25Q or Flag-Htt590–97Q). The cytoplasmic S100 fraction prepared from these cells was subjected to immunopurification with Flag-M2 agarose beads followed by peptide elution (13). Although HeLa cells are not of neuronal origin, they provide access to large amounts of material, which significantly aids in the identification of interacting proteins. Our Flag antibody purifications from the nuclear fraction (data not shown) repeatedly identified the previously reported Htt-interactors, CA150 and Tpr (14, 15). Based on Htt's abundance in the S100 fraction, we chose this fraction for the input to our purification and subsequent investigations. The peptide-eluted fraction was separated by SDS/PAGE and stained with Coomassie blue. This revealed a strong band at the appropriate molecular weight for Flag-Htt590 and a small number of nonstoichiometric copurifying proteins (Fig. 1A). The gel lanes were sectioned and subjected to in-gel tryptic digestion followed by ESI-MS/MS based peptide sequencing. Many of the distinct, abundant, and reproducibly copurifying polypeptides corresponded to the Argonaute (Ago) proteins; some were distinct to Ago1 and Ago2 isoforms [supporting information (SI) Fig. S1 A and B]. The Ago family of proteins is essential for small RNA-mediated gene silencing pathways (16). In repeated purifications, we failed to detect distinct Ago3 or Ago4 peptides. Previous Flag-Ago1 and -Ago2 purifications have reported several common copurifying proteins including Dicer (17). Repeated Flag-Htt purifications did not identify Dicer or any other factor known to be involved in si/miRNA biogenesis; thus we speculate that Htt may be involved in the effector stage of post-transcriptional gene silencing (PTGS). We used our cell lines to perform Flag immunoprecipitations followed by immunoblotting with Ago2 to confirm the data from mass spectrometry (Fig. 1B). We reproducibly observed coprecipitation of comparable amounts of endogenous Ago2 with Htt590–25Q and -97Q. We do not know whether this reflects less binding of Ago2 to Htt590–97Q or Ago2 is limiting under these conditions. We also recovered fewer peptides for Ago2 from cells expressing Htt590–97Q (Fig. S1A), suggesting that it may associate less efficiently with Ago2.
Fig. 1.
Isolation of Flag-tagged WT (25Q) and polyQ-expanded (97Q) Htt protein complexes from HeLa cell cytoplasmic S100 fraction. (A) Eluates from immunopurified Flag-Htt590 resolved by 10% SDS/PAGE and visualized by Coomassie blue staining. Mass spectrometric protein identifications are listed alongside the gel regions where the proteins were identified. (B) Verification of Ago2 as an Htt590 interactor. Immunoblot analysis of Flag immunoprecipitates from the indicated cell lysates probed with the indicated antibodies. (C) Cytoplasmic extract from HEK293T cells stably expressing Flag-Ago2 or Flag-vector (Mock) were immunoprecipitated with α-Flag M2 antibody and probed with α-Htt (HDB4E10, Abcam) or α-Flag antibody. (D) HeLa cells were cotransfected with Myc-Htt590–25Q, -97Q or deletion mutant (ΔQ or ΔP) and Flag-Ago2. Ago2 was immunoprecipitated with α-Flag antibody and the presence of Htt was determined by immunoblotting with α-Myc antibody. *, WT and mutant Myc-Htt590. NS: a nonspecific background band migrating at 100 kDa. Fig. S2D shows clearer separation of Myc-Htt590-ΔQ from the background band.
Purification of proteins associated with Ago2 from HEK293T cells stably expressing Flag-Ago2 was carried out separately and 40 unique peptides corresponding to the endogenous Htt was identified (data not shown). This interaction was confirmed by coprecipitation and immunoblotting (Fig. 1C). To further examine the Htt-Ago2 interaction, HeLa cells were transfected with Htt480–17Q and Flag-Ago2, lysed, and fractionated by glycerol gradient sedimentation. Fractions that contained both Htt480 and Ago2 were pooled and analyzed (Fig. S2A). The result indicated that Ago2 copurifies with Htt480 protein when overexpressed in cells. Furthermore, coimmunoprecipitation assays using HeLa cell lysate demonstrated that endogenous Htt and Ago2 proteins interact (Fig. S2 B and C).
We next determined the N-terminal Htt sequences required for the association with Ago2. HeLa cells were cotransfected with Flag-Ago2 and Myc-Htt590–25Q, -97Q, or with a construct deleted of polyQ or polyP sequence. Immunoprecipitation of cell lyates with α-Flag antibody demonstrated that the Htt deletion mutants did not coprecipitate with Ago2, suggesting that polyQ and polyP sequences are involved in the association of Htt with Ago2 (Fig. 1D, Fig. S2D).
Huntingtin Is Present in P Bodies.
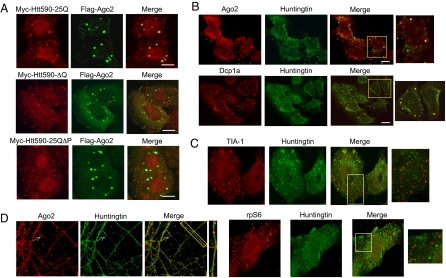
To further characterize the association of Htt with Ago2, we investigated the localization of transfected Htt and Ago2 by confocal microscopy. HeLa cells were cotransfected with Myc-Htt590–25Q and Flag-Ago2 and examined by indirect immunofluorescence (Fig. 2A Upper). The cells expressing these proteins showed distinct foci demonstrating strong colocalization of the two proteins. Interestingly, Myc-Htt590 deleted of polyQ or polyP sequence failed to colocalize with Flag-Ago2 (Fig. 2A). This finding is consistent with the coimmunoprecipitation data (Fig. 1D) and suggests that these sequences are required for targeting of Htt590 to Ago2-containing foci.
Fig. 2.
Htt associates with Ago2 and localizes to P bodies. (A) HeLa cells were transfected with indicated constructs as in Fig. 1D, fixed, and probed with rabbit α-Myc and mouse α-Flag primary antibodies. (Scale bar, 10 μm.) (B) U2OS cells were probed for endogenous Ago2, Dcp1a and Htt. (Scale bar, 20 μm.) Of 100 cells examined, 65% of foci showed Ago2 and Htt colocalization. Of 200 cells examined, 85% of foci showed Dcp1a and Htt colocalization. (C) U2OS cells were probed for endogenous TIA-1, rpS6, and Htt. Note that TIA-1 and rpS6 did not colocalize with endogenous Htt. (D) Primary rat hippocampal neurons were probed for endogenous Ago2 and Htt.
Ago proteins have been shown to localize to discrete cytoplasmic foci known as processing (P) bodies (18, 19). Proteins involved in small RNA-mediated gene silencing, translational repression, mRNA surveillance, and mRNA degradation accumulate with their bound stalled mRNAs at P bodies, where their destiny is determined (20, 21). Our colocalization data suggest that Htt may be present in P bodies. We next examined endogenous Htt and Ago2 localization in U2OS cells and found a portion of Htt to colocalize with Ago2 in discrete foci indicative of P bodies (Fig. 2B). Further, Htt colocalized with an mRNA decapping enzyme and P body marker Dcp1a (22). Similar results were obtained in neuronal N2A cells (Fig. S3) and primary rat hippocampal neurons (Fig. 2D). Interestingly, not all of the Htt foci colocalized with Ago2 and Dcp1a; this may be due to a diversity of different P body types (20, 21) and P body-like structures containing Htt. We also showed that Htt did not colocalize with TIA-1 (23) and ribosomal protein S6, two markers more likely to be associated with stress granules than P bodies. (Fig. 2C), and TIA-1 did not coprecipitate with Flag-Htt590 (Fig. 1B). These results indicate that endogenous Htt is present specifically in P bodies and thus may play a role in the post-transcriptional control of mRNA.
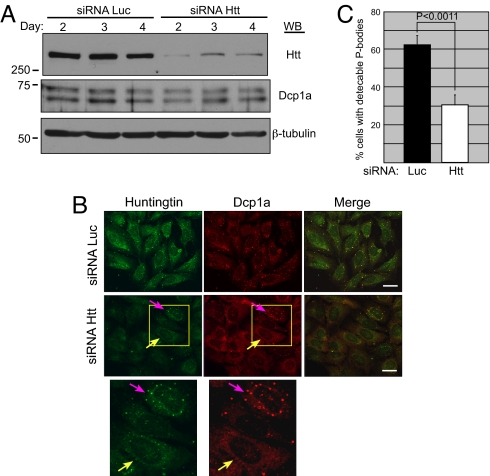
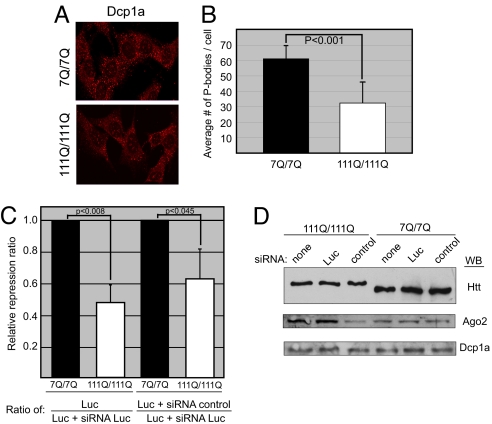
We next investigated the consequences of Htt reduction on P bodies. U2OS cells were transfected with siRNA to Htt or control luciferase (Luc). The knockdown of Htt was efficient, with no effect on total Dcp1a or β-tubulin protein levels (Fig. 3A). Indirect immunofluorescence of cells transfected with Htt siRNA demonstrated fewer P bodies than in cells transfected with Luc siRNA (Fig. 3B). Cells with Htt puncta clearly demonstrated presence of the Dcp1a P body marker (pink arrow), whereas adjacent cells with reduced Htt levels showed diffuse Dcp1a staining (yellow arrow). To quantify the effect of Htt siRNA or Luc siRNA on P bodies, the percentage of cells possessing detectable P bodies was determined. We found a 2-fold reduction in the number of P body-positive cells transfected with Htt siRNA compared with Luc siRNA (Fig. 3C). Our results echo previous findings in which depletion of P body resident proteins such as TNRC6A or LSm1–7 resulted in P body loss (20). These data led us to conclude that WT Htt plays a role in the appearance of these macromolecular assemblies and influences their stability.
Fig. 3.
Htt plays a role in P body formation/maintenance. (A) U2OS cells were transfected with siRNA to Luc or Htt and the effect on Htt, Dcp1a, and β-tubulin protein was examined by immunoblotting. Quantification of Htt protein levels showed >7-fold reduction following the siRNA treatment. (B) U2OS cells transfected with Luc or Htt siRNA were examined by indirect immunofluorescence with rabbit α-Dcp1a and mouse α-Htt antibodies. (Scale bar, 30 μm.) (C) The result of three independent Luc siRNA and Htt siRNA transfections was quantified by determining the percentage of cells with detectable P-bodies after counting >200 cells per experiment.
Huntingtin Plays a Role in Small RNA-Mediated Processes.
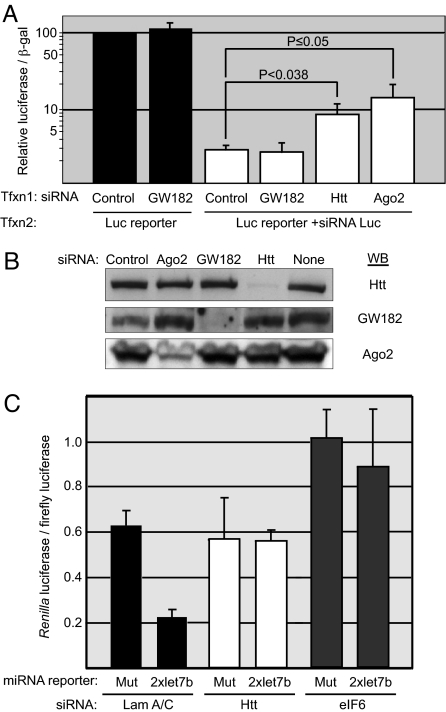
To investigate Htt's potential role in Ago-dependent siRNA gene silencing pathways, we used a Luc reporter and determined the effect of Htt knockdown on silencing of the Luc mRNA transcripts (Fig. 4A). HeLa cells were first transfected with control, GW182, Htt, or Ago2 siRNA and subsequently cotransfected with the SV40 promoter-Luc reporter plasmid and siRNA to Luc. We verified the high efficiency of knockdown by each siRNA under the same experimental conditions (Fig. 4B). Significantly, the Luc reporter activity (in the presence of Luc siRNA) in cells transfected with Htt siRNA was 3-fold greater than that of cells transfected with control siRNA. Cells transfected with Ago2 siRNA showed a 5-fold increase in reporter activity, consistent with its critical role in siRNA-mediated processes. Although depletion of GW182 has been reported to partially relieve siRNA-mediated silencing (24), we did not see an effect of GW182 depletion in our siRNA reporter assay. This may be due to the potentially redundant functions of the three GW182 paralogues present in mammalian cells. We next performed knockdown experiments of an endogenous mRNA in the presence or absence of Htt and examined the results by microscopy. We determined the effect of Htt on siRNA-mediated knockdown of host cell factor-1 (HCF-1), a transcription factor involved in cell cycle progression (25), and found that Htt is required for efficient silencing of HCF-1 in U2OS cells (Fig. S4).
Fig. 4.
Cells with reduced Htt levels show compromised RNA-induced gene silencing. (A) The effect of Htt knockdown on silencing of a Luc reporter with Luc siRNA. HeLa cells were first transfected (Tfxn1) with control, GW182, Htt, or Ago2 siRNA, and subsequently (Tfxn2) with SV40 promoter-Luc reporter plasmid, a plasmid encoding β-galactosidase, and Luc siRNA where indicated. The ratio of luciferase activity to β-galactosidase was measured and graphed on a log scale as the percentage of expression relative to the cells transfected with the control siRNA without Luc siRNA (set to 100%) (n = 16). (B) Immunoblot analysis of cell lysates from A. (C) The effect of Htt knockdown on the ability to silence a miRNA reporter by endogenous let-7b miRNA. HeLa cells were transfected with lamin A/C, Htt, or eIF6 siRNA, and a Renilla Luc reporter that contained human let-7b element or a mutant element. Normalized ratio of Renilla Luc to SV40-firefly Luc is presented. (n = 4).
To study Htt's potential contribution to miRNA pathways, we determined the effect of Htt knockdown on the ability of endogenous let-7b miRNA to silence a reporter. HeLa cells were transfected with siRNA to lamin A/C (negative control), Htt, or eIF6, and a Renilla Luc reporter that contained the human let-7b element or a mutant element (26) (Fig. 4C). Knockdown of Htt abrogated silencing by the let-7b miRNA as efficiently as knockdown of eIF6 reported (26). Collectively, these functional studies indicate that knockdown of Htt has measurable effects on RNA silencing pathways and support the idea that Htt plays a role in PTGS.
Comparison of Normal and polyQ-Expanded Huntingtin.
To investigate the potential effect of mutant Htt with an expanded polyQ on P body formation, we examined by indirect immunofluorescence immortalized mouse striatal progenitor cells expressing endogenous normal Htt (STHdhQ7/Q) and endogenous mutant Htt with 111 glutamines (STHdhQ111/Q111) (27). We found the number of P bodies in STHdhQ111/Q111 cells to be lower compared with WT cells based on probing with α-Dcp1a antibody (Fig. 5A). Quantification of the P body number in 100 cells showed that on average, STHdhQ111/Q111 cells had significantly fewer P bodies than WT cells (Fig. 5B). Furthermore, staining of primary neurons from WT or mutant HdhQ140/Q140 mice (28) with α-Dcp1a antibody and DAPI demonstrated similarly reduced number of P bodies (3-fold) in neurons of mutant HD mice (Fig. S5). In siRNA reporter assays, a strong reduction in silencing activity was consistently detected in STHdhQ111/Q111 cells compared with WT cells (Fig. 5C). The reduced number of P bodies and the decreased silencing activity in STHdhQ111/Q111 cells were not due to lower levels of Dcp1a or Ago2 protein (Fig. 5D). These results suggest that polyQ-expanded mutant Htt may interfere with P body formation/maintenance and small RNA-mediated pathways.
Fig. 5.
Mouse striatal cells expressing mutant Htt have fewer P bodies and show reduced silencing activity. (A) Indirect immunofluorescence of mouse striatal cells expressing WT Htt (STHdhQ7/Q7) or mutant Htt (STHdhQ111/Q111) probed with antibodies to Dcp1a. (B) Quantification of the number of P bodies per cell performed on 100 cells. P value (paired t test) <0.001 (C) siRNA reporter assay was performed as described for Fig. 4A. The graph compares relative repression ratios in cells expressing WT (7Q/7Q) or mutant (111Q/111Q) Htt (n = 9, Left; n = 12, Right). (D) Immunoblot analysis of cell lysates prepared for the assay shown in C.
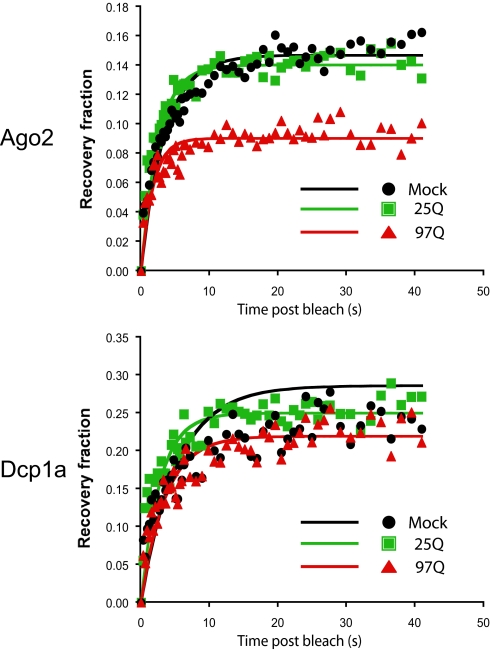
To determine whether Htt plays a role in the dynamic structure of the P body, we used fluorescence recovery after photobleaching (FRAP) analysis and investigated the effect of WT or mutant Htt on the mobility of Ago2 between the P body and the cytoplasm (Fig. 6). GFP-Ago2 was expressed in control HeLa cells or HeLa cells stably expressing Htt590–25Q or 97Q. Coexpression of RFP-Dcp1a was used to discriminate between P bodies and other Ago2-containing cytoplasmic structures. After bleaching each P body, the recovery of both Ago2 and Dcp1a was monitored and the kinetics of recovery assessed by modeling each recovery to an exponential recovery function. As reported previously, the recovery of GFP-Ago2 after bleaching was only partial, corresponding to ≈15% of the bleach depth (29). Furthermore, Ago2 recovery was unaffected by the presence of exogenous Htt (compare mock and Htt590–25Q recovery in the Ago2 graph). Interestingly, the recovery of GFP-Ago2 in the presence of mutant Htt was reduced to ≈9%. In comparison, we saw no change in the recovery of the RFP-Dcp1a to the same set of P bodies, regardless of the presence of exogenous WT or mutant Htt: the recovery of Dcp1a was ≈25% under all three conditions. In addition, when fitting the data to a simplified exponential recovery model, we saw no change in the rate of recovery of either Dcp1a or Ago2 (Table S1). These results suggest that mutant Htt does not alter the general dynamic structure of the P bodies but reduces the fraction of mobile Ago2 within each P body. Because we do not observe a similar change in a simultaneously bleached fluorophore (RFP-Dcp1a), we think the change in recovery fraction is significant, specific for Ago2, and depends on the presence of Htt590-97Q.
Fig. 6.
The fluorescence recovery of singly expressed RFP-Dcp1a or GFP-Ago2 within P bodies after photobleaching (mock, black) was compared with the fluorescence recovery of RFP-Dcp1a and GFP-Ago2 in HeLa cells expressing Htt590–25Q (green) or Htt590–97Q (red). The normalized recovery was fitted against a FRAP recovery model (solid lines). For clarity, not all points of the recovery data are plotted.
Discussion
Our comparative biochemical purifications have uncovered previously undescribed interaction partners for Htt and confirmed several previously reported. The association of Htt with Ago2 and its localization to P bodies provides evidence connecting Htt with the post-transcriptional control of gene expression and P body integrity. It is thought that P bodies are composed of a collection of subcomplexes (30). Htt can now be added to the list of proteins that interact with Agos, which include the Dcp1-Dcp2 decapping complex, GW182, and Rck/p54 (30). Recent studies have shown that P body formation is triggered as a consequence of gene silencing, but silencing can occur in the absence of microscopically visible P bodies (31, 32). Although our data do not illuminate the precise molecular role Htt plays in gene silencing, they provide compelling evidence that Htt functions as an Ago accessory factor involved in RNAi-mediated mechanisms and/or other translation repression/mRNA decay pathways associated with P bodies. Interestingly, ataxin-2, another protein subject to polyQ expansion and implicated in pathogenesis of SCA2, was reported to localize to and affect the assembly of P bodies and stress granules through an interaction with a P body component DDX6 (33). Thus, altered post-transcriptional regulation may be a common feature of polyQ expansion diseases.
Based on our immunofluorescence studies of endogenous mutant Htt in striatal cells and primary neurons, we speculate that the integrity or function of P bodies might be perturbed in HD. A recent study in yeast has reported that proteins with Q/N-rich prion-like domains contribute to the aggregation of ribonucleoproteins in P bodies (34). Thus, it is possible that normal Htt through its Q-rich domain contributes to P body assembly and that mutant Htt may affect P body formation/disassembly to alter the ability of the cell to control mRNA and protein levels. The FRAP analysis suggests that mutant Htt reduces the fraction of Ago2 that enters P bodies; this could lead to gradual changes in P body number and dynamics over time and may help to explain the long period it takes for the disease to manifest in HD patients.
The relationship between P bodies and neuronal granules is intriguing. Neuronal RNA granules are thought to mediate transport and local translational control of neuronal mRNAs that contribute to neuronal development and synaptic plasticity (35, 36). A recent study reported the presence of P body proteins in neuronal staufen-containing granules in Drosophila (37). These staufen ribonucleoprotein particles were found to contain components of RNA turnover and translational repression pathways, including those mediated by miRNAs that are present in somatic P bodies. It is thus conceivable that Htt also participates in the function of neuronal RNA granules. If so, the presence of mutant Htt may perturb mRNA processing/trafficking or local translation in critical neurons leading to the unique pathology associated with HD. Indeed, loss of miRNA processing was reported to enhance neurodegeneration in a Drosophila model of polyQ disease (38). Taken together, these studies link Htt to post-transcriptional processes. The pathogenic mechanism of HD is likely to be multilayered, involving deregulation of transcriptional and post-transcriptional regulatory pathways combined with other proposed mechanisms such as the triggering of apoptosis and mitochondrial dysfunction to give rise to progressive neurodegeneration (39–41).
Materials and Methods
Purification of Flag-Htt590.
HeLa S3 cells expressing Flag-Htt590 were established and Htt protein purified essentially as described (13). Details are provided in SI Text.
Knockdown Experiments.
U2OS cells (4.5 × 105) were plated in 60-mm plates 24 h before transfection of siRNA (100 nM final) with Dharmafect (Dharmacon). The siRNA sequences and antibodies used for immunoblotting are provided in SI Text. Antibody to human GW182 was from CytoStore (MAB-001).
Confocal Microscopy/Indirect Immunofluorescence.
Endogenous proteins were examined in U2OS and N2A cells grown to ≈70% confluence on glass cover slips. HeLa cells were used for overexpression experiments. FLAG-Ago2 was expressed from Addgene plasmid 10822 (42). Cells were fixed at room temperature with 4% (wt/vol) paraformaldehyde in PBS, permeabilized with 0.2% Triton X-100 for 20 min, followed by three PBS washes. Cells were subsequently blocked for 1 h at room temperature in 10% FBS + 0.25% saponin (Sigma) in PBS. Primary antibodies (listed in SI Text) were diluted in the blocking buffer and probed for 1 h at 37°C. Cells were washed three times with 7% fish gelatin + 0.025% saponin in PBS for 5 min each. Conjugated goat α-mouse Alexa 488 and goat α-rabbit Alexa 555 were diluted 1:250 in the blocking buffer and incubated for 1 h at 37°C. Cells were washed in PBS three times for 5 min and treated at room temperature for 15 min with Hoechst stain and washed three times with PBS before mounting with Dako. Rat hippocampal neurons were probed with α-Ago2 (laboratory of R.S.) and α-Htt antibody (Chemicon 5492). The 2166 antibody gave the same result. Fluorescence microscopy was performed with a Zeiss LSM 510 confocal microscope.
Sequential siRNA Transfections and Luciferase/β-Galactosidase Assays.
Luc reporter assays were performed after plating HeLa cells at 4 × 104 per 60-mm plate 24 h before transfection with control, GW182, Ago2, or Htt siRNA. The transfected cells were incubated for 48 h, at which time they were split equally into 12-well plates and incubated for an additional 24 h. The cells were transfected with 200 ng of SV40 promoter-Luc reporter, 20 ng of CMV-β galactosidase, 30 ng of carrier plasmid, and 315 ng of Luc siRNA where indicated. Twenty-four hours after transfection, cells were lysed and assayed for luciferase and β-galactosidase activity. The normalized (luciferase/β-galactosidase) value for each siRNA transfection was averaged and the standard deviation determined and represented as the percentage of the control siRNA transfected cells (in the absence of Luc siRNA). The data are the result of three independent siRNA transfections performed on different days. The normalized data are graphed on a log scale.
miRNA Reporter Assays.
HeLa cells were plated and transfected with siRNA as above, split equally into 12-well plates, and incubated for 24 h. For each plate, three wells were transfected with 20 ng of Renilla Luc reporter that contained two copies of the human let-7b element or a mutant element (26) and 10 ng of SV40-firefly Luc. Twenty-four hours after transfection, cells were lysed and assayed for Renilla and firefly luciferase activity.
Supplementary Material
Acknowledgments.
We thank Denise Tambasco, Benjamin Houghtaling, Jeffrey Kowalak, Kaiping Yan, Latika Khatri, Sophie Restituito, Ligang Wu, Joel Belasco, Roy Parker, Angus Wilson, Ian Mohr, Mark Philips, and members of the laboratory of N.T. for experimental help and discussions. We thank Ryo Koyama-Nasu (New York University School of Medicine), Hidesato Ogawa (Dana–Farber Cancer Institute), Yoshihiro Nakatani (Dana–Farber Cancer Institute), Jens Lykke-Andersen (University of Colorado at Boulder), Nancy Kedersha (Harvard Medical School, Boston), Thomas Tuschl (Rockefellar University, New York), Edward Chan (University of Florida), and Joan Steffan (University of California, Irvine) for reagents, and Angus Wilson for critical reading of the manuscript. This work was supported by a grant from the Mathers Charitable Foundation (N.T.) and in part by the Intramural Research Program of the National Institute of Mental Health (Grant 1 Z01 MH000274 to S.P.M.) and a grant from the National Institutes of Health (R01NS050352 to D.K.). J.N.S. was supported in part by the Vilcek Endowment Fellowship Award. Purchase of the confocal microscope was funded by a Shared Instrumentation Grant from the National Institutes of Health (Grant S10 RR017970).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800658105/DCSupplemental.
References
- 1.The Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Andrade MA, Bork P. HEAT repeats in the Huntington's disease protein. Nat Genet. 1995;11:115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- 3.Li SH, Li XJ. Huntingtin and its role in neuronal degeneration. Neuroscientist. 2004;10:467–475. doi: 10.1177/1073858404266777. [DOI] [PubMed] [Google Scholar]
- 4.Landles C, Bates GP. Huntingtin and the molecular pathogenesis of Huntington's disease. Fourth in molecular medicine review series. EMBO Rep. 2004;5:958–963. doi: 10.1038/sj.embor.7400250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harjes P, Wanker EE. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem Sci. 2003;28:425–433. doi: 10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- 6.Li SH, Li XJ. Huntingtin-protein interactions and the pathogenesis of Huntington's disease. Trends Genet. 2004;20:146–154. doi: 10.1016/j.tig.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Giorgini F, Muchowski PJ. Connecting the dots in Huntington's disease with protein interaction networks. Genome Biol. 2005;6:210. doi: 10.1186/gb-2005-6-3-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qi ML, et al. Proteome analysis of soluble nuclear proteins reveals that HMGB1/2 suppress genotoxic stress in polyglutamine diseases. Nat Cell Biol. 2007;9:402–414. doi: 10.1038/ncb1553. [DOI] [PubMed] [Google Scholar]
- 9.Perluigi M, et al. Proteomic analysis of protein expression and oxidative modification in r6/2 transgenic mice: A model of Huntington disease. Mol Cell Proteomics. 2005;4:1849–1861. doi: 10.1074/mcp.M500090-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Kaltenbach LS, et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneratio. PLoS Genet. 2007;3:e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cattaneo E, Zuccato C, Tartari M. Normal huntingtin function: An alternative approach to Huntington's disease. Nat Rev Neurosci. 2005;6:919–930. doi: 10.1038/nrn1806. [DOI] [PubMed] [Google Scholar]
- 12.Graham RK, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 13.Nakatani Y, Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 2003;370:430–444. doi: 10.1016/S0076-6879(03)70037-8. [DOI] [PubMed] [Google Scholar]
- 14.Holbert S, et al. The Gln-Ala repeat transcriptional activator CA150 interacts with huntingtin: Neuropathologic and genetic evidence for a role in Huntington's disease pathogenesis. Proc Natl Acad Sci USA. 2001;98:1811–1816. doi: 10.1073/pnas.041566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornett J, et al. Polyglutamine expansion of huntingtin impairs its nuclear export. Nat Genet. 2005;37:198–204. doi: 10.1038/ng1503. [DOI] [PubMed] [Google Scholar]
- 16.Peters L, Meister G. Argonaute proteins: Mediators of RNA silencing. Mol Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Meister G, et al. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 20.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 21.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 22.Fillman C, Lykke-Andersen J. RNA decapping inside and outside of processing bodies. Curr Opin Cell Biol. 2005;17:326–331. doi: 10.1016/j.ceb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 24.Jakymiw A, et al. Disruption of GW bodies impairs mammalian RNA interference. Nat Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- 25.Julien E, Herr W. Proteolytic processing is necessary to separate and ensure proper cell growth and cytokinesis functions of HCF-1. EMBO J. 2003;22:2360–2369. doi: 10.1093/emboj/cdg242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chendrimada TP, et al. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 27.Trettel F, et al. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet. 2000;9:2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- 28.Menalled LB, et al. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington's disease with 140 CAG repeats. J Comp Neurol. 2003;465:11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- 29.Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci USA. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson RJ, Standart N. How do microRNAs regulate gene expression? Sci STKE. 2007;2007:re1. doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- 31.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–81. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nonhoff U, et al. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell. 2007;18:1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiebler MA, Bassell GJ. Neuronal RNA granules: Movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 36.Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbee SA, et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bilen J, Liu N, Burnett BG, Pittman RN, Bonini NM. MicroRNA pathways modulate polyglutamine-induced neurodegeneration. Mol Cell. 2006;24:157–163. doi: 10.1016/j.molcel.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 39.Sugars KL, Rubinsztein DC. Transcriptional abnormalities in Huntington disease. Trends Genet. 2003;19:233–238. doi: 10.1016/S0168-9525(03)00074-X. [DOI] [PubMed] [Google Scholar]
- 40.Ross CA. Polyglutamine pathogenesis: Emergence of unifying mechanisms for Huntington's disease and related disorders. Neuron. 2002;35:819–822. doi: 10.1016/s0896-6273(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 41.Riley BE, Orr HT. Polyglutamine neurodegenerative diseases and regulation of transcription: Assembling the puzzle. Genes Dev. 2006;20:2183–2192. doi: 10.1101/gad.1436506. [DOI] [PubMed] [Google Scholar]
- 42.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.