Abstract
Molecular self-assembly is a promising approach to the preparation of nanostructures. DNA, in particular, shows great potential to be a superb molecular system. Synthetic DNA molecules have been programmed to assemble into a wide range of nanostructures. It is generally believed that rigidities of DNA nanomotifs (tiles) are essential for programmable self-assembly of well defined nanostructures. Recently, we have shown that adequate conformational flexibility could be exploited for assembling 3D objects, including tetrahedra, dodecahedra, and buckyballs, out of DNA three-point star motifs. In the current study, we have integrated tensegrity principle into this concept to assemble well defined, complex nanostructures in both 2D and 3D. A symmetric five-point-star motif (tile) has been designed to assemble into icosahedra or large nanocages depending on the concentration and flexibility of the DNA tiles. In both cases, the DNA tiles exhibit significant flexibilities and undergo substantial conformational changes, either symmetrically bending out of the plane or asymmetrically bending in the plane. In contrast to the complicated natures of the assembled structures, the approach presented here is simple and only requires three different component DNA strands. These results demonstrate that conformational flexibility could be explored to generate complex DNA nanostructures. The basic concept might be further extended to other biomacromolecular systems, such as RNA and proteins.
Keywords: icosahedron, three-dimensional, polyhedron, cryo-EM, molecular cages
Molecular self-assembly provides a bottom-up approach to the preparation of nanostructures (1–3). DNA, in particular, shows great potential to be a superb molecular system (4). In the last 20 years, DNA has been explored as building blocks for nanoconstructions, including preparation of periodic and aperiodic 2D nanopatterns (5–8) and 3D polyhedra (9–14). Most of the branched DNA structures are intrinsically flexible and are not suitable building blocks for construction of well defined geometric structures. How to overcome the conformational flexibility of branched DNA structures is a major challenge in structural DNA nanotechnology. In the last decade, a series of rigid structural motifs have been successfully engineered that lead to the rapid evolution of structural DNA nanotechnology (4). However, with more experience and knowledge, it is possible to controllably introduce the conformational flexibility to prepare complex DNA nanostructures (15). In our recent study of 3D self-assembly of DNA three-point-star tiles (16), we found that DNA tetrahedra could be readily assembled, and the tetrahedra are well behaved during sample characterizations. In contrast, DNA dodecahedra and buckyballs have significantly lower assembly yields and are prone to deformation. This phenomenon can be explained by the geometrical differences of these structures. Tetrahedra consist of triangular faces, but others do not. According to tensegrity principle, triangular faces will lead to rigid structures (11, 12, 17–20). This fact prompts us to integrate the tensegrity principle into DNA self-assembly when preparing large DNA polyhedra: DNA polyhedra should contain only triangular faces. Such structures would be expected to be rigid and resistant to deformations and will have high assembly yields. To test this hypothesis, we have designed a five-point-star motif to assemble DNA icosahedra that have 20 triangular faces and 12 five-branched vertices. With slight modification, the same motif can assemble into large molecular cages. In the latter case, the DNA tile goes an unexpected and dramatic structural change that highlights the benefit of the structural flexibility of DNA nanomotifs.
Results and Discussion
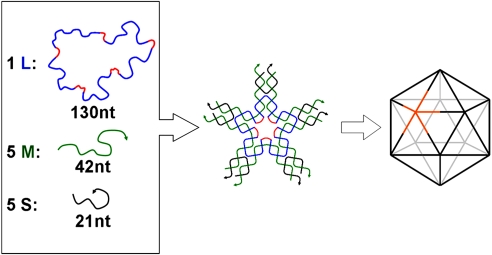
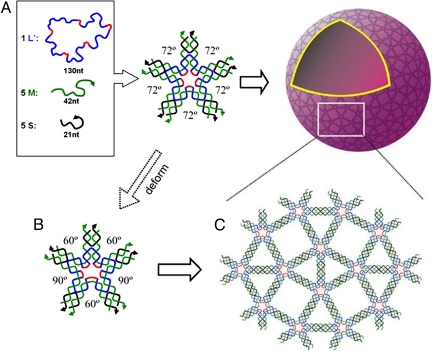
The five-point-star motif (tile) is a previously uncharacterized DNA motif. It belongs to a family of star motifs that include three-point-star motifs (15, 21), four-point-star (cross) motifs (6, 22), and six-point-star motifs (23). In icosahedra, each five-point-star tile will correspond to a five-way branched vertex (Fig. 1). The five-point-star motif contains a 5-fold rotational symmetry that goes through the center of the motif. This rotational symmetry relates the five branches of the motif and simplifies the motif. Although the motif consists of 11 strands, there are only three types: a long, repetitive, central blue-red strand (L), five identical medium green strands (M), and five identical short peripheral black strands (S). At the center of the tile, strand L contains five identical, single-stranded loops (colored red), which determine the tile flexibility. The longer the loops are, the more flexible the tile is. At the peripheral termini of each branch of the tile, there are two complementary, single-stranded overhangs, or sticky ends. Sticky-end association between tiles will lead to large structures.
Fig. 1.
Self-assembly of DNA icosahedra. Three different types of DNA single strands stepwise assemble into sticky-ended five-point-star motifs (tiles), which then further assemble into icosahedra. Each vertex in the icosahedra is a five-point-star tile; one of them is highlighted as golden. Note that the red colored central loops are 5 bases long.
In this work, DNA self-assembly is a one-pot process. When being mixed and slowly cooled from 95°C to 23°C, individual component DNA single strands first recognize each other and assemble into individual five-point-star tiles that further assemble into icosahedra through sticky-end association between the tiles. Icosahedra are the smallest closed structure for five-point-star motif without bending DNA duplexes. DNA duplexes are quite stiff at the length scales used in this work; their bending would be energetically unfavorable. Each vertex is one star tile, and each edge is composed of two associated branches from two tiles. When designing the tiles for icosahedron structures, the following factors might be important. (i) In the content of icosahedron structure, the five-point-star tiles are not planar. Instead, all component branches will bend out of the original plane by ≈32°, and the tiles become a cone shape with a 5-fold rotational symmetry. To ensure the tiles to have enough flexibility for such a degree of bending, the central single-stranded loops (red color segments in Fig. 1) are designed to be 5 bases long. (ii) Any two adjacent vertexes are separated by two parallel duplexes, whose lengths are 42 base pairs or four turns. Because of the helical nature of DNA duplexes, the repeating distance with integral number (here it is four) of helical turns will accumulate any curvature intrinsic to the DNA tiles in the same direction, which will promote the formation of closed complexes (6). (iii) Each sticky end consists of four G-C base pairs. The strong G-C base pairing imposes kinetic controls in the assembly process and is expected to favor closed complexes (24). It also ensures the stability of the final assemblies. (iv) Finally, a low DNA concentration (20 nM) is used to prevent large assemblies from forming.
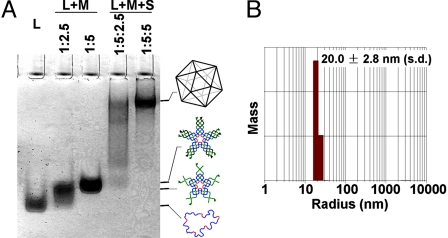
The assembled DNA icosahedron has been characterized by multiple techniques, including native PAGE, dynamic light scattering (DLS), and cryogenic electron microscopy (cryo-EM) imaging. When a correct molecular ratio is used, only one band appears on the native PAGE, which has much slower mobility than the five-point-star tile itself (Fig. 2A). It suggests that the tiles assemble into a large, well defined, molecular complex. DLS studies reveal that the DNA complex has an apparent hydrodynamic radius of 20.0 ± 2.8 nm (Fig. 2B). This value agrees well with the radius of the circumscribed sphere of the expected DNA icosahedron (19.5 nm), assuming that a DNA duplex has a pitch of 0.33 nm/base pair and a diameter of 2 nm.
Fig. 2.
Characterization of the self-assembled DNA iscosahedron. (A) Native PAGE (2.5%) analysis. The sample compositions are indicated above the gel image, and the identity of each band is suggested on the right. (B) DLS analysis of the mass distribution along the hydrodynamic radius of the DNA complexes.
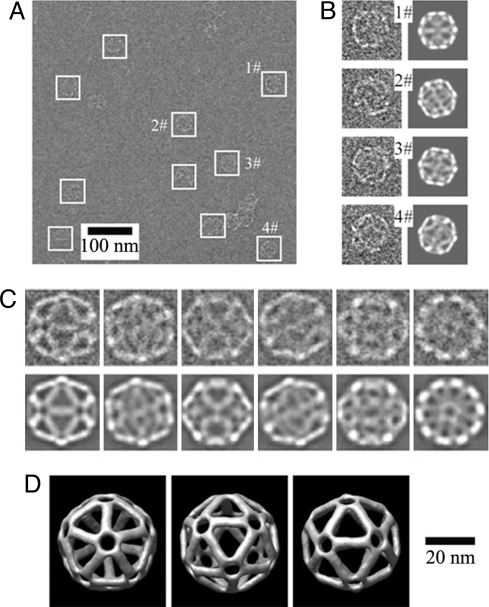
To provide direct evidence of the formation of DNA icosahedra, we have imaged the DNA samples by cryo-EM (Fig. 3). Flash-freezing is likely to keep the DNA complexes in their native conformations. Most particles observed in cryo-EM images have icosahedral shapes of the expected size. The observed diameters are ≈40 nm, nicely matching the diameter of designed icosahedron (39.0 nm). With experimentally observed particles, a DNA icosahedron structure is revealed by a technique of 3D single-particle reconstruction (25), which is a technique routinely used in studies of virus structures. The resolution of the icosahedral structure is 2.8 nm, as determined by Fourier shell correlation (25). A strong support for the reconstructed model comes from the comparison between the computed projections from model and the class averages of raw particle images with similar views [Fig. 3C and supporting information (SI) Fig. S1]. They match each other very well. Because the contrast of the DNA particles in cryo-EM is very low, it is necessary to impose the intrinsic structural symmetry (here, icosahedral symmetry) during the reconstruction to increase the resolution. Relaxed with lower symmetries (for example, C5, a 5-fold rotational symmetry), the reconstruction generates a similar structure with noisier signals (Fig. S2). It confirms that the assembled DNA complexes indeed have the icosahedral structure.
Fig. 3.
Cryogenic transimission electron microscopy (cryo-EM) analysis of DNA icosahedron. (A) A representative raw cryo-EM image. White boxes indicate the DNA particles. (B) Comparison of raw images of individual particles at a high magnification (Left) and the corresponding computer-generated model projections (Right). (C) Comparison of class average of particle images with similar views (Upper) and the corresponding computer-generated model projections (Lower). (D) Three views of the DNA icosahedron structure reconstructed from cryo-EM images.
DNA icosahedra are a class of interesting structures for their rigid geometry, high symmetry, and resemblance to spherical viral capsids. Different potential strategies have been proposed. For example, based on his recently developed concept of “DNA origami”, Paul Rothemund (26) has proposed the use of many DNA short strands to fold thousands-bases-long single DNA strands (i.e., M13 genomic DNA) into polyhedra. The idea is very interesting; however, its experimental realization remains elusive. The current work represents a successful assembly of DNA icosahedra. Moreover, the 3D DNA objects in most previous reports (9–13) are highly symmetrical in terms of DNA backbones but are not symmetrical when the DNA sequences are taken into account. Although such unique sequences provide opportunities for site-specific modifications, the assemblies require many different component DNA strands that all have unique DNA sequences. In contrast, the structure presented here is assembled out of only three unique DNA strands that can be easily synthesized and are commercially available. It has a true icosahedral symmetry, and its assembly takes advantage of component equivalence, which mimics the self-assembly of complex viral capsids in the case of T = 1 (27–29).
During assembly of large molecular cages, the DNA tiles exhibit even more complicated flexibility behaviors. Component concentration is clearly an important parameter for self-assembly. In general, high concentrations favor large complexes over small ones. This phenomenon has been observed in many systems, including DNA self-assembly. The DNA five-point-star tiles follow the same trend as well (Fig. S3). To further promote the formation of large structures, we have decreased the flexibility of the component tiles by reducing the length of the central loops to 4 bases long.
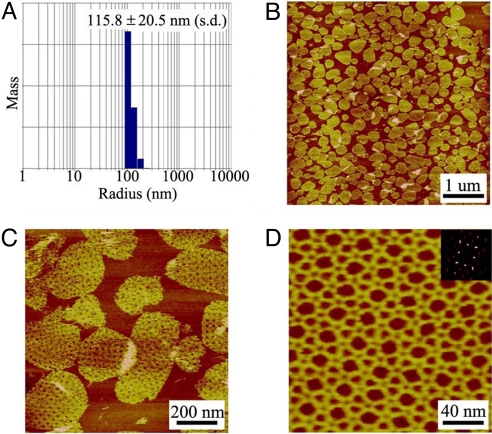
After assembly, we first used DLS to measure the hydrodynamic radius of the DNA complexes (Fig. 4A). The apparent radius (≈115 nm) significantly increased compared with that of the icosahedron samples (≈20 nm). The sample sizes have a wide distribution (± approximately 20%). For such nonuniform-sized DNA complexes, the technique of 3D single-particle reconstruction is not applicable. Thus, we characterized the DNA complexes by a more amenable technique: atomic force microscopy (AFM, Fig. 4 B–D). The DNA complexes appear as round, double-layer discs (apparent height: ≈4 nm and diameter: 200–300 nm) that are derived from DNA spherical cages. When such DNA cages adsorb onto AFM substrate (mica surfaces), they will collapse into double-layer structures because of strong electrostatic interactions between DNA and mica surfaces. At higher magnification, a tetragonal symmetry is visible. After several successive AFM scannings, the top layers are occasionally scratched out by the AFM probes and only monolayers are left. For such monolayer samples, it is possible to visualize the detailed DNA structures. The DNA assemblies show regular, periodic arrays that have a tetragonal symmetry. The experimentally observed 2D arrays cannot be simply related to the component five-point-star tiles.
Fig. 4.
Characterization of DNA cages assembled from five-point-star tiles with 4-base-long central loops. (A) DLS analysis of the assembled DNA cages. (B) A representative AFM image of the assembled DNA cages. (C) Zoom-in view. Note that the DNA structures are double-layered. (D) After the top layer is swiped away, AFM imaging reveals the detailed structures of the DNA lattices. (Inset) Corresponding Fourier transform pattern.
The planar-tiling theory predicts that objects with 5-fold rotational symmetry cannot assemble into regular (2D) lattices to completely tile a plane. It is a surprise to find that the five-point-star motif, which has a 5-fold rotational symmetry, assembles into regular 2D arrays. However, from the experimentally observed DNA 2D patterns, it is clear that the five-point-star tiles are flexible enough to deform and lose the 5-fold rotational symmetry during the assembly process. In the assemblies, each five-point-star tile involves the formation of three triangles and two squares. To adopt itself into such arrays, the five-point-star tile has to asymmetrically deform in the plane. Relative to a reference branch, two near branches bend 12° toward to the reference branch, and two far branches bend 6° from the reference branch. Theoretically, two other deformation patterns can also allow the five-point-star tiles assemble into 2D arrays (Fig. S4). They require larger bendings (12°/24° or 18°/6°, respectively) and require more energy. It is remarkable that the assembly process is so cooperative that only one deformation pattern has been observed, and the other two patterns are completely absent. Meanwhile, all branches of the tiles also have an out-of-plane bending (< 5°) to allow the formation of the overall spherical structures (Fig. 5). It is worth pointing out that the 2D crystal observed in this study is the most complicated, periodic 2D DNA array assembled from single-component tiles.
Fig. 5.
Proposed assembly process of DNA five-point-star tiles at high DNA concentrations. On the surface of the assembled DNA nanocages, DNA tiles are arranged into tetragonal 2D crystals. Compared with the free-DNA five-point-star tile, the branches in the final structure have asymmetrical bends in the molecular plane. The angles between two neighboring branches varies (three 60° and two 90°) in the final structure and are all different from the angle (72°) in the free tiles.
Conclusion
Conformational flexibility is essential for the self-assembly of five-point-star tiles in this study. However, one should not underestimate the importance of the structural rigidity. Only a fine balance between the flexibility and the rigidity allows precise control at the nanometer scale (15). For instance, five-point-star tiles with 5-base-long central loops readily assemble into icosahedra at low DNA concentration. When the loop lengths are decreased to 4 bases long, the five-point-star tiles become too stiff to assemble into icosahedra (Fig. S3). In contrast, the five-point-star tiles with 4-base-long central loops assemble into pseudo 2D crystals and further form large molecular cages. When the loop lengths are increased to 5 bases long, it is very difficult for the five-point-star tiles to assemble only into the large molecular cages and avoid producing icosahedra.
In summary, the five-point-star motif has been successfully designed and constructed. It has been used to demonstrate that DNA conformational flexibility could be exploited to assemble nanostructures that are otherwise difficult to prepare. DNA nanocages have attracted significant research efforts (9–14, 16, 30) not only because they are interesting structures, but also because they could potentially serve as encapsulation agents (31, 32), nanoreactors (33), and organizational scaffolds (34, 35).
Materials and Methods
Oligonucleotides.
DNA sequences were adapted from previous works that were originally designed by the SEQUIN (36) computer program. Central, blue-red, long-strand L (with 5-base-long loops): Agg CAC CAT CgT Agg TTT TTC TTg CCA ggC ACC ATC gTA ggT TTT TCT TgC CAg gCA CCA TCg Tag gTT TTT CTT gCC Agg CAC CAT CgT Agg TTT TTC TTg CCA ggC ACC ATC gTA ggT TTT TCT TgC C; central, blue-red, long-strand L′ (with 4-base-long loops): Agg CAC CAT CgT Agg TTT TCT TgC CAg gCA CCA TCg Tag gTT TTC TTg CCA ggC ACC ATC gTA ggT TTT CTT gCC Agg CAC CAT CgT Agg TTT TCT TgC CAg gCA CCA TCg Tag gTT TTC TTg CC; green, medium strand M: Tag CAA CCT gCC Tgg CAA gCC TAC gAT ggA CAC ggT AAC gCC; peripheral, short, black strand S: TTA CCg TgT ggT TgC Tag gCg. All oligonucleotides were purchased from IDT and purified by 10–20% denaturing PAGE.
Formation of DNA Complexes.
DNA strands were combined at indicated molecular ratios and DNA concentration in a Tris-acetic-EDTA-Mg2+ (TAE/Mg2+) buffer that contained 40 mM Tris base (pH 8.0), 20 mM acetic acid, 2 mM EDTA, and 12.5 mM magnesium acetate. The icosahedron were assembled from strands L, M, and S at DNA concentration of 20 nM (in terms of DNA tiles); large DNA cages were assembled from strands L′, M, and S at DNA concentration of 1 μM. DNA assembly involved cooling solutions from 95°C to room temperature (≈23°C) over 48 h. After assembly, the DNA samples were then directly used for AFM imaging and DLS studies. For cryo-EM imaging of the icosahedron, the sample was concentrated to ≈2 μM with Microcon YM-30 (30 kDa) Centrifugal Filter Units before flash-freezing.
Nondenaturing PAGE.
Nondenaturing gels containing polyacrylamide (19:1 acrylamide/bisacrylamide) were run on a FB-VE10–1 electrophoresis unit (FisherBiotech) at 4°C (80 V, constant voltage). The running buffer was TAE/Mg2+ buffer. Before electrophoresis, the DNA samples were concentrated with Microcon YM-30 (30 kDa) Centrifugal Filter Units to ≈200 nM if the sample concentrations were <200 nM. After electrophoresis, the gels were stained with Stains-All (Sigma) and scanned.
AFM Imaging.
A drop of 1.5 μl of DNA solution was spotted onto freshly cleaved mica surface (Ted Pella, Inc.), and incubated for 10 s to allow DNA to absorb onto the substrate. Then 50 μl of TAE/Mg2+ buffer was placed on the top of the DNA sample. The imaging was performed at 23°C in tapping mode on a Multimode AFM with Nanoscope IIIa controller (Veeco) with NP-S tips (Veeco) in a fluid cell. The tip–surface interaction was minimized by optimizing the scan set-point to the highest possible value.
DLS.
DNA sample solutions (12 μl) were measured by DynaPro 99 (Protein Solutions/Wyatt) with laser wavelength of 824 nm at 23°C.
Cryo-EM Imaging.
A drop of 3 μl of concentrated DNA solution was pipetted onto a Quantifoil grid. Then, the grid was blotted and immediately plunge-frozen into ethane slush cooled by liquid nitrogen. The data were recorded by using a Gatan 4 k × 4 k CCD camera in a Philips CM200 transmission electron microscope with field-emission gun operating at 200 kV accelerating voltage. To enhance the image contrast, underfocuses in the range of 2–4 μm were used to record the images. The calibrated magnification used for DNA icosahedrons was ×52,260, resulting in pixel sizes of 2.87 Å.
Single-Particle Reconstruction.
Thee-dimensional reconstructions of the DNA icosahedron used the single-particle image-processing software EMAN (25). The initial models were built by using 100 randomly selected raw particles. The initial orientation of individual particles was randomly assigned within the corresponding asymmetry unit of the icosahedron. Eight hundred eighty-four particles were used for the single-particle reconstruction. Ninety-nine reference projections in the icosahedron asymmetric unit were generated in the icosahedral asymmetric unit with an angular interval of 2°. A projection-matching algorithm was then used to determine the center and orientation of raw particles in the iterative refinement. The icosahedron symmetry was imposed during the reconstruction. The map resolution was determined to be at 2.8 nm by using the Fourier shell correlation (0.5 threshold criterion) of two 3D maps independently built from half datasets. Control reconstructions without imposing any symmetry or with lower symmetries imposed were performed to check that particles indeed had the icosahedral symmetry. Final 3D maps were visualized by using UCSF Chimera software (37).
Supplementary Material
Acknowledgments.
This work was supported by National Science Foundation Grant CCF-0622093, National Institutes of Health (NIH) Grant R21EB007472, and through the NIH Roadmap for Medical Research Grant PN2EY018230. AFM and DLS studies were carried out in the Purdue Laboratory for Chemical Nanotechnology. The cryo-EM images were taken in the Purdue Biological Electron Microscopy Facility, and the Purdue Rosen Center for Advanced Computing provided the computational resource for the 3D reconstructions.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. H.Y. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803841105/DCSupplemental.
References
- 1.Hamley IW. Nanotechnology with soft materials. Angew Chem Int Ed. 2003;42:1692–1712. doi: 10.1002/anie.200200546. [DOI] [PubMed] [Google Scholar]
- 2.Lehn JM. Toward complex matter: Supramolecular chemistry and self-organization. Proc Natl Acad Sci USA. 2002;99:4763–4768. doi: 10.1073/pnas.072065599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reinhoudt DN, Crego-Calama M. Synthesis beyond the molecule. Science. 2002;295:2403–2407. doi: 10.1126/science.1069197. [DOI] [PubMed] [Google Scholar]
- 4.Seeman NC. DNA in a material world. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 5.Winfree E, Liu FR, Wenzler LA, Seeman NC. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 6.Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science. 2003;301:1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- 7.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 8.Rothemund PWK, Papadakis N, Winfree E. Algorithmic self-assembly of DNA Sierpinski triangles. PloS Biol. 2004;2:2041–2053. doi: 10.1371/journal.pbio.0020424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen JH, Seeman NC. Synthesis from DNA of a molecule with the connectivity of a cube. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 10.Zhang YW, Seeman NC. Construction of a DNA-truncated octahedron. J Am Chem Soc. 1994;116:1661–1669. [Google Scholar]
- 11.Shih WM, Quispe JD, Joyce GF. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 12.Goodman RP, et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 13.Aldaye FA, Sleiman HF. Modular access to structurally switchable 3D discrete DNA assemblies. J Am Chem Soc. 2007;129:13376–13377. doi: 10.1021/ja075966q. [DOI] [PubMed] [Google Scholar]
- 14.Zimmerman J, Cebulla MPJ, Monninghoff S, von Kiedrowski G. Self-assembly of a DNA dodecahedron from 20 trisoligonucleotides with C3h linkers. Angew Chem Int Ed. 2008 doi: 10.1002/anie.200702682. [DOI] [PubMed] [Google Scholar]
- 15.He Y, Mao CD. Balancing flexibility and stress in DNA nanostructures. Chem Commun. 2006;9:968–969. doi: 10.1039/b513962g. [DOI] [PubMed] [Google Scholar]
- 16.He Y, et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 17.Ingber DE. Tensegrity: The architectural basis of cellular mechanotransduction. Annu Rev Physiol. 1997;59:575–599. doi: 10.1146/annurev.physiol.59.1.575. [DOI] [PubMed] [Google Scholar]
- 18.Motro R. Tensegrity: Structural Systems for the Future. Boston: Butterworth–Heinemann; 2006. [Google Scholar]
- 19.Liu D, Wang MS, Deng ZX, Walulu R, Mao CD. Tensegrity: Construction of rigid DNA triangles with flexible four-arm DNA junctions. J Am Chem Soc. 2004;126:2324–2325. doi: 10.1021/ja031754r. [DOI] [PubMed] [Google Scholar]
- 20.Scheffler M, Dorenbeck A, Jordan S, Wustefeld M, von Kiedrowski G. Self-assembly of trisoligo-nucleotidyls: The case for nano-acetylene and nano-cyclobutadiene. Angew Chem Int Edit. 1999;38:3312–3315. [PubMed] [Google Scholar]
- 21.He Y, Chen Y, Liu HP, Ribbe AE, Mao CD. Self-assembly of hexagonal DNA two-dimensional (2D) arrays. J Am Chem Soc. 2005;127:12202–12203. doi: 10.1021/ja0541938. [DOI] [PubMed] [Google Scholar]
- 22.He Y, et al. Sequence symmetry as a tool for designing DNA nanostructures. Angew Chem Int Ed. 2005;44:6694–6696. doi: 10.1002/anie.200502193. [DOI] [PubMed] [Google Scholar]
- 23.He Y, Tian Y, Ribbe AE, Mao CD. Highly connected two-dimensional crystals of DNA six-point-stars. J Am Chem Soc. 2006;128:15978–15979. doi: 10.1021/ja0665141. [DOI] [PubMed] [Google Scholar]
- 24.Ke YG, Liu Y, Zhang JP, Yan H. A study of DNA tube formation mechanisms using 4-, 8-, and 12-helix DNA nanostructures. J Am Chem Soc. 2006;128:4414–4421. doi: 10.1021/ja058145z. [DOI] [PubMed] [Google Scholar]
- 25.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 26.Rothemund PWK. In: Nanotechnology: Science and Computation. Chen JH, Jonoska N, Rozenberg G, editors. Heidelberg: Springer; 2006. pp. 3–21. [Google Scholar]
- 27.Casjens S. Virus Structure and Assembly. Sudbury, MA: Jones and Bartlett; 1985. [Google Scholar]
- 28.Caspar DLD, Klug A. Physical principles in construction of regular viruses. Cold Spring Harb Symp. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 29.Twarock R, Hendrix RW. Crosslinking in viral capsids via tiling theory. J Theor Biol. 2006;240:419–424. doi: 10.1016/j.jtbi.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 30.Goodman RP, et al. Reconfigurable, braced, three-dimensional DNA nanostructures. Nat Nanotechnol. 2008;3:93–96. doi: 10.1038/nnano.2008.3. [DOI] [PubMed] [Google Scholar]
- 31.Comellas-Aragones M, et al. A virus-based single-enzyme nanoreactor. Nat Nanotechnol. 2007;2:635–639. doi: 10.1038/nnano.2007.299. [DOI] [PubMed] [Google Scholar]
- 32.Erben CM, Goodman RP, Turberfield AJ. Single-molecule protein encapsulation in a rigid DNA cage. Angew Chem Int Ed. 2006;45:7414–7417. doi: 10.1002/anie.200603392. [DOI] [PubMed] [Google Scholar]
- 33.Douglas T, Young M. Host–guest encapsulation of materials by assembled virus protein cages. Nature. 1998;393:152–155. [Google Scholar]
- 34.Wang Q, Lin TW, Tang L, Johnson JE, Finn MG. Icosahedral virus particles as addressable nanoscale building blocks. Angew Chem Int Ed. 2002;41:459–462. doi: 10.1002/1521-3773(20020201)41:3<459::aid-anie459>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 35.Duckworth BP, et al. A universal method for the preparation of covalent protein-DNA conjugates for use in creating protein nanostructures. Angew Chem Int Ed. 2007;46:8819–8822. doi: 10.1002/anie.200701942. [DOI] [PubMed] [Google Scholar]
- 36.Seeman NC. De novo design of sequences for nucleic-acid structural-engineering. J Biomol Struct Dyn. 1990;8:573–581. doi: 10.1080/07391102.1990.10507829. [DOI] [PubMed] [Google Scholar]
- 37.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.