Abstract
Natural killer (NK) cells constitute a subpopulation of lymphocytes that develop from precursors in the bone marrow (BM), but the transcriptional regulation of their development and maturation is only beginning to be understood, in part due to their relatively rare abundance, especially of developmental subsets. Using a mouse model in which NK cells are arrested at an immature stage of development, and a gene expression profiling approach, we uncovered transient normal NK cell expression of a homeobox transcription factor (TF) family, called Distal-less (Dlx), which had been primarily implicated in murine CNS, craniofacial, limb, and skin development. Our studies demonstrate that Dlx1, Dlx2, and Dlx3 are transiently expressed in immature Mac-1lo NK cells within the BM, with Dlx3 being the predominantly expressed member. These genes are expressed in a temporally regulated pattern with overlapping waves of expression, and they display functional redundancy. Expression is extinguished in fully mature splenic NK cells, and persistent expression of Dlx genes leads to functionally immature NK cells arrested at the Mac-1lo stage. Whereas conventional splenic NK cells develop but are arrested at an immature stage, there appears to be a complete failure to develop CD127+ thymic NK cells when Dlx genes are persistently expressed. We also observed that T and B cells fail to develop in the context of persistent Dlx1 expression. Thus, these studies indicate that Dlx TFs play a functional role in lymphocyte development.
Keywords: Dlx, NK, lymphocyte, gene expression
Natural killer (NK) cells represent a subset of lymphocytes, distinct from T and B cells, with important functions in the host defense against virus-infected and malignant target cells (1, 2). They are unique in that they do not require prior sensitization for target killing and cytokine production. Despite these important functions, their development is just beginning to be understood.
Adult splenic NK cells develop in the bone marrow (BM), arising from a common T and NK cell precursor (3). Development occurs in an ordered progression toward a fully functional NK cell that then enters the peripheral circulation (4). Commitment to the NK cell lineage from the T/NK cell precursor appears to be first marked by the expression of the IL-2/IL-15 receptor β chain (CD122), and as such, the CD122+CD3−NK1.1− cell has also been called the NK cell precursor (NKP) (5). After acquisition of NK1.1 expression, there appears to be regulated expression of several other cell surface markers. In the final stages of conventional splenic NK cell development, the up-regulation of Mac-1 (CD11b/CD18 or integrin-αM) expression coincides with functional maturation (3, 4, 6), with the majority of circulating NK cells being Mac-1hi. Recent data indicate that a sublineage of NK cells develops and matures in the thymus (7). These thymic NK cells express the receptor for IL-7 (CD127+) and, unlike splenic NK cells, critically depend on IL-7 receptor signaling during development (8). As with splenic NK cells, the development of thymic NK cells is incompletely understood at the molecular level.
The regulation of development from NK progenitors to fully mature NK cells involves transcription factors (TFs), including T-bet (Tbx21), GATA-3 (Gata3), and Id2 (Id2), that have important roles in other aspects of lymphopoiesis, (8–13). Attempts to identify novel TFs regulating NK cell development by gene expression profiling have been difficult, because immature NK cells are a heterogenous population of rare cells at different stages of development that are just beginning to be defined.
Herein, we took advantage of a mouse that displays a defect in NK cell maturation (14). This transgenic mouse contains the Ly49A cDNA under control of the granzyme A promoter. It manifests a selective defect in NK cell development that is related to integration of the transgene into the Atf2 locus on chromosome 2, but not to Atf2 gene deficiency or to expression of Ly49A (15). These NK-deficient (NKD) mice have a reduced number of NK cells in the periphery, all of which are functionally immature with poor target killing and cytokine production. Furthermore, unlike splenic NK cells in WT mice, which typically have a Mac-1hi phenotype, the few peripheral splenic NK cells in NKD mice display a Mac-1lo phenotype, and there is an accumulation of similar cells in the NKD BM. Thus, the functional and phenotypic characteristics of NK cells from NKD mice appear to result from a developmental arrest.
IL-15 is required for NK cell development, and it fosters proliferation (16), consistent with its putative role in constitutive proliferation at an immature stage of NK cell differentiation. However, crossing the NKD mice to IL-15 transgenic (IL15tg) mice, which overexpress IL-15, did not overcome the NKD developmental block. Rather, there was dramatic accumulation of the functionally immature Mac-1lo splenic NK cells, consistent with an expansion of NK cells normally observed at this stage of development (4, 17). These NK cells are uniformly Mac-1lo, similar to those from NKD mice, whereas NK cells from IL15tg mice mature normally and are Mac-1hi. Because the double transgenic (NKDxIL15tg) mice have an abundance of NK cells apparently arrested at a discrete stage of development, we reasoned that these cells would provide a unique opportunity to study a relatively homogeneous population of immature NK cells and facilitate the identification of novel TFs important in NK cell development.
In general, TFs in lymphocyte development are often involved in disparate developmental processes where the TFs regulate different sets of genes depending on the context. For example, both GATA-3 and Id2 are involved in the development of the nervous system in addition to lymphocytes (18–20). TFs shared between immune and neural development are also exemplified by homeodomain-containing TFs, such as the paired box TF Pax5. This TF is critically important in the development of B cells and the midbrain/hindbrain regions of the CNS (21). Thus, it is likely that there are other TFs, involved in lymphocyte development, that are also involved in the developmental processes of other systems.
With these rationales, we used a gene expression profiling approach with NKDxIL15tg NK cells to identify TFs that may be involved in NK cell development. We focused on homeobox TFs, because they are known to be involved in the development and differentiation of a variety of organ systems. Herein, we show that NK cell development and differentiation involves the homeobox TF family called Distal-less (Dlx), which had been primarily defined in terms of murine CNS, craniofacial, limb, and skin development (22). We found that Dlx genes are transiently expressed during immature stages of NK cell development, and their persistent expression results in developmental arrest of splenic NK cells at an immature stage and a complete failure to develop CD127+ thymic NK cells. Moreover, persistent Dlx expression resulted in a defect in T and B cell development. Thus, these studies provide insight into the regulation of lymphocyte development in general.
Results
Gene Expression Profiling of NK Cells from NKDxIL15tg Mice.
We isolated NK1.1+CD3− cells to high purity by FACS from the individual spleens of NKDxIL15tg mice and control IL15tg mice. For another source of immature NK cells, we isolated NK1.1+CD3− cells to high purity by FACS from the BM of NKDxIL15tg mice. Gene expression profiling was performed using genome-wide oligonucleotide microarrays (see Methods). Unsupervised hierarchical clustering of the data grouped all of the NKDxIL15tg samples (BM and spleen) together. As expected, the NKDxIL15tg samples had lower transcript expression of Itgam (Mac-1), consistent with the Mac-1lo phenotype of the NK cells from these mice [supporting information (SI) Fig. S1]. In general, there was also good concordance of results between replicate samples in each group, as validated by subsequent expression analysis (see below, and data not shown).
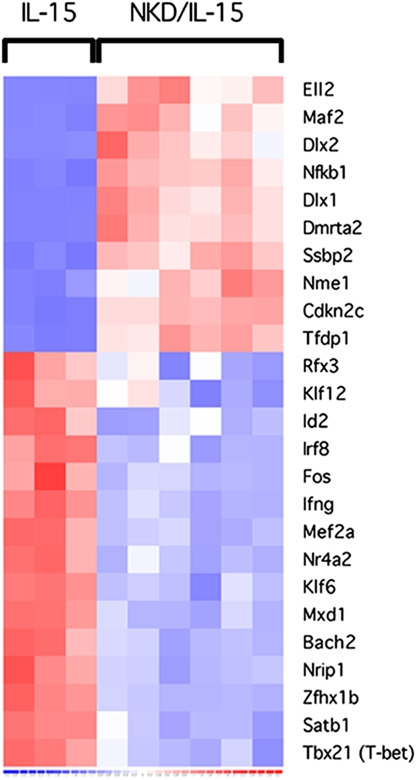
A comparison of the NKDxIL15tg samples to the IL15tg samples was focused on a list of genes having transcription regulator activity (Fig. 1 and Table S1). Consistent with previous studies showing that Tbx21 and Id2 become up-regulated during NK cell maturation (9, 11), the expression levels of these genes were lower (2.5- and 2-fold, respectively) in the NKDxIL15tg samples. However, Gata3 expression, which has also been described to increase as NK cells mature (10), was not differentially expressed. Among the most differentially expressed TF genes, two of them, Dlx1 and Dlx2, were notable for their abundance. These members of the Distal-less homeobox TF family had 46- and 23-fold increases in expression in the NKDxIL15tg samples, respectively.
Fig. 1.
Gene expression profiling of NK cells from NKDxIL15tg and IL15tg mice. NK1.1+CD3− cells were purified by FACS from the spleens and BM of NKDxIL15tg mice and from the spleens from IL15tg mice. Gene expression was assessed by genome-wide microarrays. Each chip represented pooled samples independently sorted from 3 mice. Heat map represents expression of genes from the gene ontology list of transcription regulators. GEO accession no. GSE10365 (www.ncbi.nlm.nih.gov/projects/geo).
Inasmuch as Dlx1 and Dlx2 belong to a family of related genes (Dlx1-6), we investigated expression of other Dlx genes. However, expression of Dlx3-6 was not detected in any of the samples, even though probe sets for these members were present on the microarrays. Confirmation of the differences in expression of Dlx1 and Dlx2 was obtained by quantitative Taqman PCR of cDNA made from NK cells that were FACS-purified from NKDxIL15tg and IL15tg mice (data not shown). In addition, abundant transcripts from both genes were detected in NK1.1+CD3− cells sorted from spleens of NKD (single transgenic) mice, whereas NK1.1+CD3− cells sorted from the spleens of WT C57BL/6 (B6) mice had no detectable Dlx1 or Dlx2 transcript. These findings indicate that the differences in expression of Dlx1 and Dlx2, observed in the microarray data, were not a consequence of IL-15-mediated NK cell expansion in the NKDxIL15tg mice (data not shown). Because Dlx1 and Dlx2 encode homeodomain-containing TFs that are often important in many developmental processes and were among the most differentially expressed TFs, we chose to explore further the possible role of these genes in NK cell development.
Dlx1, Dlx2, and Dlx3 Genes Are Expressed During Normal NK Cell Development.
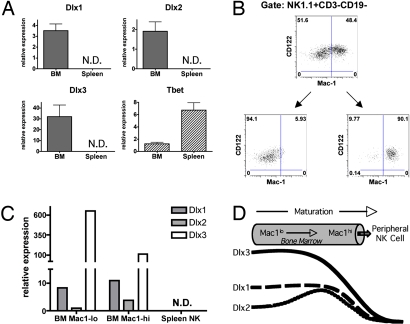
To determine whether Dlx genes are expressed during normal NK cell development, we examined their expression in immature and mature NK cells from WT B6 mice. NK1.1+CD3− cells in the BM represent a heterogeneous population of NK cells at different stages of development, and hence, these cells are, collectively as a group, more immature than splenic NK cells. NK1.1+CD3− cells were sorted to high purity from the BM and the spleen, and gene expression was examined by Taqman quantitative RT-PCR (qRT-PCR) (Fig. 2A). In concordance with the genechip data, we found Dlx1 and Dlx2 to be expressed in the BM NK cells. We also found these immature WT NK cells to express another family member, Dlx3, but, as with the NKDxIL15tg NK cells, we did not find expression of Dlx4-6 by Taqman analysis. More importantly, the expression of Dlx1, Dlx2, and Dlx3 was undetectable in the WT splenic NK cells. This is in contrast to the TF Tbx21, which is expressed more highly in the splenic, compared with BM, NK cells, consistent with prior description of Tbx21 expression in NK cell development (9). Thus, Dlx1, Dlx2, and Dlx3 are expressed at an immature stage of normal NK cell development, and expression is down regulated with final maturation.
Fig. 2.
Dlx genes are expressed in immature NK cells. (A) NK1.1+CD3− cells were sorted from the BM and spleens of B6 mice, and expression of Dlx family members and T-bet (normalized to Hprt1 expression) was assessed by qRT-PCR. N.D., not detected. (B) Mac-1lo and Mac-1hi NK cells (CD122+NK1.1+CD3−CD19−) were sorted to high purity from the BM of Rag2−/− mice, and (C) qRT-PCR was used to assess the expression of Dlx1, Dlx2, and Dlx3 in these NK cell populations, and in NK cells sorted from the spleen. N.D., not detected. (D) Immature NK cells express Dlx1, Dlx2, and Dlx3 in a temporally regulated manner.
To more precisely determine the BM stage in which Dlx genes are expressed, we next sorted Mac-1lo and Mac-1hi NK cells (CD122+NK1.1+CD3−CD19−) from the BM of Rag2−/− mice to high purity (Fig. 2B). Rag2−/− mice were used to enhance the degree of purity during the sorting of minor populations in the BM. We assessed the expression of Dlx1, Dlx2, and Dlx3 by qRT-PCR in both of these populations and compared it to the expression of these genes in NK cells sorted from the spleen. Again, none of the Dlx genes was expressed in the splenic NK cells. In the BM, however, Dlx1, Dlx2, and Dlx3 were expressed to different degrees in the Mac-1lo and Mac-1hi populations (Fig. 2C). Dlx3 was expressed at a much higher level (>100-fold) than either Dlx1 or Dlx2, and the expression of Dlx3 was primarily detected in the Mac-1lo population of NK cells, consistent with this gene being expressed at an early stage of development. Dlx1 was expressed early in the Mac-1lo NK cell population but was also detected in the Mac-1hi BM NK cell population. Dlx2, however, was expressed primarily in the Mac-1hi BM NK cells. These data suggest a temporally regulated expression pattern of Dlx1, Dlx2, and Dlx3, with overlapping waves of expression, during NK cell development in the BM (Fig. 2D), which is reminiscent of the transient expression patterns of Dlx homeobox genes during other developmental processes (22).
NK Cell Development Appears Normal in the Absence of Dlx1 and Dlx2.
Although the predominantly expressed Dlx family member in the immature NK cells is Dlx3, assessment of Dlx3 loss of function on NK cell development was not readily addressable, because Dlx3−/− mice die at embryonic day 8.5 (E8.5), before onset of hematopoiesis, precluding fetal liver transplantation (23). To examine the requirement for Dlx1 in NK cell development, we analyzed Dlx1−/− knockout mice after backcrossing onto the B6 background. Because these knockout mice live only to ≈4 weeks (24), NK cell development was examined in 4-week-old knockout mice. The phenotypic and functional analysis did not reveal any defect (data not shown). Because NK cell maturation could have been affected by the terminal illness, we also analyzed NK cells in irradiated mice that underwent either BM transplantation (at donor age 4 weeks) or fetal liver transplantation (at donor E17) with Dlx1−/− knockout derived BM cells at least 8–12 weeks posttransplantation. Again, NK cell development appeared to be unperturbed (data not shown).
Next, we assessed the requirement for both Dlx1 and Dlx2 in NK cell development by the study of Dlx1−/−Dlx2−/− double-knockout mice backcrossed onto the B6 background. Because Dlx1−/−Dlx2−/− double-knockout mice die at birth (25), fetal liver transplantation was performed at E17 into lethally irradiated congenic CD45.1 (Ly5.1) WT recipient mice. NK cell development was then studied 8–12 weeks posttransplantation. We found that the percentage and absolute number of NK cells developing from Dlx1−/−Dlx2−/− donor fetal liver progenitor cells did not differ significantly from those derived from heterozygous or WT fetal liver progenitor cells (Fig. S2A). In addition, the knockout-derived NK cells had normal expression of Mac-1 and CD43, resembling WT mature splenic NK cells, and they produced IFN-γ upon stimulation by immobilized anti-NK1.1 in vitro at levels equivalent to IFN-γ production by heterozygous and WT NK cells (Fig. S2B and data not shown). Thus, NK cell development does not seem to be impaired in the absence of both Dlx1 and Dlx2, but given our findings of redundant expression of the Dlx family members and that Dlx3 is the most highly expressed family member in immature WT NK cells, it is possible that Dlx3 could function alone in the absence of Dlx1 and Dlx2 during NK cell development.
Persistent Expression of Dlx Arrests NK Cell Development at an Immature Stage.
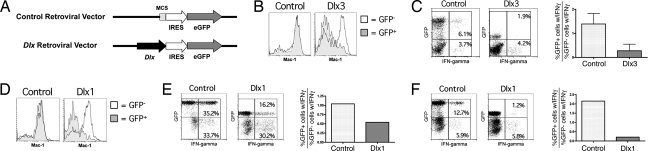
The absence of Dlx gene expression in mature splenic NK cells suggested that suppression of these genes is required for full NK cell maturation. To address this hypothesis, we studied Dlx3, which was the predominantly expressed Dlx family member in the BM NK cell population (Fig. 2C). We cloned Dlx3 into a bicistronic retroviral vector containing an internal ribosomal entry site (IRES) for the expression of enhanced GFP (Fig. 3A). The recombinant virus derived from this vector was then used to infect in vitro BM cells prepared from B6 mice. These BM cells were then transferred into lethally irradiated B6 mice, and NK cell development was analyzed 8–12 weeks posttransplantation. Persistent Dlx3 expression, tracked by the expression of GFP, conferred low Mac-1 expression on splenic NK cells, compared with the GFP− NK cells, which displayed a normal Mac-1hi phenotype within the same mouse (Fig. 3B). This effect was not due to the expression of GFP itself or to the retroviral infection, because transduction with the control virus, lacking Dlx3, did not result in a Mac-1lo phenotype. Persistent expression of Dlx3 also resulted in functionally immature NK cells, in that they produced IFN-γ poorly upon stimulation by anti-NK1.1 plate-bound antibody in vitro (Fig. 3C). Thus, persistent expression of Dlx3 alone is sufficient to result in NK cells that appear to be arrested at an immature stage as measured by both surface marker phenotype and functional capacity.
Fig. 3.
Persistent expression of Dlx genes arrests NK cell development. (A) PMXs-IG retrovirus constructs. (B) PMXs-IG retrovirus was used to transduce Dlx3 into BM progenitors, which were transplanted into irradiated B6 recipients. Mac-1 expression was measured by FACS of splenocytes from recipients 8–12 weeks postreconstitution, gating on NK1.1+CD3−CD19− lymphocytes. Retrovirus-transduced NK cells are marked by GFP expression. Results are representative of two independent experiments. (C) Splenocytes from recipients of BM transduced with either the control or the Dlx3-expressing retrovirus were stimulated by plate-bound anti-NK1.1 antibodies. IFN-γ production was measured by bivalent IFN-γ catch antibody secretion assay. Results are representative of two independent experiments. (D) Dlx1-transduced BM progenitors were transplanted into irradiated B6 recipients, and Mac-1 expression was measured as in 4B. Results are representative of three independent experiments. Splenocytes from recipients of BM transduced with either the control or the Dlx1 expressing retrovirus were stimulated by YAC-1 target cells in vitro (E) or by plate-bound anti-NK1.1 antibodies (F). IFN-γ production was measured as in Fig. 4C.
Inasmuch as functional redundancy was suggested by the absence of phenotype in the Dlx1−/−Dlx2−/− double-knockout mice, we further explored this possibility with the persistent expression strategy. We found that Dlx1 transduction also led to NK cells arrested at an immature stage. Specifically, the Dlx1-transduced NK cells were Mac-1lo in surface marker phenotype, and they produced IFN-γ poorly upon stimulation in vitro (Fig. 3 D–F). Thus, these data suggest that functional redundancy between the Dlx family members can occur during NK cell development, similar to what has been described of these TFs in other developmental processes (26).
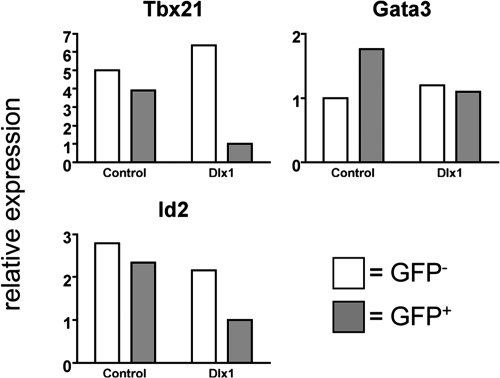
We next examined the expression of TFs, known to be involved in NK cell maturation, in the Dlx1-transduced NK cells. We sorted GFP+ and GFP− cells from the CD122+NK1.1+CD3−CD19− population in the spleens of the Dlx1-transduced BM recipient mice (Fig. S3) and examined expression of Tbx21, Gata3, and Id2 by qRT-PCR (Fig. 4). Tbx21 expression was much lower in the Dlx1-transduced NK cells (GFP+) compared with the nontransduced NK cells (GFP−). This is consistent with the defect in NK cell maturation observed in Tbx21−/− mice (9). There was also a lower expression of Id2 in the GFP+ NK cells, but there was no difference in Gata3 expression. These findings are strikingly similar to the gene expression data seen in the NKDxIL15tg mice (Fig. 1, Table S1) and implicate Dlx1 in the regulation of Tbx21 and Id2 expression.
Fig. 4.
Dlx1 transduction results in decreased Tbx21 and Id2 expression by NK cells. NK1.1+CD3−CD19− splenocytes from recipients of BM transduced with either the control or the Dlx1-expressing retrovirus were sorted into GFP+ and GFP− populations (Fig. S3), and qRT-PCR was performed to measure expression of Tbx21, Gata3, and Id2 (normalized to Hprt1 expression).
Dlx Expression in BM Progenitors Inhibits Development of Thymic NK, T, and B Cells.
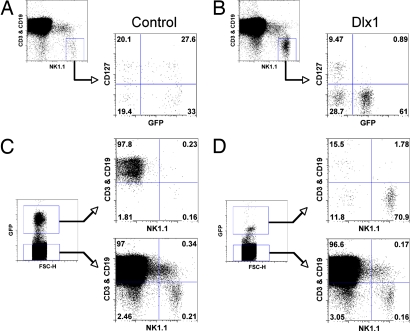
The recent description of thymic NK cells, which have been characterized as being CD127+Mac-1lo and to express Gata3 highly (8), led us to consider whether they were affected by Dlx expression. Because a substantial percentage of lymph node NK cells are thymic NK cells in WT mice (27), we examined lymph nodes from Dlx1-transduced BM transplant recipients for CD127 expression. Surprisingly, persistent Dlx1 expression, tracked by GFP expression, led to complete absence of CD127+ NK cells (Fig. 5 A and B). This is in contrast to the GFP− population within the same mouse, which expressed CD127 on >20% of the NK cells. These observations indicate that persistent Dlx1 expression prevents the development of thymic NK cells in addition to arresting the maturation of conventional splenic NK cells.
Fig. 5.
Dlx1 expression inhibits development of thymus-derived NK, T, and B cells. BM progenitor cells were transduced with either Dlx1-expressing retrovirus or control retrovirus, and irradiated mice were reconstituted with transduced BM cells as in Fig. 3. ≈8–12 weeks postreconstitution, NK cells (NK1.1+CD3−CD19−) from lymph nodes obtained from control (A) or Dlx1 (B) retrovirus-transduced BM recipients were examined for CD127 expression. Retrovirus-transduced NK cells are marked by GFP expression. Lymph node lymphocytes from control (C) or Dlx1 (D) retrovirus-transduced BM recipients were examined for CD3, CD19 expression, and NK1.1 expression.
Interestingly, when Dlx1 was persistently expressed in BM hematopoietic progenitors, there was also a striking failure to develop T or B cells in the periphery (Fig. 5 C and D). In the thymus of the Dlx1-transduced BM recipients, only a few GFP+ cells were observed, making their analysis difficult. However, these cells appeared to be negative for CD4 and CD8 expression, suggesting that the developmental arrest of the T cells was at the CD4−CD8− double-negative (DN) stage (data not shown). To explore this developmental block, we sorted CD4−CD8− DN, CD4+CD8+ double-positive (DP), CD4+ single-positive (SP), and CD8+ SP populations from the thymus of WT B6 mice. Expression of the Dlx genes was assessed in these populations by qRT-PCR, and we found Dlx1 to be expressed primarily in the DP population (Fig. S4). We also observed Dlx1 to be expressed at a low level in the DN population, but it was not detected in the SP populations. In contrast to this transient temporally regulated expression of Dlx1, Dlx2-6 expression was not readily detected in the thymocyte subsets. Thus, these data indicate that Dlx genes appear to have a functional role in the development of other lymphocyte lineages in addition to NK cells.
Discussion
The Dlx family was originally identified in Drosophila as a single gene (Dll) and found to be important in proximodistal patterning in limb development. The vertebrate orthologs are comprised of six family members that are expressed in a tissue- and spatiotemporal-specific manner during development (22). These genes have been shown to be critically important in the development of a number of different structures and tissues, including the nervous system, craniofacial skeleton, limbs, and skin (28, 29). However, essentially no prior information was available concerning their role in hematopoiesis or lymphopoiesis.
In this study, we show that Dlx1, Dlx2, and Dlx3 are expressed in immature WT NK cells in a temporally regulated manner. Dlx3 appears to be the earliest expressed Dlx during development and to be decreasing in expression, whereas the other Dlx genes are still abundantly expressed as maturation proceeds, suggesting that Dlx3 expression may be extinguished before the others. This may explain why Dlx3 was not found to be expressed in genechip analysis of the arrested NKDxIL-15tg NK cells that express high levels of Dlx1 and Dlx2.
We found that with developing WT NK cells, persistent expression of either Dlx1 or Dlx3 in vivo results in splenic NK cells that are arrested at a Mac-1lo functionally immature stage. The similar effects of persistent expression of Dlx1 and Dlx3 are consistent with the redundant functions of these TFs as regulators of differentiation in neural, epidermal and craniofacial development (22, 26). Our analyses of Dlx1−/− mice and mice that were hematopoietically reconstituted with Dlx1−/−Dlx2−/− fetal liver cells are also consistent with the possibility of functional redundancy. Dlx1 alone and Dlx1 and Dlx2 loss of function revealed no appreciable effect on NK cell development, suggesting that Dlx3, which is expressed at a higher level than either Dlx1 or Dlx2 in immature NK cells, may be able to compensate for the loss of these genes.
Within the Dlx family, paralogs have highly conserved homeodomains, and as such, it is not surprising they can function redundantly in many situations (22). As with Hox proteins, it seems that Dlx proteins have both shared and paralog-specific target genes, and their subtle specificities can be revealed by compound deletions of different family members (26, 30). Studies of Dlx3−/− mice are currently underway; however, this analysis is complicated by the fact that these mice die at E8.5 from placental failure, precluding fetal liver transplantation studies (23). As such, these studies will require other approaches, such as complementation of blastocysts isolated from Rag−/−γc−/− mice (which do not develop T, B, or NK cells) with knockout-derived embryonic stem cells. Thus, in light of the issues with redundancy between Dlx family members, it may also be necessary to study Dlx1−/−Dlx2−/−Dlx3−/− triple-knockout mice by blastocyst complementation to assess loss-of-function effects on NK cell development.
In our gain-of-function studies with Dlx transduction, it is important to note that target gene specificity by homeobox TF families is defined by their specific DNA-binding homeodomains and cofactors (30). For example, a chimeric protein of the homeobox TF Ultrabithorax, in which the homeodomain has been switched with that of Antennapedia, can substitute for Antennapedia in cell fate determination (31). Given this level of target gene specificity by homeodomains, Dlx homeodomains are unlikely to affect genes regulated by another family of distinct homeodomain-containing TFs, and thus, it seems unlikely that persistent expression of Dlx genes in our studies resulted in non-specific effects.
Although the mechanism by which Dlx genes affect NK cell development is unclear, our studies suggest that the Dlx genes may regulate the expression of other TFs (such as Tbx21 and Id2 but not Gata3) that are involved in NK cell development and maturation (9–11, 13). Our studies also indicate that Dlx TFs play a role in the development of other lymphocyte lineages. Specifically, we observed that T and B cell development is severely perturbed when Dlx1 is persistently expressed from the BM progenitor stage. We also found that the recently described thymic NK cell sublineage fails to develop when Dlx1 is persistently expressed. Because a deficiency in IL-7 signaling in mice leads to severe lymphopenia, resulting from a failure to develop T, B, and thymic (CD127+) NK but not conventional NK cells (8, 32–35), one potential explanation for our observations could be that Dlx1 negatively regulates IL-7Rα expression. Interestingly, we found that Dlx1 is transiently expressed almost exclusively during the DP stage of normal T cell development. This is the stage at which IL-7Rα expression is normally repressed in the thymus (36, 37), again suggesting that Dlx1 may negatively regulate IL-7Rα expression and raising the intriguing possibility that Dlx1 may have a role in thymocyte selection or T/NK cell lineage commitment. Studies to determine whether Dlx1 regulates IL-7Rα expression directly or indirectly are underway.
Although our study describes the previously uncharacterized functional role of Dlx genes in lymphocyte development, the expression of these genes in hematopoietic cells has been reported (38–40). For example, BP1, an isoform of Dlx4, is expressed in early erythroid development and is down-regulated during differentiation (41). This gene appears to repress β-globin expression, and thus its down-regulation has been suggested to be necessary for terminal erythroid differentiation (41–43), akin to what we have observed with Dlx1 and Dlx3 in NK cell development. Thus, it is likely that the Dlx family may play an important role in the differentiation of many hematopoietic lineages, in addition to NK and T cells, and that the repression of these genes is associated with maturation.
Methods
Antibodies and FACS.
Antibodies to NK1.1 (PK136), CD3 (2C11), CD19 (1D3), Mac-1 (M1/70), CD122 (TM-β1), and Ly5.2 (1O4) were obtained from BD PharMingen. Purification of cells by FACS was performed at Washington University in both the Siteman Cancer Center and the Department of Pathology and Immunology core facilities.
Mice.
C57BL/6 (WT B6) mice and B6 mice with the Ly5.1 antigen were purchased from National Cancer Institute and Taconic, respectively. NKD and NKDxIL15tg mice were described (15, 17). IL15tg mice on the B6 background were kindly provided by M. Caligiuri at Ohio State University, Columbus (44). NK (NK1.1+CD3−) cells were purified from 8- to 12-week-old IL15tg and NKDxIL15tg for the microarray experiments. Rag2−/− mice on the B6 background were obtained from Taconic. Dlx1−/− and Dlx1−/−Dlx2−/− mice (25) were backcrossed to the B6 background more than six generations using a marker-assisted breeding strategy. All mice were used in accordance with institutional guidelines for animal experimentation.
Gene Expression Profiling.
NK1.1+CD3− cells were sorted from the individual BM and spleens of nine NKDxIL15tg mice and from the individual spleens of nine IL15tg mice. Total RNA was prepared by the TRIzol method according to the manufacturer's protocol, and target synthesis for hybridization to Affymetrix 430 v2.0 GeneChips was performed with two rounds of linear amplification by the Washington University Siteman Cancer Center Microarray Core Facility. Target synthesis was performed for each of the 18 splenic NK cell RNA samples (nine NKDxIL15tg and nine IL15tg) and each of the nine BM NK cell RNA samples from the NKDxIL15tg mice. Then, three target samples were pooled and hybridized to each chip, resulting in three chips for the BM samples and three for each set of spleen samples. Data analysis was performed using dChip software from the Harvard School of Public Health (http://biosun1.harvard.edu/complab/dchip). The chips were normalized to the chip with the median intensity, and expression values were then assigned for each probe set. Gene expression assays were performed by qRT-PCR, using Taqman primer/probe sets and real-time PCR instruments from Applied Biosystems.
Retroviral Transduction and Hematopoietic Reconstitution by BM Transplantation.
The pMXs-IRES-GFP (PIG) retroviral vector (a gift from T. Kitamura, University of Tokyo, Tokyo) was used for these studies. The coding sequences of Dlx1 and Dlx3 were PCR-amplified from NKDxIL15tg NK cell RNA and fetal mouse RNA, respectively. The amplicons were cloned into the PIG vectors and used to transduce BM hematopoietic precursors from B6 mice for reconstitution of lethally irradiated (9.5 Gy) B6 mice, as described (45). Analysis of recipient mice was performed between 8 and 12 weeks postreconstitution.
Fetal Liver Transplantation.
Timed matings were performed, and pregnant dames were killed at E17. Fetal livers were harvested from each embryo, single-cell suspensions were made by using a sterile technique, and the cells were injected into the tail veins of lethally irradiated (9.5 Gy) Ly5.1 B6 congenic mice (5–10 million cells per mouse). Genotyping by PCR was performed on DNA prepared from each fetus (SI Methods).
Cytokine Stimulation Assays.
Stimulation of NK cells was performed as described, with slight modification (45). Splenocytes (107 ml−1) were incubated with an equal volume of Yac-1 tumor targets (106 ml−1) for 30–60 min and then further incubated in the presence of brefeldin A for an additional 5–8 h. Alternatively, splenocytes (107 ml−1) were incubated in six-well plates precoated with anti-NK1.1 monoclonal antibody ascites at a 1:30,000 dilution. IFN-γ production by CD3−CD19−NK1.1+ cells was measured by using the IFN-γ bivalent antibody secretion assay by Miltenyi Biotec per the manufacturer's instructions.
Supplementary Material
Acknowledgments.
We thank M. Cooper, A. French, P. Klekotka, and B. Plougastel for critical review of the paper. Work in the Yokoyama laboratory is supported by the Howard Hughes Medical Institute and by grants from the National Institutes of Health. We also thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes–Jewish Hospital in St. Louis, MO, for the use of the High Speed Cell Sorter Core, and the Multiplexed Gene Analysis Core. The Siteman Cancer Center is supported in part by an National Cancer Institute Cancer Center Support Grant P30 CA91842. Genotyping of the Dlx knockout mice was provided by the Speed Congenics Facility of the Rheumatic Diseases Core Center Grant P30 AR048335.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805205105/DCSupplemental.
References
- 1.French AR, Yokoyama WM. Natural killer cells and viral infections. Curr Opin Immunol. 2003;15:45–51. doi: 10.1016/s095279150200002x. [DOI] [PubMed] [Google Scholar]
- 2.Gorelik E, et al. Role of NK cells in the control of metastatic spread and growth of tumor cells in mice. Int J Cancer. 1982;30:107–112. doi: 10.1002/ijc.2910300118. [DOI] [PubMed] [Google Scholar]
- 3.Yokoyama WM, Kim S, French AR. The dynamic life of natural killer cells. Annu Rev Immunol. 2004;22:405–429. doi: 10.1146/annurev.immunol.22.012703.104711. [DOI] [PubMed] [Google Scholar]
- 4.Kim S, et al. In vivo developmental stages in murine natural killer cell maturation. Nat Immunol. 2002;3:523–528. doi: 10.1038/ni796. [DOI] [PubMed] [Google Scholar]
- 5.Rosmaraki EE, et al. Identification of committed NK cell progenitors in adult murine bone marrow. Eur J Immunol. 2001;31:1900–1909. doi: 10.1002/1521-4141(200106)31:6<1900::aid-immu1900>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 6.Di Santo JP. Natural killer cell developmental pathways: A question of balance. Annu Rev Immunol. 2006;24:257–286. doi: 10.1146/annurev.immunol.24.021605.090700. [DOI] [PubMed] [Google Scholar]
- 7.Di Santo JP, Vosshenrich CA. Bone marrow versus thymic pathways of natural killer cell development. Immunol Rev. 2006;214:35–46. doi: 10.1111/j.1600-065X.2006.00461.x. [DOI] [PubMed] [Google Scholar]
- 8.Vosshenrich CA, et al. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat Immunol. 2006;7:1217–1224. doi: 10.1038/ni1395. [DOI] [PubMed] [Google Scholar]
- 9.Townsend MJ, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 10.Samson SI, et al. GATA-3 promotes maturation, IFN-gamma production, and liver-specific homing of NK cells. Immunity. 2003;19:701–711. doi: 10.1016/s1074-7613(03)00294-2. [DOI] [PubMed] [Google Scholar]
- 11.Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J Exp Med. 2007;204:1119–1130. doi: 10.1084/jem.20061959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikawa T, et al. Commitment to natural killer cells requires the helix-loop-helix inhibitor Id2. Proc Natl Acad Sci USA. 2001;98:5164–5169. doi: 10.1073/pnas.091537598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokota Y, et al. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- 14.Kim S, et al. In vivo natural killer cell activities revealed by natural killer cell-deficient mice. Proc Natl Acad Sci USA. 2000;97:2731–2736. doi: 10.1073/pnas.050588297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S, et al. Arrested natural killer cell development associated with transgene insertion into the Atf2 locus. Blood. 2006;107:1024–1030. doi: 10.1182/blood-2005-04-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper MA, et al. In vivo evidence for a dependence on interleukin 15 for survival of natural killer cells. Blood. 2002;100:3633–3638. doi: 10.1182/blood-2001-12-0293. [DOI] [PubMed] [Google Scholar]
- 17.French AR, et al. Chronic lymphocytosis of functionally immature natural killer cells. J Allergy Clin Immunol. 2007;120:924–931. doi: 10.1016/j.jaci.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 18.Nardelli J, et al. Expression and genetic interaction of transcription factors GATA-2 and GATA-3 during development of the mouse central nervous system. Dev Biol. 1999;210:305–321. doi: 10.1006/dbio.1999.9278. [DOI] [PubMed] [Google Scholar]
- 19.Jogi A, et al. Modulation of basic helix-loop-helix transcription complex formation by Id proteins during neuronal differentiation. J Biol Chem. 2002;277:9118–9126. doi: 10.1074/jbc.M107713200. [DOI] [PubMed] [Google Scholar]
- 20.Wang S, et al. A role for the helix-loop-helix protein Id2 in the control of oligodendrocyte development. Neuron. 2001;29:603–614. doi: 10.1016/s0896-6273(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 21.Urbanek P, et al. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 1994;79:901–912. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 22.Panganiban G, Rubenstein JL. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129:4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- 23.Morasso MI, et al. Placental failure in mice lacking the homeobox gene Dlx3. Proc Natl Acad Sci USA. 1999;96:162–167. doi: 10.1073/pnas.96.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cobos I, et al. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat Neurosci. 2005;8:1059–1068. doi: 10.1038/nn1499. [DOI] [PubMed] [Google Scholar]
- 25.Qiu M, et al. Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: Mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev Biol. 1997;185:165–184. doi: 10.1006/dbio.1997.8556. [DOI] [PubMed] [Google Scholar]
- 26.Depew MJ, Simpson CA, Morasso M, Rubenstein JL. Reassessing the Dlx code: The genetic regulation of branchial arch skeletal pattern and development. J Anat. 2005;207:501–561. doi: 10.1111/j.1469-7580.2005.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huntington ND, Vosshenrich CA, Di Santo JP. Developmental pathways that generate natural-killer-cell diversity in mice and humans. Nat Rev Immunol. 2007;7:703–714. doi: 10.1038/nri2154. [DOI] [PubMed] [Google Scholar]
- 28.Qiu M, et al. Null mutation of Dlx-2 results in abnormal morphogenesis of proximal first and second branchial arch derivatives and abnormal differentiation in the forebrain. Genes Dev. 1995;9:2523–2538. doi: 10.1101/gad.9.20.2523. [DOI] [PubMed] [Google Scholar]
- 29.Eisenstat DD, et al. DLX-1, DLX-2, and DLX-5 expression define distinct stages of basal forebrain differentiation. J Comp Neurol. 1999;414:217–237. doi: 10.1002/(sici)1096-9861(19991115)414:2<217::aid-cne6>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 30.Joshi R, et al. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131:530–543. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mann RS, Hogness DS. Functional dissection of Ultrabithorax proteins in D. melanogaster. Cell. 1990;60:597–610. doi: 10.1016/0092-8674(90)90663-y. [DOI] [PubMed] [Google Scholar]
- 32.Grabstein KH, et al. Inhibition of murine B and T lymphopoiesis in vivo by an anti-interleukin 7 monoclonal antibody. J Exp Med. 1993;178:257–264. doi: 10.1084/jem.178.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peschon JJ, et al. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med. 1994;180:1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Freeden-Jeffry U, et al. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He YW, Malek TR. Interleukin-7 receptor alpha is essential for the development of gamma delta + T cells, but not natural killer cells. J Exp Med. 1996;184:289–293. doi: 10.1084/jem.184.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sudo T, et al. Expression and function of the interleukin 7 receptor in murine lymphocytes. Proc Natl Acad Sci USA. 1993;90:9125–9129. doi: 10.1073/pnas.90.19.9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazzucchelli R, Durum SK. Interleukin-7 receptor expression: Intelligent design. Nat Rev Immunol. 2007;7:144–154. doi: 10.1038/nri2023. [DOI] [PubMed] [Google Scholar]
- 38.Woodside KJ, et al. Expression of Dlx and Lhx family homeobox genes in fetal thymus and thymocytes. Gene Expr Patterns. 2004;4:315–320. doi: 10.1016/j.modgep.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 39.Haga SB, et al. BP1, a new homeobox gene, is frequently expressed in acute leukemias. Leukemia. 2000;14:1867–1875. doi: 10.1038/sj.leu.2401912. [DOI] [PubMed] [Google Scholar]
- 40.Ferrari N, et al. DLX genes as targets of ALL-1: DLX 2,3,4 down-regulation in t(4;11) acute lymphoblastic leukemias. J Leukocyte Biol. 2003;74:302–305. doi: 10.1189/jlb.1102581. [DOI] [PubMed] [Google Scholar]
- 41.Chase MB, et al. BP1, a homeodomain-containing isoform of DLX4, represses the beta-globin gene. Mol Cell Biol. 2002;22:2505–2514. doi: 10.1128/MCB.22.8.2505-2514.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu S, et al. Distinct functions of two isoforms of a homeobox gene, BP1 and DLX7, in the regulation of the beta-globin gene. Gene. 2001;278:131–139. doi: 10.1016/s0378-1119(01)00716-8. [DOI] [PubMed] [Google Scholar]
- 43.Mpollo MS, et al. BP1 is a negative modulator of definitive erythropoiesis. Nucleic Acids Res. 2006;34:5232–5237. doi: 10.1093/nar/gkl680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blaser BW, et al. Donor-derived IL-15 is critical for acute allogeneic graft-versus-host disease. Blood. 2005;105:894–901. doi: 10.1182/blood-2004-05-1687. [DOI] [PubMed] [Google Scholar]
- 45.Kim S, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.