Abstract
Signal peptides (SPs) are critical for protein transport across cellular membranes, have a highly conserved structure, and are cleaved from the mature protein upon translocation. Here, we report that naturally occurring mutations in the SP of the adhesive, tip-associated subunit of type 1 fimbriae (FimH) are positively selected in uropathogenic Escherichia coli. On the one hand, these mutations have a detrimental effect, with reduced FimH transport across the inner membrane, fewer FimH and fimbriae expressed on the bacterial surface, and decreased bacterial adhesion under flow conditions. On the other hand, the fimbriae expressed by the mutants are significantly longer on average, with many fimbriae able to stretch to >20 μm in length. More surprisingly, the SP mutant bacteria display an increased ability to resist detachment from the surface upon a switch from high to low flow. This functional effect of longer fimbriae highlights the importance of the nonadhesive fimbrial rod for adhesive function. Also, whereas bacterial adhesion to bladder epithelial cells was preserved in most mutants, binding to and killing by human neutrophils was decreased, providing an additional reason the SP mutations are relatively common among uropathogenic strains. Thus, this study demonstrates how mutations in an SP, while decreasing transport function and not affecting the final structure of the translocated protein, can lead to functional gains of the extracellular organelles that incorporate the protein and overall adaptive changes in the organism's fitness.
Keywords: bacterial pathogenesis, fimbrial morphology, protein transport, shear force
Protein transport across membranes is a fundamental process essential to all forms of life. Most transported proteins contain an amino-terminal region, termed the signal peptide (SP), which directs and facilitates membrane transport of the nascent protein and, then, in most cases is cleaved off, releasing the mature protein. The basic structural characteristics of SPs are highly conserved with three regions important for function: a positively charged amino terminus, a hydrophobic core, and a hydrophilic carboxyl terminus (1–3). Although natural variation in SPs has been described for proteins from a variety of species, an adaptive benefit of the mutations is unlikely or is poorly understood (4–9).
In bacteria, SP-mediated membrane transport is common for proteins comprising fimbriae, hair-like surface structures that confer an adhesive phenotype to target cells or surfaces. Type 1 fimbriae of Escherichia coli are a model of the most common fimbrial class (I) of Gram-negative bacteria and are built of proteins transported across the inner (cytoplasmic) membrane via the general secretory pathway into the periplasm. During this process the SPs of the fimbrial subunits are cleaved. Fimbrial assembly is initiated at the outer membrane usher/chaperone (FimD/FimC) complex by the tip subunit, FimH, which is also the adhesive protein and consists of two domains, the mannose-binding lectin and fimbria-incorporating pilin domains. After additional minor subunits (FimG and FimF) are added, hundreds of the major subunit (FimA) are added to build the fimbrial rod in a spiral fashion (10, 11). Whereas another class I fimbrial system (P fimbriae) has a terminator protein that controls fimbrial length by preventing further rod extension (12), there is no well defined mechanism for controlling type 1 fimbrial length. Expression of type 1 fimbriae is regulated via an on/off mechanism controlled by an invertible promoter that initiates transcription of the entire fimAICDFGH operon (13).
Type 1 fimbriae are expressed by >90% of both commensal and pathogenic E. coli (14), and are a critical virulence factor for uropathogenic E. coli (15). Knocking out FimH decreases bacterial colonization of the mouse bladder (16), and FimH vaccination prevents urinary tract infections (UTIs) in mouse and primate models (17, 18). Although FimH is important for adherence to uroepithelial cells, it also increases bacterial susceptibility to the host immune response by triggering lectinophagocytosis by neutrophils (19).
The FimH adhesin mediates shear-enhanced binding via an allosteric catch bond mechanism, whereby tensile force converts FimH from a low- to high-affinity conformation that involves separation of the lectin and pilin domains (20). The nonadhesive fimbrial rod might also be functionally significant for the mechanics of binding. It was shown that the helical rod uncoils under increasing tensile force and recoils upon a drop in force, possibly maintaining optimal force on the FimH-mannose bond during changes in shear stress (21).
In uropathogenic strains the FimH adhesin often has adaptive point mutations in the mature protein that increase the ability of FimH to bind strongly under low shear stress typical of the urinary tract. However, additional mutations are present in the FimH SP in a sizeable portion of the E. coli population. The functional effects of these SP mutations remain unknown.
Here, we show that mutations in the FimH SP decrease FimH transport across the inner membrane, resulting in fewer fimbriae and decreased surface accumulation under flow. However, the fimbriae are also much longer. This increased length can explain the paradoxical phenomenon of enhanced maintenance of adhesion upon a shift from high to low flow, highlighting the functional importance of the nonadhesive fimbrial rod. Additionally, SP mutations result in decreased bacterial binding to and killing by neutrophils. These findings suggest that partial loss-of-function mutations in the FimH SP confer gain-of-function phenotypes that could be advantageous for uropathogenic E. coli.
Results
Signatures of Positive Selection in the FimH SP.
Phylogenetic analysis of fimH sequences from E. coli isolates revealed six nonsynonymous (amino acid replacement) mutations in the fimH signal sequence (Fig. 1A). Threonine at position −6 was converted to asparagine (T-6N) four times independently, as the alleles with this mutation are phylogenetically distinct and the fimH sequences do not show signs of intragenic recombination by the Recombination Detection Program (22). Nonsynonymous mutations also occur at position −10, where valine was converted to isoleucine (V-10I) and alanine (V-10A). These findings and a lack of synonymous (silent) changes in the fimH signal sequence indicate positive selection in the FimH SP (23).
Fig. 1.
Mutations in FimH SP and predicted effects. (A) fimH DNA tree indicating SP mutations. (B) Alignment of the four resulting SP variants, with SP regions denoted by N (amino terminus), H (hydrophobic core), and C (carboxyl terminus). (C) Average core hydrophobicity of FimH SP variants plotted on a graph depicting results from Doud et al. (3).
The FimH SP mutations are observed significantly more often in E. coli urine isolates in the setting of cystitis or pyelonephritis, likely uropathogenic strains (29 of 212 strains, 13.7%), than in E. coli fecal or vaginal isolates from healthy individuals, likely nonpathogenic strains (5 of 136 strains, 3.7%; Fisher's exact test P = 0.0016). No associations were detected between uropathogenicity and phylogenetic background or mutations in the FimH mature protein (data not shown), suggesting that FimH SP mutations are independently associated with uropathogenic strains.
Decreased Transport Activity of the SP Mutants.
Both mutated SP amino acid positions are located in the hydrophobic core region, which spans positions −5 to −13 as predicted by the SignalP hidden Markov model (24) (Fig. 1B). Doud et al. (3) showed that the average hydrophobicity of an SP core region correlates with membrane transport efficiency, as measured by preprotein processing. The average hydrophobicities of the SP core regions of the WT and mutant FimH alleles were calculated by using the GES scale (25) and plotted on a graph depicting results from the Doud et al. study (3), revealing that the mutations decrease the hydrophobicity of the core region to an extent that is predicted to decrease processing of the preprotein (Fig. 1C).
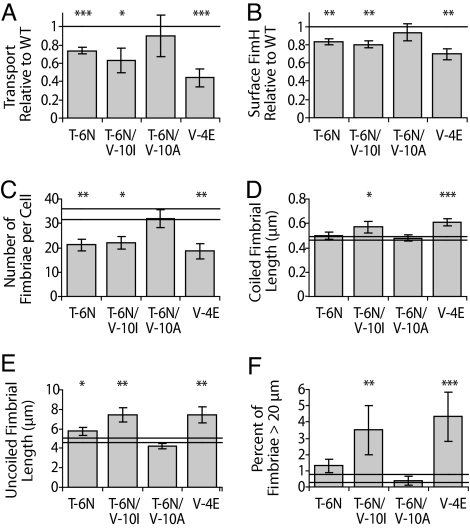
FimH SP-mediated inner membrane transport activity was measured via translational fusions to alkaline phosphatase, a reporter gene that is only active once secreted into the periplasmic space (26). Consistent with the computational prediction, the T-6N and T-6N/V-10I variants significantly decreased transport relative to WT (by 26% and 37%, respectively; Fig. 2A). The V-10I mutation in isolation also resulted in a decrease relative to WT (data not shown). However, inconsistent with the computational prediction, the T-6N/V-10A variant did not significantly change transport relative to WT.
Fig. 2.
Mutations that decrease FimH SP function reduce fimbrial number and increase fimbrial length. (A) FimH SP transport activity measured by units of alkaline phosphatase activity. (B) Surface FimH measured by flow cytometry. (C and D) Average (C) number and (D) length of type 1 fimbriae measured on electron micrographs. (E and F) Uncoiled fimbrial length (E) and percent of fimbriae >20 μm (F) remaining attached to the AFM cantilever at the end of a pull. All error bars represent SEM. Double horizontal lines represent WT ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001
For further studies several synthetic mutations were tested to find an SP alteration with a partial, but more dramatic, effect on membrane transport than the naturally occurring SP mutations. Among them, V-4E, which introduces an acidic residue in the basic amino terminus and causes a 56% decrease in transport efficiency relative to WT (Fig. 2A), was included in subsequent analyses.
SP Mutations Decrease the Number but Increase the Length of Surface-Expressed Fimbriae.
Decreased transport measured above proved to correspond with decreased surface expression of FimH measured by flow cytometry (with 17%, 20%, and 30% decreases relative to WT for T-6N, T-6N/V-10I, and V-4E, respectively; Fig. 2B). Consistent with these results, directly counting the number of type 1 fimbriae from 14–20 electron micrographs per strain revealed significantly fewer fimbriae per cell (37%, 37%, and 45% fewer fimbriae than WT for T-6N, T-6N/V-10I, and V-4E, respectively; Fig. 2C). Sample electron micrographs are in supporting information (SI) Figs. S1–S6.
We also measured fimbrial lengths from the electron micrographs and found that the T-6N/V-10I and V-4E mutants had significantly longer fimbriae than WT (19% and 27% longer, respectively; Fig. 2D). Atomic force microscopy (AFM) was used as an additional method of fimbrial length measurement by pulling fimbriae via FimH attached to a cantilever and determining the length of uncoiled fimbriae. Average lengths were 20%, 56%, and 55% greater than WT for the T-6N, T-6N/V-10I, and V-4E mutants, respectively. At the end of a subset of pulls, some cantilever-attached fimbriae failed to detach because their uncoiled length was greater than the extent of the instrument (20 μm). Such fimbriae were not included in the length measurements, but the T-6N/V-10I and V-4E variants had a significantly greater number of fimbriae that failed to detach than WT (Fig. 2F), indicating that the average lengths in Fig. 2E are underestimated to a greater extent for these mutants.
SP Mutations Result in Decreased Attachment Under Flow.
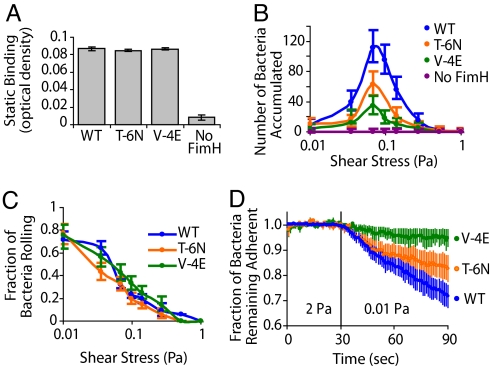
When bacterial binding to a mannose-BSA-coated surface was tested under static conditions, no difference between WT and mutant strains was detected (Fig. 3A). However, in the presence of shear stress generated by fluid flow, the accumulation rate of mutant bacteria was significantly decreased relative to WT, corresponding to the decreased number of surface FimH (Fig. 3B). All strains demonstrated a shear-enhanced, catch bond mode of binding that peaked at the same shear stress (0.07 Pa; Fig. 3B) and a similar rate of shear-induced conversion from a loosely attached, rolling mode of adhesion to a firmly bound, stationary mode (Fig. 3C).
Fig. 3.
Static and dynamic bacterial adhesion to mannose-coated surfaces. Bacteria express FimH with different SPs or have no FimH. (A) Static binding by growth assay. Error bars represent SEM. (B) Surface accumulation of bacteria over 5 min at different shear stresses. Error bars represent the Poisson distribution 95% C.I. (C) Fraction of bacteria rolling on the surface relative to surface accumulated bacteria, which includes rolling and stationary. Error bars represent the binomial distribution 95% C.I. (not included where counts ≤1, which occurred only at the highest two shear levels). (D) Maintenance of adhesion after bacteria were bound to the surface statically, then subjected to high (2 Pa) and low (0.01 Pa) flow. Error bars at each second represent the SEM of three independent experiments. At the 90-s time point, a t test comparing WT and V-4E is significant (P = 0.015).
To test whether SP mutant bacteria exhibit increased detachment rates under flow conditions, bacteria that were attached to the surface were subjected to high shear stress (2 Pa). For all variants, there was no detectable detachment during 30 s of observation under high shear stress (Fig. 3D; 0–30 s).
SP Mutations Result in Sustained Strong Binding upon a Drop in Shear Stress.
Catch bonds not only switch from low- to high-affinity binding under increased shear stress, but also switch back from high- to low-affinity binding upon decreased shear stress, resulting in detachment. To test whether the mutations affect detachment after a drop in shear stress, the adherent bacteria that were subjected above to high shear stress (2 Pa) were abruptly switched to low shear stress conditions of 0.01 Pa. As expected, bacteria with WT FimH SP gradually detached from the surface after the drop in shear stress (Fig. 3D; 30–90 s). Surprisingly, the mutants detached from the surface more slowly. Most significant were the V-4E mutant bacteria, of which 95% remained attached and stationary after 1 min of low shear stress versus 72% of WT and 83% of the T-6N mutant. Thus, despite having fewer fimbriae, the mutant bacteria are more capable than WT of sustaining a strong binding mode upon a switch from high to low shear stress.
SP Mutations Do Not Dramatically Affect Binding to Bladder Epithelial Cells but Decrease Killing by Neutrophils.
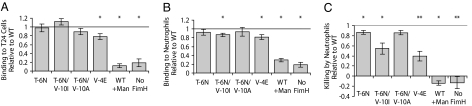
In vitro binding to T24 bladder epithelial cells was compared for FimH SP mutants versus WT. None of the naturally occurring mutants bound significantly differently to bladder epithelial cells than WT. Only the synthetic mutant, V-4E, showed a significant difference with a 22% decrease (Fig. 4A). Binding to human neutrophils was tested and found to be decreased from the WT level for the T-6N/V-10I and V-4E mutants (15 and 19% decreases, respectively; Fig. 4B). Bacterial killing by human neutrophils was also compared for all variants, and several mutants were more resistant to killing; the T-6N, T-6N/V-10I, and V-4E mutants were killed significantly less than WT, with 15%, 45%, and 60% decreases in killing, respectively (Fig. 4C).
Fig. 4.
Interactions with bladder epithelial cells and human neutrophils. WT was additionally tested in the presence of soluble mannose (+Man). (A) Adhesion to T24 bladder epithelial cells. (B) Binding to neutrophils. (C) Killing by neutrophils. For each strain, fraction killed was measured as one minus the fraction of bacteria surviving incubation with neutrophils relative to a growth control, then the ratio to WT was taken. All error bars represent SEM. *, P < 0.05; **, P < 0.01.
Positive Selection of SP Mutations in Another Type 1 Fimbrial Subunit, FimG.
Phylogenetic analysis of signal sequence mutations in all genes of the fimAICDFGH operon revealed an abundance of nonsynonymous (n = 6) relative to synonymous (n = 0) mutations in fimG, the minor tip subunit that is added immediately after FimH during fimbriae biogenesis (Table 1). This finding suggests positive selection for replacements in the FimG SP. The FimG SP mutations are predicted to decrease transport function based on amino acid properties and the SignalP hidden Markov model (26). When the occurrence of FimH SP mutations was analyzed in strains sequenced for FimG, the vast majority of the mutant FimH SP alleles were found to occur in strains that also have FimG SP mutations predicted to decrease transport (Table S1). In contrast, most strains with FimH SP mutations have the phylogenetically primary SP haplotype of the other fim genes.
Table 1.
Signal sequence analysis of type 1 fimbrial cluster genes
| Feature | Gene |
||||||
|---|---|---|---|---|---|---|---|
| fimA | fimI | fimC | fimD | fimF | fimG | fimH | |
| Function | Rod subunit | Unknown | Chaperone | Usher | Tip subunit | Tip subunit | Adhesin |
| No. of sequences analyzed | 365 | 179 | 215 | 20 | 87 | 102 | 365 |
| SP length, aa | 23 | 19 | 36 | 37 | 22 | 23 | 21 |
| #NS/#S | 6/12 | 2/4 | 6/3 | 3/6 | 2/4 | 6/0 | 6/0 |
#NS/#S, number of nonsynonymous to synonymous signal sequence mutations.
Discussion
This study demonstrates that naturally occurring FimH SP mutations are loss-of-function in nature, as they diminish the translocation efficiency of FimH across the inner membrane, leading to fewer fimbriae expressed on the bacterial surface and decreased binding under flow conditions. Paradoxically, the mutations also result in a gain-of-function phenotype, an increase in the adhesive strength upon a sudden drop in flow, that is likely caused by the increased fimbrial length. In addition, the mutations lead to an increased resistance to killing by neutrophils. Both of these gain-of-function phenotypes could lead to increased uropathogenicity of FimH SP mutant bacteria.
E. coli has acquired the T-6N mutation at least four times, which is strong evidence of positive selection. Consistent with the computational prediction, the T-6N mutation decreased transport of the reporter gene alkaline phosphatase. The two other mutations, V-10I and V-10A, occurred at the same position and on the T-6N background, which is also evidence of positive selection. Although both mutations at position −10 decrease hydrophobicity, V-10I resulted in a further loss-of-function, whereas V-10A had a compensatory effect to T-6N. The error in computational prediction regarding the V-10A mutation suggests there are other factors in addition to hydrophobicity important for function. Although the natural mutations that decrease transport have a moderate effect (within 26–37%), even subtle changes in fitness can have profound adaptive effects under physiologic conditions over many generations. However, to ease experimental dissection of the effects of the natural mutations, we constructed an additional mutation in the SP, V-4E, with a more significant loss-of-function effect than the natural mutations.
Decreased transport of FimH across the inner membrane resulted in fewer FimH on the bacterial surface and fewer fimbriae, as one would expect because FimH initiates fimbriae formation. At the same time, however, fimbrial length measurements by EM and AFM showed longer fimbriae for the mutants. Although it is unknown whether a mechanism exists for controlling type 1 fimbrial length, studies here indicate that type 1 fimbrial length is determined, at least in part, by the ratio of FimA to FimH in the periplasm. Although the FimH SP mutations decrease transport of FimH, they are not expected to affect transport of FimA. As a result, FimA:FimH in the periplasm would be increased, translating into more FimA incorporated per individual fimbria and consequently longer fimbriae.
The fact that under static conditions bacterial binding to mannose was not affected by SP mutations is likely because the number of FimH-mannose bonds formed was high enough in all strains to resist postincubation bacterial removal by washing. Under various levels of shear stress, however, SP mutants adhered significantly less than WT. This lower accumulation rate is likely caused by decreased on rates of attachment of mutant bacteria, with fewer adhesins translating into fewer opportunities to initiate adhesion under the fast dynamics of bond formation under flow. At the same time, fewer adhesins did not affect the adhesion mode (i.e., rolling vs. stationary) of surface-attached bacteria, suggesting that the lowered attachment rates have a negligible effect on maintaining bacterial attachment to the surface under continuous flow, likely because this step does not require as fast of a bond-formation rate as during adhesion initiation. A similar difference in the dependence of adhesion initiation and maintenance on fast on-rates was noted previously (27).
Despite the lowered accumulation rate, the shear-enhanced, catch bond properties of the FimH adhesin do not appear to be affected by the SP mutations, based on the similarity in the level of shear stress where surface accumulation peaks and in the conversion rates from rolling to stationary under increased shear. The FimH catch bond is allosteric in nature, with separation of the lectin and pilin domains under shear-induced tensile force resulting in a shift of the mannose-binding site from a low- to high-affinity conformation (20). Although the details of the conformational link between the interdomain configuration and the binding site are unclear, the allosteric properties of FimH are strongly supported by the presence in FimH of an integrin-like ligand-induced binding site epitope for monoclonal antibodies and by single-molecule studies using AFM (28). This catch bond mechanism was shown to provide a means for resistance of bacterial adhesion to soluble inhibitors and for surface spreading in biofilm formation (29).
One of the characteristics of the FimH catch bond is its reversibility upon a drop in shear stress, whereby in the absence of a strong tensile force, interaction of the domains is restored and the binding site shifts from the high- to low-affinity configuration (30, 31). Consequently, firmly bound bacteria detach from the surface when shear changes from high to low. Surprisingly, this rate of detachment after a drop in shear stress was significantly reduced in the SP mutant bacteria. In other words, paradoxically, a decreased number of adhesins is accompanied by an increased strength of bacterial adhesion. Because the functional properties of individual FimH adhesins do not appear to be affected by SP mutations, the increased adhesion could be attributed to the mutants' longer fimbriae. In a recent study, type 1 fimbriae were shown to uncoil under high tensile force and recoil back upon a drop in force (21). This recoiling is proposed to maintain optimal force on the FimH-mannose bond, slowing FimH's shift from the high- to low-affinity conformation. This could be a mechanism for resisting bacterial detachment from the surface in environments where shear stress frequently fluctuates. The increased adhesion of the SP mutants relative to WT upon a drop in shear stress could be caused by longer uncoiled fimbriae taking more time to recoil, increasing the bond lifetimes. Also, longer fimbriae might allow for farther uncoiling before additional fimbriae bind the surface, especially considering the mutants' decreased surface FimH. When the flow is decreased and the fimbriae recoil, the mutants could be anchored between fimbriae pulling in opposite directions, maintaining force on the FimH bonds and preventing the bacteria from detaching (illustrated in Fig. S7). Regardless of the underlying mechanism, this is an unexpected gain-of-function effect of loss-of-function SP mutations. It also suggests that the fimbrial rod has a direct impact on adhesive function and that its length is a finely adapted property optimized for the shear-enhanced, catch bond mechanism of FimH-mediated adhesion.
It is possible that sustaining strong binding upon an abrupt decrease in shear stress could provide an advantage to bacteria in certain environments with oscillating shear stress, such as compartments of the urinary tract. There could also be other physiologic consequences of FimH SP mutations. Capsules have been shown to block adhesion (32) and, if a thicker capsule provides an advantage under certain conditions, longer fimbriae could preserve the adhesive phenotype. Also, the surface of bacteria and certain target cells could have similar electric charges, and longer fimbriae could accomplish adhesion more efficiently under cell–cell repulsive conditions. Additional studies are needed to evaluate the many potential adaptive benefits of longer fimbriae.
Some direct evidence for a possible advantage of SP mutations in the urinary tract was obtained from the studies with bladder epithelial cells and human neutrophils. The natural SP mutations do not affect in vitro adhesion to bladder epithelial cells under the assay conditions, which were static. Because a static or low shear stress environment is likely typical within the bladder between urinations, the mutants' fewer fimbriae may not result in significantly decreased tropism to the bladder uroepithelium. However, the synthetic mutation significantly decreased uroepithelial cell binding, suggesting that if the natural mutations had a stronger effect they, too, would decrease binding. In contrast, both the synthetic and a naturally occurring SP mutant bound significantly less efficiently to human neutrophils. One reason for the difference in binding to bladder epithelial cells and neutrophils could be the assay conditions, which in the case of neutrophil binding included intermittent, low shear stress from sample inversion (once every 10 min). Under physiological conditions in the urinary tract, interactions between bacteria and neutrophils are also likely to occur under some shear stress, considering encounters in the liquid (urine) phase and active movement of defense cells along the uroepithelium surface. Another possible explanation for the small difference in binding to uroepithelial cells and neutrophils is a structural or quantitative difference in the mannosylated receptors on the different cell types. Decreased mutant binding to neutrophils could be the reason for their increased resistance to neutrophil-mediated killing, although the role of their longer fimbriae in reducing killing cannot be excluded. Neutrophils migrate to and across bladder epithelium at the first sign of infection to phagocytose and kill bacteria. The increased survival to human neutrophil-mediated killing exhibited by mutants with fewer and longer fimbriae could be highly advantageous.
While possibly benefiting E. coli survival in and colonization of the urinary tract, the FimH SP mutations could be detrimental to E. coli in other niches and/or in the long term. Indeed, fimH alleles with SP mutations appear to circulate only for a short time from an evolutionary perspective, as they do not accumulate silent changes. In this respect, the mutant SP alleles are similar to mutant mature protein alleles that enhance FimH's ability to bind at low shear conditions and are also selected in uropathogenic E. coli (23, 33). A tradeoff of mature protein mutations could be a reduced resistance to adhesion inhibition by salivary proteins in the course of oropharyngeal colonization as a part of the fecal-oral transmission of E. coli (16). A tradeoff of SP mutations could be decreased adhesion under shear stress created by flow of high viscosity fluids bathing the oropharyngeal or other mucosal surfaces. Interestingly, the allele with the V-10A compensatory mutation is carried by a nonpathogenic, fecal isolate, supporting the overall negative effect of SP loss-of-function mutations for commensal ecology.
Finally, we found evidence for positive selection in the SP of FimG, the tip-associated subunit added immediately after FimH in the course of fimbrial biogenesis. The phenotype of the wild strains with FimH SP mutations is likely to be enhanced by loss-of-function FimG SP mutations, suggesting that these mutations are coselected for the same phenotype. Although SP mutations were found in other fimbrial subunits as well, evidence for positive selection in these genes is weak, and FimH mutant alleles occur predominantly with the primary, nonmutated forms of these genes. Thus, there is no evidence for compensatory effects between FimH SP mutations and such mutations in other subunits, including the major subunit FimA. Beyond type 1 fimbriae, it is possible that SP mutations could be positively selected in other fimbriae or surface organelles, providing a common mechanism for regulating membrane transport and surface expression of structural proteins.
Materials and Methods
Strains and Plasmids.
Strains are listed in Table S2. fimH signal sequence-alkaline phosphatase (phoA) fusions were constructed in pJDT3 (34), and the plasmids were introduced into ΔphoA strain CC191 (35) to generate strains LS4-4-20 (WT FimH SP), LS4-4-1 (T-6N), LS4-4-27 (T-6N/V-10I), LS3-31-2 (T-6N/V-10A), and LS4-4-8 (V-4E). Strain CC191pJDT3 has pJDT3 without the fimH signal sequence. For the morphology, adhesion, and neutrophil studies we used strains based on fimH-null UTI isolate CI10-9 (16) with pPKL9, which contains the positive type 1 fimbrial regulator, FimB (33). fimH alleles, all with the same mature protein coding sequence and different signal sequences, were introduced into CI10-9 on pBeloBAC11 (New England Biolabs), generating strains LS76 (WT FimH SP), LS60 (T-6N), LS68 (T-6N/V-10I), LS154 (T-6N/V-10A), and LS141 (V-4E). Thus, the strains are isogenic other than the FimH SP. LSpBB has pBeloBAC11 without fimH. For all experiments the strains were grown in super broth statically overnight at 37°C with appropriate antibiotics and then washed with PBS. To control for possible inconsistencies in gene expression, two separate strains were constructed for each SP allele and tested in parallel for all experiments. No discrepancies between corresponding strains were found and the results were combined to enhance statistical resolution.
Phylogenetic Tree Building.
fim gene sequences, including those from publicly available and in-house sequences, were used to construct maximum-likelihood DNA trees by using PAUP* 4.0b (36).
Alkaline Phosphatase Assay.
Assays were performed on strains CC191pJDT3, LS4-4-20, LS4-4-1, LS4-4-27, LS3-31-2, and LS4-4-8, and units of activity were calculated as described (37) (see also SI Text).
Flow Cytometry.
Flow cytometry was performed essentially as described (38), labeling FimH on LS76, LS60, LS68, LS154, LS141, and LSpBB using primary antibodies against FimH and FITC-conjugated secondary antibodies (see also SI Text).
EM.
Grids were prepared as described (39). They were examined with a Philips 410 transmission electron microscope, and digital images were taken by operators blind to strain identity. Numbers and lengths of type 1 fimbriae were determined by using AlphaEaseFC software and were reproducible by two individuals blind to strain identity. The number of cells analyzed per strain were: 20 WT, 17 T-6N, 14 T-6N/V-10I, 15 T-6N/V-10A, and 15 V-4E.
AFM.
Surface and tip functionalization with mannosylated protein (bovine ribonuclease B), bacterial adherence to the surface, and cantilever spring constant calibration were performed as described (21). All pulls were performed at 2 μm/s over 20 μm using the same cantilever. Uncoiled fimbrial length was measured as the distance from the bacterial surface to the location of FimH bond rupture, which is detected by a characteristic spike in force (21). The number of areas interrogated for each strain were: 47 WT, 39 T-6N, 12 T-6N/V-10I, 20 T-6N/V-10A, and 20 V-4E.
Binding to Mannose-Coated Surfaces.
d-mannose-BSA (V-Labs) and strains LSpBB, LS76, LS60, and LS141 were used. Static binding was performed as described (40). Bacterial surface accumulation over 5 min and fraction rolling were measured as described (29). For detachment experiments, bacteria at 2 × 109 cells/ml were allowed to briefly bind statically to the flow chamber surface. Flow with PBS–BSA buffer alone was applied at 0.27 Pa to remove free-floating bacteria. Flow was applied at 2 Pa for 1 min to promote high-affinity binding and fimbrial uncoiling. Then, flow was decreased to 0.01 Pa and the percentage of bacteria that remained attached at each second relative to the average number attached over the last 30 s of the 2-Pa section was calculated.
Adhesion to T24 Uroepithelial Cells.
Strains LS76, LS60, LS68, LS154, LS141, and LSpBB were tested as described (41) (see also SI Text).
Isolation of Human Neutrophils.
According to a protocol approved by the University of Washington Human Subjects Division, human neutrophils were isolated from venous blood of volunteers under informed consent as described (42) (see also SI Text).
Neutrophil Bactericidal and Adherence Assays.
Killing and binding of strains LS76, LS60, LS68, LS154, LS141, and LSpBB were measured as described (43) (see also SI Text).
Statistical Analysis.
Unless otherwise noted, statistical analyses were performed by using ANOVA and, if significant, posthoc two-sided Welch two-sample t tests comparing each mutant with WT.
Supplementary Material
Acknowledgments.
We thank James Ronald, Steve Moseley, and Colin Manoil for helpful discussions and members of E.V.S.'s laboratory for donating blood. L.S.R. was supported by the National Institutes of Health Medical Scientist Training Program, a National Science Foundation Integrative Graduate Education and Research Traineeship, and the University of Washington University Initiatives Fund. This work was supported by National Institutes of Health Grants 5R01AI045820-07 and 2R01AI050940-05 (to E.V.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803158105/DCSupplemental.
References
- 1.Von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983;133:17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- 2.Nesmeyanova MA, et al. Positively charged lysine at the N terminus of the signal peptide of the Escherichia coli alkaline phosphatase provides the secretion efficiency and is involved in the interaction with anionic phospholipids. FEBS Lett. 1997;403:203–207. doi: 10.1016/s0014-5793(97)00052-5. [DOI] [PubMed] [Google Scholar]
- 3.Doud SK, Chou MM, Kendall DA. Titration of protein transport activity by incremental changes in signal peptide hydrophobicity. Biochemistry. 1993;32:1251–1256. doi: 10.1021/bi00056a008. [DOI] [PubMed] [Google Scholar]
- 4.Feng J, et al. High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci Lett. 2006;409:10–13. doi: 10.1016/j.neulet.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 5.Kiraly O, et al. Signal peptide variants that impair secretion of pancreatic secretory trypsin inhibitor (SPINK1) cause autosomal dominant hereditary pancreatitis. Hum Mutat. 2007;28:469–476. doi: 10.1002/humu.20471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mukherjee O, et al. HDDD2 is a familial frontotemporal lobar degeneration with ubiquitin-positive, tau-negative inclusions caused by a missense mutation in the signal peptide of progranulin. Ann Neurol. 2006;60:314–322. doi: 10.1002/ana.20963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guarnieri M, et al. Point mutations upstream of hepatitis B virus core gene affect DNA replication at the step of core protein expression. J Virol. 2006;80:587–595. doi: 10.1128/JVI.80.2.587-595.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilder JA, et al. Contrasting modes of natural selection acting on pigmentation genes in the Drosophila dunni subgroup. J Exp Zool B Mol Dev Evol. 2004;302:469–482. doi: 10.1002/jez.b.21012. [DOI] [PubMed] [Google Scholar]
- 9.Atherton JC, et al. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 10.Nishiyama M, et al. Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD. EMBO J. 2005;24:2075–2086. doi: 10.1038/sj.emboj.7600693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Remaut H, et al. Donor-strand exchange in chaperone-assisted pilus assembly proceeds through a concerted beta strand displacement mechanism. Mol Cell. 2006;22:831–842. doi: 10.1016/j.molcel.2006.05.033. [DOI] [PubMed] [Google Scholar]
- 12.Verger D, Miller E, Remaut H, Waksman G, Hultgren S. Molecular mechanism of P pilus termination in uropathogenic Escherichia coli. EMBO Rep. 2006;7:1228–1232. doi: 10.1038/sj.embor.7400833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abraham JM, Freitag CS, Clements JR, Eisenstein BI. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:5724–5727. doi: 10.1073/pnas.82.17.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagberg L, et al. Adhesion, hemagglutination, and virulence of Escherichia coli causing urinary tract infections. Infect Immun. 1981;31:564–570. doi: 10.1128/iai.31.2.564-570.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connell I, et al. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci USA. 1996;93:9827–9832. doi: 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sokurenko EV, et al. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc Natl Acad Sci USA. 1998;95:8922–8926. doi: 10.1073/pnas.95.15.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langermann S, et al. Vaccination with FimH adhesin protects cynomolgus monkeys from colonization and infection by uropathogenic Escherichia coli. J Infect Dis. 2000;181:774–778. doi: 10.1086/315258. [DOI] [PubMed] [Google Scholar]
- 18.Poggio TV, La Torre JL, Scodeller EA. Intranasal immunization with a recombinant truncated FimH adhesin adjuvanted with CpG oligodeoxynucleotides protects mice against uropathogenic Escherichia coli challenge. Can J Microbiol. 2006;52:1093–1102. doi: 10.1139/w06-065. [DOI] [PubMed] [Google Scholar]
- 19.Ofek I, Goldhar J, Keisari Y, Sharon N. Nonopsonic phagocytosis of microorganisms. Annu Rev Microbiol. 1995;49:239–276. doi: 10.1146/annurev.mi.49.100195.001323. [DOI] [PubMed] [Google Scholar]
- 20.Yakovenko O, et al. Fimh forms catch bonds that are enhanced by mechanical force due to allosteric regulation. J Biol Chem. 2008;283:11596–11605. doi: 10.1074/jbc.M707815200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forero M, Yakovenko O, Sokurenko EV, Thomas WE, Vogel V. Uncoiling mechanics of Escherichia coli type I fimbriae are optimized for catch bonds. PLoS Biol. 2006;4:e298. doi: 10.1371/journal.pbio.0040298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin DP, Williamson C, Posada D. RDP2: Recombination detection and analysis from sequence alignments. Bioinformatics. 2005;21:260–262. doi: 10.1093/bioinformatics/bth490. [DOI] [PubMed] [Google Scholar]
- 23.Sokurenko EV, et al. Selection footprint in the FimH adhesin shows pathoadaptive niche differentiation in Escherichia coli. Mol Biol Evol. 2004;21:1373–1383. doi: 10.1093/molbev/msh136. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen H, Krogh A. Prediction of signal peptides and signal anchors by a hidden Markov model. Proc Int Conf Intell Syst Mol Biol. 1998;6:122–130. [PubMed] [Google Scholar]
- 25.Engelman DM, Steitz TA, Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 26.Hoffman CS, Wright A. Fusions of secreted proteins to alkaline phosphatase: An approach for studying protein secretion. Proc Natl Acad Sci USA. 1985;82:5107–5111. doi: 10.1073/pnas.82.15.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nilsson LM, et al. The cysteine bond in the Escherichia coli FimH adhesin is critical for adhesion under flow conditions. Mol Microbiol. 2007;65:1158–1169. doi: 10.1111/j.1365-2958.2007.05858.x. [DOI] [PubMed] [Google Scholar]
- 28.Tchesnokova V, et al. Integrin-like allosteric properties of the catch-bond forming FimH adhesin of E. coli. J Biol Chem. 2008;283:7823–7833. doi: 10.1074/jbc.M707804200. [DOI] [PubMed] [Google Scholar]
- 29.Anderson BN, et al. Weak rolling adhesion enhances bacterial surface colonization. J Bacteriol. 2007;189:1794–1802. doi: 10.1128/JB.00899-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aprikian P, et al. Interdomain interaction in the FimH adhesin of Escherichia coli regulates the affinity to mannose. J Biol Chem. 2007;282:23437–23446. doi: 10.1074/jbc.M702037200. [DOI] [PubMed] [Google Scholar]
- 31.Thomas WE, Nilsson LM, Forero M, Sokurenko EV, Vogel V. Shear-dependent “stick-and-roll” adhesion of type 1 fimbriated Escherichia coli. Mol Microbiol. 2004;53:1545–1557. doi: 10.1111/j.1365-2958.2004.04226.x. [DOI] [PubMed] [Google Scholar]
- 32.Hanna A, Berg M, Stout V, Razatos A. Role of capsular colanic acid in adhesion of uropathogenic Escherichia coli. Appl Environ Microbiol. 2003;69:4474–4481. doi: 10.1128/AEM.69.8.4474-4481.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sokurenko EV, Courtney HS, Maslow J, Siitonen A, Hasty DL. Quantitative differences in adhesiveness of type 1 fimbriated Escherichia coli due to structural differences in fimH genes. J Bacteriol. 1995;177:3680–3686. doi: 10.1128/jb.177.13.3680-3686.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mdluli KE, Treit JD, Kerr VJ, Nano FE. New vectors for the in vitro generation of alkaline phosphatase fusions to proteins encoded by G+C-rich DNA. Gene. 1995;155:133–134. doi: 10.1016/0378-1119(94)00909-c. [DOI] [PubMed] [Google Scholar]
- 35.Lee E, Manoil C. Mutations eliminating the protein export function of a membrane-spanning sequence. J Biol Chem. 1994;269:28822–28828. [PubMed] [Google Scholar]
- 36.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 2003. Version 4. [Google Scholar]
- 37.Michaelis S, Inouye H, Oliver D, Beckwith J. Mutations that alter the signal sequence of alkaline phosphatase in Escherichia coli. J Bacteriol. 1983;154:366–374. doi: 10.1128/jb.154.1.366-374.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Humphries AD, et al. The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Mol Microbiol. 2003;48:1357–1376. doi: 10.1046/j.1365-2958.2003.03507.x. [DOI] [PubMed] [Google Scholar]
- 39.Hahn E, et al. Exploring the 3D molecular architecture of Escherichia coli type 1 pili. J Mol Biol. 2002;323:845–857. doi: 10.1016/s0022-2836(02)01005-7. [DOI] [PubMed] [Google Scholar]
- 40.Sokurenko EV, Chesnokova V, Doyle RJ, Hasty DL. Diversity of the Escherichia coli type 1 fimbrial lectin. Differential binding to mannosides and uroepithelial cells. J Biol Chem. 1997;272:17880–17886. doi: 10.1074/jbc.272.28.17880. [DOI] [PubMed] [Google Scholar]
- 41.Zunino P, et al. Proteus mirabilis fimbriae (PMF) are important for both bladder and kidney colonization in mice. Microbiology. 2003;149:3231–3237. doi: 10.1099/mic.0.26534-0. [DOI] [PubMed] [Google Scholar]
- 42.Voyich JM, DeLeo FR. Host–pathogen interactions: Leukocyte phagocytosis and associated sequelae. Methods Cell Sci. 2002;24:79–90. doi: 10.1023/a:1024154200702. [DOI] [PubMed] [Google Scholar]
- 43.Nazareth H, Genagon SA, Russo TA. Extraintestinal pathogenic Escherichia coli survives within neutrophils. Infect Immun. 2007;75:2776–2785. doi: 10.1128/IAI.01095-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.