Abstract
Misfolding and subsequent aggregation of endogeneous proteins constitute essential steps in many human disorders, including Alzheimer and prion diseases. In most prion protein-folding studies, the posttranslational modifications, the lipid anchor in particular, were lacking. Here, we studied a fully posttranslationally modified cellular prion protein, carrying two N-glycosylations and the natural GPI anchor. We used time-resolved FTIR to study the prion protein secondary structure changes when binding to a raft-like lipid membrane via its GPI anchor. We observed that membrane anchoring above a threshold concentration induced refolding of the prion protein to intermolecular β-sheets. Such transition is not observed in solution and is membrane specific. Excessive membrane anchoring, analyzed with molecular sensitivity, is thought to be a crucial event in the development of prion diseases.
Keywords: FTIR, membrane anchoring, prion protein, protein aggregation, secondary structure
Conversion of host-encoded prion protein (PrP) from its cellular form, PrPC, into an infectious isoform, PrPSc, is the molecular event underlying prion diseases (1). The transition from PrPC to PrPSc can be regarded as a posttranslational refolding process without any covalent modification (2), but leading to different physicochemical properties. It leads to an increase in β-sheet structure, insolubility, and partial resistance against digestion with proteinase K (3–6). This conversion has been investigated in vitro, predominantly by using recombinant PrP (recPrP), expressed in Escherichia coli, (7–10). However, the eucaryotic PrPC is posttranslationally modified, carrying two N-glycosylations and a GPI anchor. The latter attaches PrP to the cell membrane (11). Like many other GPI-anchored proteins, PrPC is enriched in specific membrane microdomains called rafts (12, 13).
To understand the mechanism underlying the structural transition from PrPC to PrPSc, the knowledge of both structures is necessary. Determination of the infectious PrPSc conformation is hampered by its insolubility and structural heterogeneity. Structural models were obtained from electron microscopic studies of two-dimensional crystals of PrP, which were prepared from infectious prions (14). The three-dimensional structure of recPrP as a model for PrPC has been solved by NMR-spectroscopic analyses of nearly all prion-susceptible species (15–17). With only minor variation, recPrP exhibits a C-terminal globular domain, consisting of three α-helices and a small antiparallel β-sheet. The N terminus is highly flexible and lacks a well-defined structure. These structural analyses were carried out with recPrP purified from prokaryotic expression systems. More recently, the one-dimensional NMR spectrum of natural PrPC from bovine brain, carrying the two N-glycosylations but lacking the GPI-anchor, has been reported. No significant structural differences between anchorless PrPC and recPrP in solution were found (18).
PrPC in vivo is anchored to the cell membrane. Thus, the effect of membrane binding on the structure of PrP is naturally of interest. Recent studies used recPrP with synthetic membrane anchors and followed the secondary structure upon lipid contact by using either UV-CD spectroscopy (19) or FTIR measurements (20). Both studies concluded that the structure of recPrP, with a synthetic membrane anchor, is identical upon lipid contact to the structure of anchorless PrP in lipid-free solutions. Very recently, it has been reported that the interaction of anchorless recPrP with lipids can evoke a conformational transition (21, 22). However, our present approach continuously follows the secondary structural changes of native, fully posttranslationally modified PrPC upon membrane anchoring. Our preparation is much closer to the in vivo situation than those of the earlier studies. Native PrPC, carrying the two N-glycosylations and the natural GPI anchor, purified from an eukaryotic transgenic cell line (23, 24), was anchored to a solidly supported, raft-like membrane in a buffered environment. In an earlier study, we exploited the alterations of surface plasmon resonance to analyze the lipid anchoring kinetics (25). We showed that PrPC binds with high affinity to raft-like lipid bilayers by the insertion of the natural GPI-anchor into the membrane. Here, we monitored the secondary structure changes of PrPC during its membrane anchoring using time-resolved FTIR spectroscopy (trFTIR). Recently, we established a novel deconvolution approach specific for the prion protein, which allows an unequivocal determination of secondary structural changes based on amide I band shifts by calibration with NMR data (26). In the present study, we exposed a solid-supported lipid membrane on an ATR (attenuated total reflection) crystal to PrPC under physiological buffer conditions. In agreement with the earlier studies of artificial membrane anchors, PrPC at lower concentrations adapted the same secondary structure on the membrane as recPrP did in solution NMR studies. At higher protein concentrations in the solution, we observed a spectral shift, which indicated a depletion of random coil and the formation of intermolecular β-sheets. Increasing the local concentration of membrane-anchored PrPC seems to induce a conformational transition accompanied by di- or oligomerization of PrPC. This is a possible explanation for the atypical pathologies found in transgenic mice overexpressing prion protein (27), as well as the first step on the pathway to the disease-associated conformation of PrP. We propose that membrane anchoring of an excess of prion protein is the structural prerequisite in the development of prion diseases.
Results
Binding of PrPC to Lipid Bilayers.
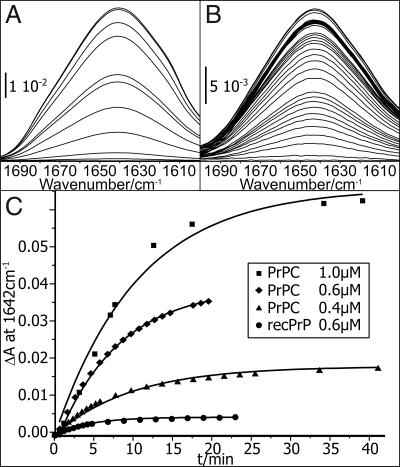
A raft-containing lipid bilayer was formed by the spontaneous fusion of small unilamellar lipid vesicles with the hydrophilic surface of a germanium ATR crystal. The crystal was mounted in a flow cell. Lipids and proteins were administered in buffer, followed by excessive washing. In the ATR technique, an evanescent IR wave probes a sample in the immediate proximity of the ATR crystal surface, penetrating only <1 μm into an aqueous sample with exponentially decaying intensity. This evanescent wave is similar to that in the surface plasmon resonance approach. The formation of a stable lipid bilayer on the surface of the ATR crystal prior protein application was also followed by trFTIR. The evolution of the lipid C-H stretching vibration extinction bands at 3,050–2,800 cm−1 were monitored [supplemental information (SI) Fig. S1]. Because of the low solubility of PrPC under physiological conditions, PrPC was diluted from the stock solution at pD 4 in citrate-buffered saline (CBS) at pD 6 immediately before the measurement. PrPC in CBS was then attached to the lipid membrane in a circulating flow within the ATR cell. Binding of PrPC to the membrane was continuously recorded, particularly tracing the amide I absorption band. The frequency of the amide I band is sensitive to the protein structure and represents the C = O stretching vibration of the protein backbone. The observed amide I bands are shown in Fig. 1 as a function of time. The gain in extinction is due to increased local PrPC concentration on the membrane. Different concentrations of PrPC in the bulk solution led to different equilibrium surface concentrations (Fig. 1C). The specificity of membrane anchoring was shown with anchorless and unglycosylated recombinant full-length PrP (recPrP). This control expressed only very low affinity for the model membrane (Fig. 1C). This clearly demonstrates the anchoring of PrPC to the tethered raft-like lipid bilayer. Binding kinetics, observed with ATR-trFTIR, are slower than observed in the SPR experiments (25). The increase of the amide I band observed in the ATR experiments reflects not solely binding. It is superimposed with diffusion and streaming processes on the 1,500-mm2 ATR surface, until a homogeneous PrPC distribution on the membrane was achieved. The extinction ascents observed in Fig. 1C are typical for these anchoring experiments, as seen with other proteins.
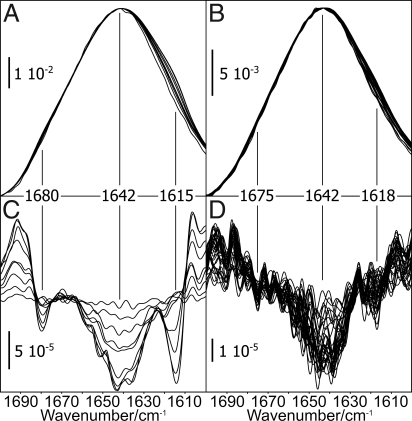
Fig. 1.
Membrane binding of PrPC and recPrP. The evolution of the amide I band displays the progression of membrane anchoring of PrPC. (A and B) A 1.0 μM protein solution (A) yields a higher final extinction, thus a higher final surface concentration than 0.4 μM (B). (C) The amplitudes at 1,642 cm−1 are plotted against time. The data points correspond to the different amplitudes in A and B, respectively. The very low adhesion of anchorless recPrP even at 0.6 μM (C) demonstrates specific membrane contact of PrPC via its GPI anchor.
Next, we addressed whether PrPC experiences structural alterations upon membrane anchoring. The frequencies of the amide I extinction band are very sensitive to protein secondary structure. A deconvolution determines the secondary structure composition in a manner similar to CD spectroscopy. In contrast, IR can benefit from the selective surface sensitivity of an ATR setup. IR bands clearly distinguish between α-helices, which absorb ≈1,660–1,645 cm−1, and β-sheets, which absorb in the quite different region of ≈1,635–1,615 cm−1. However, as in CD spectroscopy, deconvolution of the amide I band is experimentally underdetermined. Therefore, we have implemented an approach in which the deconvolution is calibrated with the established NMR structure (PDB 1B10) (28). The calibration discriminates the useful deconvolution from several possible solutions. This approach has already been described in detail (26). It provides deconvolution of the prion protein amide I band into its secondary structural components. In this case, it provided the secondary structure with an uncertainty of ≈4%. We are aware that this accuracy is obtained only in this specific case and cannot be generalized. The calibration has to be done for each protein individually. However, once the contributions of the individual secondary structural elements to the amide I band are determined, the relative changes of the secondary structure during membrane binding can be traced with an uncertainty of <5%.
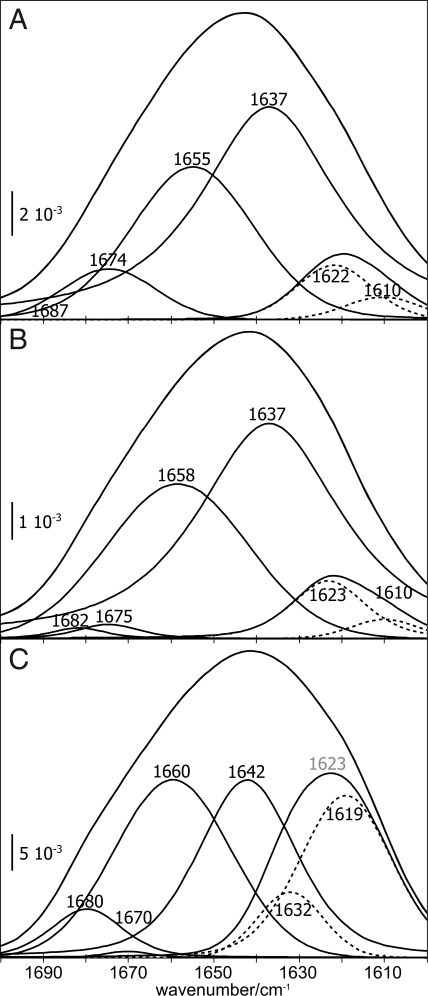
Fig. 2 A and C present the deconvolution of the amide I bands of PrPC after achieving a binding equilibrium (compare Fig. 1 A and B). The secondary structure data using 0.4 μM PrPC (9% β-sheet, 57% random coil, 28% α-helix and 7% β-turn, Fig. 2A) agree with the data of free, soluble recPrP within the expected error of 4% (11% β-sheet, 56% random coil, 27% α-helix and 6% β-turn, Fig. 3A). Incubation of the raft-like membrane with 1 μM PrPC initially preserved the solution secondary structure within the expected error (10% β-sheet, 58% random coil, 31% α-helix, and 1% β-turn, Fig. 2B). However, continued incubation, implying an increased local concentration at the lipid bilayer, decreased the random coil band from 58% to 31%, whereas the β-sheet increased from 10% to 37% in the equilibrium (Fig. 2C, Table 1). The α-helix persisted, and the β-turns decreased negligibly. We deconvolved the β-sheet band at 1,623 cm−1 further, into an intramolecular β-sheet band at higher frequencies and an intermolecular β-sheet band at lower frequencies. The intermolecular band indicates interacting sheets from two different proteins as schematically shown in Fig. 6. That showed a considerable increase at 1619 cm−1 in equilibrium. 80% of the 1,623-cm−1 band indicated intermolecular β-sheet structure. In summary, we observed a decrease of random coil during membrane anchoring. This transition is not observed in solution under the same conditions. To the same extent, intermolecular β-sheets were formed with increased concentration of PrPC. These transitions were not observed at 0.4 μM PrPC, whereas recPrP collapsed drastically upon membrane contact (Fig. 3B). Only a small fraction of the anchorless protein bound to the surface, as indicated by the low-intensity amide I band. The β-sheet disappeared, and β-turns were largely reduced. The amide band consisted mostly of random coil and α-helix with a 60:40 distribution. It was accompanied by a blue shift of the helix-assigned band from 1,654 to 1,662 cm−1. This high frequency is common in proteins with extended and extraordinarily stable helices, such as bacteriorhodopsin or myoglobin. It suggests that the membrane induced a stabilized helix formation in recPrP when attached to the membrane compared to the solution state.
Fig. 2.
The amide I bands of PrPC shift upon membrane anchoring. (A and B) The spectral deconvolution of the amide I band at 0.4 μM PrPC after 34 min incubation with the lipid membrane (A) resembles the one of 1.0 μM PrPC after 3 min (B). (C) After 34 min incubation time at 1.0 μM PrPC, substantial band shifts toward oligomeric β-structure were evident. Bands between 1690–1678 cm−1 were assigned to antiparallel β-sheets, 1,676–1,663 cm−1 to β-turns, 1,662–1,645 cm−1 to α-helices, 1,644–1,635 cm−1 to random coil, and 1,634–1,610 cm−1 to β-sheets. This region was subdivided (dashed bands) approximately at 1,620 cm−1, with the low-frequency component representing intermolecular β-structure. The labels depict deconvolution results, regardless of component intensity.
Fig. 3.
Membrane binding of anchorless recPrP does not conserve its structure. The band shifts between recPrP in solution (A) and attached to the membrane (B) indicate an increase of α-helix accompanied with a blue shift of the assigned band, which indicates stabilization.
Table 1.
Determination of secondary structure by calibrated FTIR spectra evaluation
| Secondary structure | NMR | recPrP |
SHaPrPC |
|||
|---|---|---|---|---|---|---|
| A | B | C | D | E | ||
| α-Helix | 30 | 27 | 40 | 28 | 31 | 31 |
| β-Sheet | 4 | 11 | 0 | 9 | 10 | 37 |
| β-Turns | 7 | 6 | 0 | 7 | 1 | 1 |
| Random coil | 60 | 56 | 60 | 57 | 58 | 31 |
NMR calibration data was derived from STRIDE analysis (40) of PDB file 1B10. Residues not included in the PDB file were assumed as random coil. Here, we compare recPrP in solution (A) and membrane bound (B) with native SHaPrPC at 0.4 μM bulk concentration after 34 min incubation (C), and at 1.0 μM after 3.2 min (D) and 34 min (E). Values given are percentages.
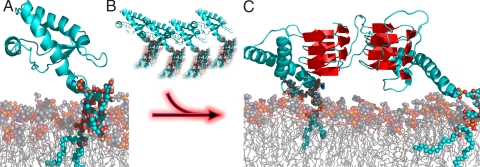
Fig. 6.
Concentration-dependent secondary structure changes upon membrane anchoring. (A) PrPC bound to the raft-like lipid bilayer exhibits the same secondary structure (at lower concentrations) as anchorless recPrP in solution in NMR studies (PDP 1AG2). (B) An increased concentration of PrPC at the membrane leads to a structural transition toward intermolecular beta sheet. (C) Schematic illustration of intermolecular β-sheet. This dimerization could well be the initial step on the pathway of the conversion into PrPSc.
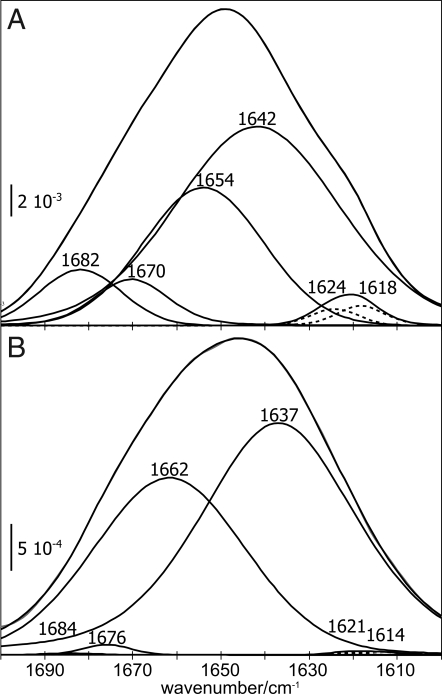
To exclude a deconvolution artifact, we also analyzed the second derivative of the amide I bands (Fig. 4). This was intended as a clear-cut method for the characterization of band shifts without any assumptions at all. In full agreement with the deconvolution, it confirms shifts at 1,680, 1,642, and 1,615 cm−1 for the 1 μM PrPC assay. At 0.4 μM PrPC, band shifts did not exceed the noise level to influence the secondary structure analysis.
Fig. 4.
Band shifts due to an increased PrPC concentration at the membrane. A decrease in the second derivative of a spectrum refers to an actual increase of extinction in the absorbance band. (A and B) The amide I bands, as recorded during membrane anchoring, were normalized to identical amplitudes at 1,642 cm−1. (C) Band shifts were identified at 1,680 and 1,642 cm−1 for 1.0 μM PrPC. (D) The according analysis of the 0.4 μM assay indicated an earliest onset of the discussed structural conversion after 34 min incubation.
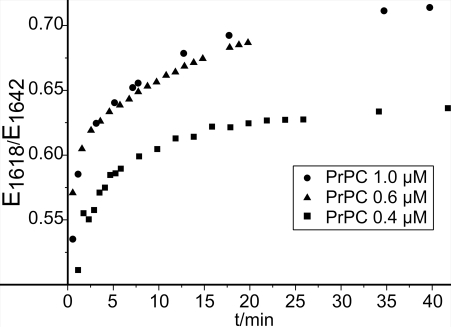
The concentration-normalized changes in absorbance at 1,618 cm−1 (Fig. 5) display the formation of intermolecular β-sheets with increasing concentration of PrPC at the lipid bilayer. At 0.4 μM PrPC, spectra indicated equilibrium within 15 min. A PrPC concentration above 0.6 μM resulted in increased formation of β-sheets. Within the measuring time of 40 min no equilibrium was reached. No structural difference between 0.6 μM and 1.0 μM PrPC could be observed. This indicates that the transition exhibited a switch-like behavior, which commenced at a threshold of ≈0.6 μM concentration in the bulk phase. It appeared insensitive to further concentration increase.
Fig. 5.
Development of membrane binding and refolding with time. The ratio of extinction values at 1,618 and 1,642 cm−1 indicates a relation between membrane anchoring and refolding. The ratio increased until 15 min for the 0.4-μM assay, reaching equilibrium. At higher protein concentrations, further membrane anchoring resulted in refolding without reaching equilibrium within 40 min.
Discussion
In the present study, we observed the specific membrane binding and accumulation of native, fully glycosylated PrPC on the membrane due to hydrophobic interactions between its GPI anchor and the membrane lipids. We showed that upon anchoring to the membrane, native PrPC initially assumes a structure similar to recPrP in solution. However, a PrPC concentration above a particular threshold at the membrane leads to a decrease of random coil and to the formation of intermolecular β-sheets. Consequently, dimers or oligomers of PrPC are formed on the membrane. Indeed, dimers of PrPC and recPrP have been observed in solution in several studies (7, 29).
In vivo PrPC is localized in lipid rafts. These microdomains within the cell membrane are known to generate high local concentrations of specific proteins (30, 31), which increases the opportunity for intermolecular interactions. In our studies, only the lipid moiety of rafts could be established; other proteins were not present. In our earlier binding studies, significantly higher binding of PrPC to raft-like lipids was shown as compared to other lipids, but it was pointed out that binding of PrPC was not exclusive to rafts (25). The formation of PrPC clusters was described in cell-culture studies (32–35). For example, high-density PrPC clusters on the surface of primary culture neurons were observed (32). Therefore, a high local PrPC concentration is assumable under physiological conditions.
The results of the present work are summarized in the scheme shown in Fig. 6. The structure of membrane-bound PrPC at lower concentrations agrees with the structural model derived from NMR studies on recPrP. Above a threshold of local concentration of membrane-bound PrPC, random coil structure decreased and intermolecular β-sheets were formed. This facilitates intermolecular contacts and could induce di- or probably oligomerization. The structure shown is only meant schematically. Actually, Eisenberg and colleagues showed that peptide fragments of Aβ, yeast prion protein sup35, and other proteins form amyloid cross-β spines with intermolecular β-strands (36). Furthermore, very similar vibrational features, as measured by ATR-FTIR, have been observed during fibrilization of the tau protein (37).
Although regular cross-β spines are characteristic of the structure of the pathological isoform and not, as in our case, of the cellular protein, intermolecular β-strands can be formed by PrPC molecules on the membrane surface if a threshold population is exceeded. In that respect, PrP might be particularly susceptible to conversion into the pathological isoform. Herewith, we present molecular evidence for a prion-disease mechanism based on PrP accumulation. This accumulation may be due to genetically caused overexpression or to a different, as yet unknown trigger. One might even speculate that the presentation of PrPC in the β-sheet containing structure on the outer cell surface is the molecular basis for why prion diseases are transmissible, whereas other protein misfolding diseases are not.
Materials and Methods
Unless indicated otherwise, all chemicals and solutions were obtained from Sigma Aldrich, Munich, Germany.
Purification of PrPC.
Chinese hamster ovary (CHO) cell lines overexpressing PrPC of the Syrian Golden hamster sequence were established by Dr. S. B. Prusiner's group (23). PrPC derived from this cell line carries both the two N-glycosylations and the C-terminally attached GPI anchor. Purification of PrPC was achieved by two affinity chromatographic steps: An immobilized metal chelating affinity chromatography (IMAC) using the intrinsic property of PrPC to bind copper ions preceded an immunopurification using the antibody 3F4 covalently coupled to proteinG, carried out as described (24). Purified protein was concentrated in Centricon tubes with a cutoff of 10 kDa (Millipore), and the buffer was changed to 1 mM NaOAc pD4 in D2O.
Purification of recPrP (23–231).
Full-length recombinant Syrian hamster PrP [recPrP(23–231)] expressed in E. coli was purified as described (38). We oxidized the disulfide bonds by incubation in 1 mM glutathione at room temperature overnight before reversed-phase HPLC.
Preparation of Small Unilamellar Vesicles for Surface Fusion.
All lipids used in this study were natural lipids purchased from Avanti Polar Lipids. Stock solutions in chloroform were stored at −20°C. For each experiment, lipids were dried under a stream of nitrogen and evacuated for 15 h. Dried lipids were hydrated to a concentration of 200 μg/ml in 10 mM sodium citrate, pD 6, and 137 mM NaCl for 1 h, and were then sonicated in a 250-W analogous Branson sonifier equipped with a high-efficiency cup resonator at 70% output power, water-bath thermostabilized at 37°C. Raft-like vesicles were of the following lipid composition: dimyristoyl phosphatidylcholine (DMPC), sphingomyelin (brain), cerebroside (brain), and cholesterol in a molar ratio of 2:1:1:2 (39).
FTIR Measurement.
A trapezoid germanium internal reflection element (IRE) (52 × 20 × 2 mm3, A. C. M.) was used in a vertical, variable angle ATR setup (Specac), in a Bruker IFS 66 FTIR spectrometer equipped with a liquid nitrogen cooled mercury cadmium telluride (MCT) detector. The IRE was cleaned with sulfuric acid, rinsed thoroughly with distilled water, and incubated in a plasma cleaner (Harrick) prior to use. The setup was adjusted to a 45° incidence angle, yielding 25 internal reflections. The IRE was held in a customized, stream-optimized flow-through cuvette with non-grease silicone sealings. Dead volume of the system was 1,600 μl. Flux control was provided by a digital peristaltic pump (Ismatec), set to 1.0 ml/min. The membrane was put up on the IRE surface by rinsing 200 μl of a 100 μg/ml solution through the cuvette system until a stable lipid signal was observed (Fig. S1). Excess lipid was rinsed out with buffer until the spectra were stable again (Fig. S1B). Then the protein solution was introduced into the system in the same way, and excess protein was again rinsed out with excess amounts of buffer.
The spectra were continuously recorded, accumulating 256 full interferograms prior Fourier transformation. Datapoints each 0.32 cm−1 were achieved with an instrument resolution of 2 cm−1 and a factor 4 zero filling. Extinction spectra were calculated according to Lambert-Beer′s law with single channel spectra of blank, buffer-surrounded model membranes as reference. After water vapor correction, the spectra were smoothed with a Fourier self deconvolution, eliminating noise with bands thinner than 4 cm−1 full-width-half-height (FWHH). Contributions of amino acid side chains to the amide I′ and II′ area (1,700–1,400 cm−1) were eliminated by subtraction of calculated residue spectra.
To estimate the protein secondary structure fractions, a calibrated amide I band decomposition was performed as described (26). Briefly, amide I bands of recPrP and PrPC in the different states were decomposed into Cauchy curves. The decomposition, per se an experimentally underdetermined problem, requires the definition of initialization parameters. Of the possible parameter sets, one was selected that resulted in (i) a high quality band decomposition for all bands and (ii) a minimum deviation of the secondary structure fractions from the calibration data. In this study, a STRIDE secondary structure analysis (40) of the three-dimensional NMR structure of recSHaPrP(90–231), PDB 1B10 (28), served for this purpose. Residues not contained in the structure file were considered as random coil. Band integrals determined the fraction of the position-dependently assigned structure, regarding the overall amide I integral. Band positions were assigned to secondary structures as commonly found in the literature (26).
Supplementary Material
Acknowledgments.
We thank David Rumschitzky for helpful discussions and critical reading of the manuscript and Steffen Wolf for help in preparation of Fig. 5. D.R. and K.G. thank Manfred Eigen for enlightening discussions about the prion protein in the Klosters-Winterseminar. This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG), EU-Network of Excellence (NeuroPrion), and the Virtual Institute (VH-VI-013).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804721105/DCSupplemental.
References
- 1.Prusiner SB. Molecular biology of prion diseases. Science. 1991;252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 2.Stahl N, et al. Structural studies of the scrapie prion protein using mass spectrometry and amino acid sequencing. Biochemistry. 1993;32:1991–2002. doi: 10.1021/bi00059a016. [DOI] [PubMed] [Google Scholar]
- 3.McKinley MP, Bolton DC, Prusiner SB. A protease-resistant protein is a structural component of the scrapie prion. Cell. 1983;35:57–62. doi: 10.1016/0092-8674(83)90207-6. [DOI] [PubMed] [Google Scholar]
- 4.Caughey BW, et al. Secondary structure analysis of the scrapie-associated protein PrP 27–30 in water by infrared spectroscopy. Biochemistry. 1991;30:7672–7680. doi: 10.1021/bi00245a003. [DOI] [PubMed] [Google Scholar]
- 5.Pan KM, et al. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Safar J, Roller PP, Gajdusek DC, Gibbs CJ., Jr Thermal stability and conformational transitions of scrapie amyloid (prion) protein correlate with infectivity. Protein Sci. 1993;2:2206–2216. doi: 10.1002/pro.5560021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jansen K, et al. Structural intermediates in the putative pathway from the cellular prion protein to the pathogenic form. Biol Chem. 2001;382:683–691. doi: 10.1515/BC.2001.081. [DOI] [PubMed] [Google Scholar]
- 8.Leffers KW, et al. Assembly of natural and recombinant prion protein into fibrils. Biol Chem. 2005;386:569–580. doi: 10.1515/BC.2005.067. [DOI] [PubMed] [Google Scholar]
- 9.Caughey B, et al. Interactions and conversions of prion protein isoforms. Adv Protein Chem. 2001;57:139–169. doi: 10.1016/s0065-3233(01)57021-7. [DOI] [PubMed] [Google Scholar]
- 10.Baskakov IV, Legname G, Baldwin MA, Prusiner SB, Cohen FE. Pathway complexity of prion protein assembly into amyloid. J Biol Chem. 2002;277:21140–21148. doi: 10.1074/jbc.M111402200. [DOI] [PubMed] [Google Scholar]
- 11.Stahl N, Borchelt DR, Hsiao K, Prusiner SB. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell. 1987;51:229–240. doi: 10.1016/0092-8674(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 12.Naslavsky N, Stein R, Yanai A, Friedlander G, Taraboulos A. Characterization of detergent-insoluble complexes containing the cellular prion protein and its scrapie isoform. J Biol Chem. 1997;272:6324–6331. doi: 10.1074/jbc.272.10.6324. [DOI] [PubMed] [Google Scholar]
- 13.Gilch S, Kehler C, Schatzl HM. The prion protein requires cholesterol for cell surface localization. Mol Cell Neurosci. 2006;31:346–353. doi: 10.1016/j.mcn.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Govaerts C, Wille H, Prusiner SB, Cohen FE. Evidence for assembly of prions with left-handed beta-helices into trimers. Proc Natl Acad Sci USA. 2004;101:8342–8347. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donne DG, et al. Structure of the recombinant full-length hamster prion protein PrP(29–231): The N terminus is highly flexible. Proc Natl Acad Sci USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riek R, Hornemann S, Wider G, Glockshuber R, Wuthrich K. NMR characterization of the full-length recombinant murine prion protein, mPrP(23–231) FEBS Lett. 1997;413:282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 17.Lopez GF, Zahn R, Riek R, Wuthrich K. NMR structure of the bovine prion protein. Proc Natl Acad Sci USA. 2000;97:8334–8339. doi: 10.1073/pnas.97.15.8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornemann S, Schorn C, Wuthrich K. NMR structure of the bovine prion protein isolated from healthy calf brains. EMBO Rep. 2004;5:1159–1164. doi: 10.1038/sj.embor.7400297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eberl H, Tittmann P, Glockshuber R. Characterization of recombinant, membrane-attached full-length prion protein. J Biol Chem. 2004;279:25058–25065. doi: 10.1074/jbc.M400952200. [DOI] [PubMed] [Google Scholar]
- 20.Hicks MR, et al. Synthesis and structural characterization of a mimetic membrane-anchored prion protein. FEBS J. 2006;273:1285–1299. doi: 10.1111/j.1742-4658.2006.05152.x. [DOI] [PubMed] [Google Scholar]
- 21.Wang F, et al. Lipid interaction converts prion protein to a PrP(Sc)-like proteinase K-resistant conformation under physiological conditions. Biochemistry. 2007;46:7045–7053. doi: 10.1021/bi700299h. [DOI] [PubMed] [Google Scholar]
- 22.Re F, et al. Prion protein structure is affected by pH-dependent interaction with membranes: A study in a model system. FEBS Lett. 2008;582:215–220. doi: 10.1016/j.febslet.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Blochberger TC, et al. Prion protein expression in Chinese hamster ovary cells using a glutamine synthetase selection and amplification system. Protein Eng. 1997;10:1465–1473. doi: 10.1093/protein/10.12.1465. [DOI] [PubMed] [Google Scholar]
- 24.Elfrink K, Riesner D. In: Methods and Tools in Biosciences: Techniques in Prion Research. Lehmann S, Grassi F, editors. Basel: Birkhaeuser; 2004. pp. 4–15. [Google Scholar]
- 25.Elfrink K, Nagel-Steger L, Riesner D. Interaction of the cellular prion protein with raft-like lipid membranes. Biol Chem. 2007;388:79–89. doi: 10.1515/BC.2007.010. [DOI] [PubMed] [Google Scholar]
- 26.Ollesch J, Kuennemann E, Glockshuber R, Gerwert K. Prion protein a-to-β transition monitored by time-resolved Fourier transform infrared spectroscopy. Appl Spectrosc. 2007;61:1025–1031. doi: 10.1366/000370207782217680. [DOI] [PubMed] [Google Scholar]
- 27.Westaway D, et al. Degeneration of skeletal muscle, peripheral nerves, and the central nervous system in transgenic mice overexpressing wild-type prion proteins. Cell. 1994;76:117–129. doi: 10.1016/0092-8674(94)90177-5. [DOI] [PubMed] [Google Scholar]
- 28.James TL, et al. Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc Natl Acad Sci USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer RK, et al. A monomer-dimer equilibrium of a cellular prion protein (PrPC) not observed with recombinant PrP. J Biol Chem. 2000;275:38081–38087. doi: 10.1074/jbc.M007114200. [DOI] [PubMed] [Google Scholar]
- 30.Silvius JR. Partitioning of membrane molecules between raft and non-raft domains: Insights from model-membrane studies. Biochim Biophys Acta. 2005;1746:193–202. doi: 10.1016/j.bbamcr.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Michel V, Bakovic M. Lipid rafts in health and disease. Biol Cell. 2007;99:129–140. doi: 10.1042/BC20060051. [DOI] [PubMed] [Google Scholar]
- 32.Madore N, et al. Functionally different GPI proteins are organized in different domains on the neuronal surface. EMBO J. 1999;18:6917–6926. doi: 10.1093/emboj/18.24.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taraboulos A, Raeber AJ, Borchelt DR, Serban D, Prusiner SB. Synthesis and trafficking of prion proteins in cultured cells. Mol Biol Cell. 1992;3:851–863. doi: 10.1091/mbc.3.8.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brugger B, et al. The membrane domains occupied by glycosylphosphatidylinositol-anchored prion protein and Thy-1 differ in lipid composition. J Biol Chem. 2004;279:7530–7536. doi: 10.1074/jbc.M310207200. [DOI] [PubMed] [Google Scholar]
- 35.Paar C, Wurm S, Pfarr W, Sonnleitner A, Wechselberger C. Prion protein resides in membrane microclusters of the immunological synapse during lymphocyte activation. Eur J Cell Biol. 2007;86:253–264. doi: 10.1016/j.ejcb.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Sawaya MR, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 37.von Bergen M, et al. Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif ((306)VQIVYK(311)) forming beta structure. Proc Natl Acad Sci USA. 2000;97:5129–5134. doi: 10.1073/pnas.97.10.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mehlhorn I, et al. High-level expression and characterization of a purified 142-residue polypeptide of the prion protein. Biochemistry. 1996;35:5528–5537. doi: 10.1021/bi952965e. [DOI] [PubMed] [Google Scholar]
- 39.Schroeder R, London E, Brown D. Interactions between saturated acyl chains confer detergent resistance on lipids and glycosylphosphatidylinositol (GPI)-anchored proteins: GPI-anchored proteins in liposomes and cells show similar behavior. Proc Natl Acad Sci USA. 1994;91:12130–12134. doi: 10.1073/pnas.91.25.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.