Abstract
The genetic impacts of hybridization between native and introduced species are of considerable conservation concern, while the possibility of reticulate evolution affects our basic understanding of how species arise and shapes how we use genetic data to understand evolutionary diversification. By using mitochondrial NADH dehydrogenase subunit 2 (ND2) sequences and 467 amplified fragment-length polymorphism nuclear DNA markers, we show that the introduced white sucker (Catostomus commersoni) has hybridized with two species native to the Colorado River Basin—the flannelmouth sucker (Catostomus latipinnis) and the bluehead sucker (Catostomus discobolus). Hybrids between the flannelmouth sucker and white sucker have facilitated introgression between the two native species, previously isolated by reproductive barriers, such that individuals exist with contributions from all three genomes. Most hybrids had the mitochondrial haplotype of the introduced white sucker, emphasizing its pivotal role in this three-way hybridization. Our findings highlight how introduced species can threaten the genetic integrity of not only one species but also multiple previously reproductively isolated species. Furthermore, this complex three-way reticulate (as opposed to strictly bifurcating) evolution suggests that seeking examples in other vertebrate systems might be productive. Although the present study involved an introduced species, similar patterns of hybridization could result from natural processes, including stream capture or geological formations (e.g., the Bering land bridge).
Keywords: conservation, hybridization, native fish, reticulate evolution
Hybridization can facilitate evolutionary novelty and diversification (1–5) or act as a force driving genetic homogenization or extinction (6–10). Hybridization stemming from nonnative species introductions is a known threat to the genetic integrity and persistence of native species. Well documented examples of the genetic threat to native species from introduced species include the threat to cutthroat trout (Oncorhynchus clarki) from introduced rainbow trout (Oncorhynchus mykiss) in the western United States (11, 12) and the virtual genetic elimination of the gray duck (Anas superciliosa) by introduced mallard ducks (Anas platyrhynchos) in New Zealand (13). The native fishes of the western United States, in particular, face an enormous threat because of pervasive translocations of nonnative fishes into different drainages where hybridization with native fishes has become a common occurrence (9).
The fishes of the Colorado River drainage have been greatly affected by habitat changes such as flow regulation and water diversion projects, as well as by introduced species that compete with, prey on, or hybridize with them (9). The white sucker (Catostomus commersoni) is widespread in eastern North America and is native to Wyoming east of the Continental Divide (14) but did not occur in the Colorado River Basin, west of the Continental Divide, at the time of settlement by Europeans. It was likely introduced near the beginning of the 20th century (14). Flannelmouth suckers (Catostomus latipinnis) and bluehead suckers (Catostomus discobolus) are native to the drainages of the Colorado River, west of the Continental Divide, and commonly occur sympatrically. In the brief time since their introduction, white suckers have become widespread and abundant throughout the Colorado River Basin. Moreover, hybridization appears to have become rampant between the introduced white sucker and the native flannelmouth sucker and represents an urgent conservation concern for native fishes in the drainage (15).
Recent studies suggest that reticulate evolution may be more common than previously suspected (1, 4, 16). A more reticulate process affects many aspects of evolutionary understanding, from the interpretation of genetic data to inferences about trait evolution and evolutionary novelty. Our interest was stimulated by field observations suggesting that initial hybridization between white suckers and flannelmouth suckers might be extending to hybridization of either or both species, or hybrids among them, with bluehead suckers. Although multispecies reticulate evolution has been studied extensively in plants (17), we are aware of no empirical examples in vertebrates, where the well documented examples (e.g., ref. 18) refer to pairs rather than trios of species.
To characterize patterns of hybridization, we analyzed nuclear and mitochondrial DNA in native bluehead suckers and flannelmouth suckers, introduced white suckers, and presumed hybrids among these three species, as assessed in the field by morphological intermediacy. Because of the possibility of reticulation and other complexities, better understanding of both speciation and hybridization results from using multiple genetic markers (17). Our study focused on the Muddy Creek drainage of the Colorado River, west of the Continental Divide in south-central Wyoming. We also obtained samples of white suckers from the Laramie River, east of the Continental Divide, where it is native and where the bluehead sucker and flannelmouth sucker do not occur. In addition, we used published mitochondrial NADH dehydrogenase subunit 2 (ND2) sequences of the Utah sucker (Catostomus ardens; ref. 19). Our goals were to (i) assess patterns of genetic divergence among the introduced white sucker and the native bluehead sucker and flannelmouth sucker, (ii) determine the genetic characteristics of morphologically judged hybrids to assess the nature and extent of hybridization among the two native and one introduced species, and (iii) examine the potential for introgression between the two native species mediated by hybridization with the introduced white sucker.
Results and Discussion
Assessment of pure versus hybrid phenotype occurred in the field, judged by variation in seven morphological characters that separate the three species (14), including mouthpart morphology. The bluehead sucker, in particular, has a distinctive and unique scraping ridge on the mouth (14). We generated nuclear amplified fragment length polymorphism (AFLP; ref. 20) markers for 161 individuals at 467 polymorphic loci by using three selective primer combinations. Fragment profiles were 96% similar for eight individual fish subjected to the AFLP procedure twice, indicating high reproducibility of the AFLP data. We also generated sequences from the mitochondrial ND2 for a subset of 46 fish representing all three species and their hybrids, including three longnose suckers (Catostomus catostomus) for outgroup purposes. There were 18 distinct ND2 haplotypes, including one in white sucker, two in flannelmouth sucker, two in longnose sucker, and three in bluehead sucker (GenBank accession nos. EU652910–EU652917). We also included 10 haplotypes from Utah suckers, sampled over a wide geographic region (19).
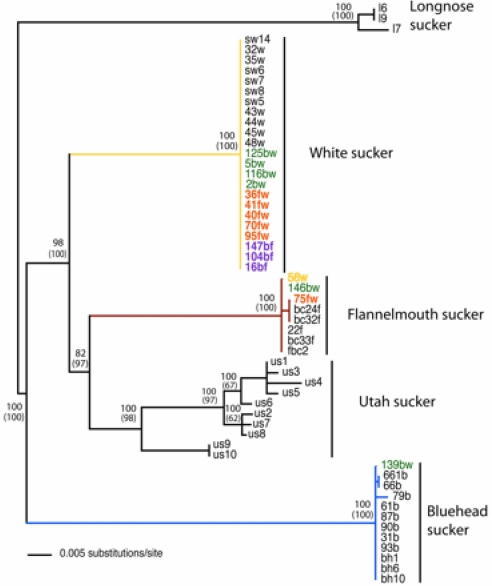
Several lines of evidence support a closer phylogenetic relationship between white sucker and flannelmouth sucker than between either of these species and bluehead sucker. A neighbor-joining dendrogram based on Nei and Li's (21) restriction-site distances across 467 AFLP loci showed clear separation of the bluehead sucker from the other two species but intermediate and complex patterns in hybrids (Fig. 1). We also examined AFLP fragment frequencies for “fixed” differences, here defined as a differential of ≥0.97 in frequency between species. Between flannelmouth sucker and white sucker, only two loci showed fixed differences. In contrast, 15 loci were fixed between bluehead sucker and flannelmouth sucker, and 21 were fixed between bluehead sucker and white sucker. Furthermore, AFLP loci showing fixed differences among the parental species were present at intermediate frequencies in hybrids. Clustering in the neighbor-joining tree showed that flannelmouth suckers and white suckers were more similar genetically than either was to the bluehead sucker, a pattern also shown by values of FST (bluehead vs. flannelmouth = 0.38; bluehead vs. white = 0.40; flannelmouth vs. white = 0.28) and Nei's distance (bluehead vs. flannelmouth = 0.25; bluehead vs. white = 0.26; flannelmouth vs. white = 0.17).
Fig. 1.
Unrooted neighbor-joining tree constructed from Nei and Li's restriction site distance from AFLP genotypes. The 34 bluehead suckers formed a monophyletic unit (a single wedge of blue). The 25 bluehead–white hybrids (green) occurred as a fairly cohesive unit, kept from being monophyletic only by three bluehead–flannelmouth hybrids (purple). Although most white suckers formed a fairly large monophyletic group (yellow wedge) that included all 14 white suckers from the Laramie River, a few white suckers from Muddy Creek were interspersed with flannelmouth–white hybrids (orange). The 38 flannelmouth samples (red) were interspersed with two flannelmouth–white hybrids (orange) and three flannelmouth–bluehead hybrids (purple).
Bayesian and parsimony-based phylogenetic analyses of mitochondrial ND2 sequences provided strong support for distinct clades corresponding to the parental species (Fig. 2), with substantial sequence divergence between them (bluehead vs. flannelmouth = 13.1%; bluehead vs. white = 12.5%; flannelmouth vs. white = 8.5%). Utah sucker, white sucker, and flannelmouth sucker formed a clade that was monophyletic with respect to the more distantly related bluehead sucker (Fig. 2). All white suckers in our study had the same ND2 haplotype (Fig. 2). The lone white sucker haplotype is consistent with the possibility of a small founding population of Muddy Creek white suckers derived from nearby rivers east of the Continental Divide, including the Laramie River. Hybrid fish had mostly (12/16) white sucker ND2 haplotypes, indicating that hybridization usually involved female white suckers. Nonetheless, one flannelmouth–white hybrid had a flannelmouth ND2 haplotype, one bluehead–white hybrid had a bluehead ND2 haplotype, and one white sucker had a flannelmouth haplotype, indicating introgression of mitochondrial DNA in both directions during hybridization.
Fig. 2.
Bayesian phylogenetic tree with a molecular clock enforced, based on mitochondrial ND2 sequence variation for white suckers, flannelmouth suckers, bluehead suckers, and their hybrids. Values at the nodes represent posterior probability values from the Bayesian analysis, and those in parentheses indicate bootstrap support based on 1,000 replicates in a parsimony analysis. The colors of the branches in the tree match the colors used for the other figures, with yellow for white suckers, red for flannelmouth suckers, and blue for bluehead suckers. Hybrids are identified at the branch tips by their colored highlighting, also as in the previous figure, with orange for flannelmouth–white hybrids, green for bluehead–white hybrids, and purple for bluehead–flannelmouth hybrids.
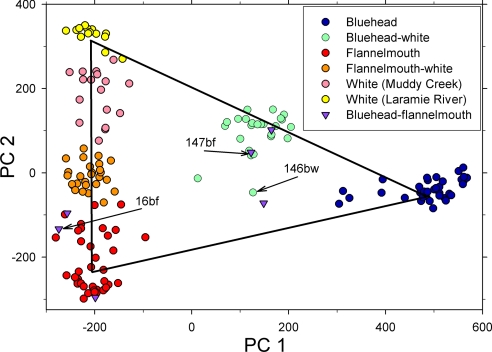
Additional AFLP analyses demonstrated that phenotype was generally a reliable guide to nuclear DNA genotype for all three parental species and their hybrids. Based on AFLP fragment frequencies, each individual received three likelihood scores (summing to 1) for membership in each of the three parental species by using a procedure analogous to an assignment test (22). We used principal components analysis to reduce the dimensionality of the likelihood scores; the first component captured 73% of the variance, and the second captured 25%. Thus, two-dimensional triangular plotting (Fig. 3) of likelihood (with the parental species at the apices of the triangle) led to virtually no loss of information content. In the likelihood triangle plot of Fig. 3, individuals representing the three parental species formed tight, nonoverlapping clusters, and hybrids occurred in clusters intermediate between parental species.
Fig. 3.
Likelihood plot from fragment frequencies at 467 loci, with colors representing field-judged phenotype and spatial placement representing genetic assignment. Axis labels PC 1 and PC 2 refer to principal components 1 and 2. Note that we distinguish here between white suckers from Muddy Creek (hybridization possible) and white suckers from the Laramie River (hybridization not possible). Hybrid phenotypes would be expected to fall along the line connecting the centroids of the three species (apices of the triangle); that is, they should be genetically intermediate between the parental species. Phenotype reliably indicated genotype, with the exception of the bluehead–flannelmouth (purple) hybrids; only one of those six hybrids was close to the expected position along the line between the appropriate apices. Furthermore, all three phenotypically bluehead–flannelmouth hybrids that we sequenced had white sucker haplotypes. We had AFLP genotypes for two of those fish, 147bf and 16bf, which are marked with arrows. The results suggest that flannelmouth sucker and bluehead sucker rarely hybridize directly, hybrids may even be inviable, and that viable hybrids may require a more complex backcrossing series involving white sucker. We conclude that the introduced white sucker and its subsequent hybrids may act as a bridge for introgression between the two previously reproductively isolated native species. Note also that a fish in the center of the triangle likelihood plot (146bw), characterized phenotypically and with AFLP as a bluehead–white hybrid, had a flannelmouth sucker haplotype, supporting the idea of a multiorigin genome (termed “muttsucker”).
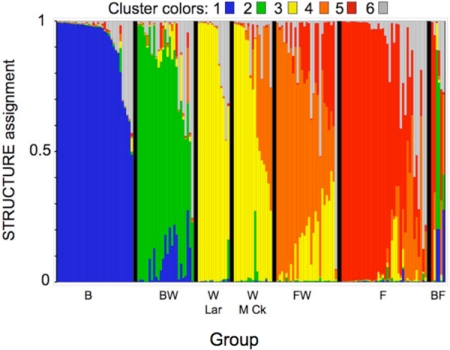
We used the Bayesian procedure in the program STRUCTURE (23, 24) to infer the number of genotypic clusters in the AFLP dataset and to assign every individual a probability of membership in each of the clusters. By using the method of Evanno et al. (25), we found a strong modal peak for seven genotypic clusters (K = 7). Five of the seven clusters were clearly interpretable—the three parental species plus the bluehead–white and flannelmouth–white hybrids. For these five clusters, most individuals had majority assignment (>0.5) to the cluster appropriate for their phenotype. The two remaining clusters had majority assignment for only eight fish, most of which were outliers from their respective groupings in the likelihood triangle plot (Fig. 3). Fish in every sampled taxon had various (usually nonmajority) amounts of assignment to either or both of these two remaining clusters. We therefore interpreted these clusters as denoting “other” and lumped them into a single Cluster 6 (represented by gray in the program STRUCTURE bar plot of Fig. 4). The STRUCTURE results (Fig. 4) were highly concordant with those from the likelihood triangle plot (Fig. 3) and with phenotypic judgments made in the field, indicating that hybrids are both genetically and phenotypically intermediate between the parental species.
Fig. 4.
Output from assignment procedure of program STRUCTURE for 161 suckers of seven groups (B, bluehead; BW, bluehead–white hybrids; W Lar, white suckers from the Laramie River; W M Ck, white suckers from Muddy Creek; FW, flannelmouth–white hybrids; F, flannelmouth; BF, bluehead–flannelmouth hybrids). The 161 individuals are aligned along the x axis, with the groups separated by black bars. Colors represent the proportion of assignment (y axis) of each individual to six clusters determined by STRUCTURE. In general, the clusters (colors) correspond well with the seven groups (groups of bars). For example, most bluehead suckers (B) fell in Cluster 1 (blue), with a few also receiving partial assignment to Cluster 6 (gray). Note that whereas white suckers from the Laramie River (W Lar, no hybridization) were assigned only to Clusters 3 (yellow) and 6 (gray), many white suckers from Muddy Creek (M Ck) were assigned to Cluster 4 (orange), the flannelmouth–white cluster. Also note that the six bluehead–flannelmouth hybrids (BF) showed by far the most intermingling of multiple, often approximately equivalent, cluster assignments within individuals, supporting the idea that they represent multigenome muttsuckers.
Several lines of evidence suggested that white suckers and flannelmouth suckers have hybridized extensively in Muddy Creek, leading to virtually continuous variation between the extremes of the parental species. The likelihood triangle plot (Fig. 3) showed a continuous spectrum of variation from white suckers to flannelmouth suckers from Muddy Creek. On the one hand, white suckers sampled from the Laramie River east of the Continental Divide, where no opportunity exists for hybridization with bluehead suckers or flannelmouth suckers, formed the cluster closest to the white sucker apex in the upper left of the likelihood triangle plot (Fig. 3). On the other hand, white suckers from Muddy Creek had genotypes strongly suggesting an admixture of flannelmouth sucker genetic material, as illustrated by their position closer to the flannelmouth sucker apex in the likelihood triangle plot (Fig. 3; see also Figs. 1 and 4).
The STRUCTURE results (Fig. 4) added further evidence for a continuous spectrum of variation between white suckers and flannelmouth suckers. All of the white suckers from the Laramie River (W Lar) had majority assignment to Cluster 3 (white). In contrast, white suckers from Muddy Creek (W M Ck) were less unambiguously assigned, with 6 of 17 given majority or near-complete assignment to Cluster 4 (flannelmouth–white) or Cluster 6 (other). Furthermore, one of the five Muddy Creek white suckers sequenced (56w) had a flannelmouth sucker ND2 haplotype (Fig. 2). Flannelmouth–white hybrids (FW) were mostly assigned to Cluster 4 (flannelmouth–white), with some individuals also receiving partial assignment to Clusters 3 (white) and 5 (flannelmouth). Finally, the flannelmouth sucker sample included five individuals with majority assignment to Cluster 6 (other) as well as several individuals with partial assignment to a smattering of all of the other clusters, with an emphasis on Cluster 4 (flannelmouth–white) and Cluster 6 (other). Thus, analyses based on both nuclear and mitochondrial DNA data showed a continuous spectrum of genetic variation between flannelmouth suckers and white suckers, suggesting an existing or incipient hybrid swarm.
Hybridization has also occurred between white suckers and bluehead suckers, although backcrossing appears to be far more limited. For example, bluehead–white hybrids formed a discrete intermediate cluster in the likelihood triangle plot (Fig. 3). Furthermore, bluehead suckers from Ringdahl Reservoir, where they occur in the absence of the opportunity for hybridization, were completely intermixed with Muddy Creek bluehead suckers in both the AFLP neighbor-joining dendrogram and the likelihood triangle plot (Figs. 1 and 3). In STRUCTURE, all but one of the bluehead suckers (B) had majority assignment to Cluster 1 (the bluehead cluster). That most of the bluehead–white hybrids formed a fairly distinct, intermediate grouping (Figs. 1 and 3) suggests that hybridization between bluehead sucker and white sucker rarely proceeds beyond the F1 generation.
In addition to the extensive hybridization between white suckers and flannelmouth suckers, several lines of evidence indicated that fish now occur with ancestors from all three parental species (termed “muttsuckers”). Bluehead–flannelmouth hybrids were rare and were anomalous genetically in that all but one showed evidence of white sucker nuclear DNA based on their placement in the likelihood triangle plot (Fig. 3). That is, only one bluehead–flannelmouth hybrid was close to the expected position along the line connecting the flannelmouth sucker and bluehead sucker apices in Fig. 3. Furthermore, a number of other fish showed evidence of three-way hybridization, as demonstrated by their position in the interior of the likelihood triangle plot (Fig. 3). Inspection of the STRUCTURE bar plot (Fig. 4) shows that most bluehead–flannelmouth hybrids, and a smattering of other fish, had assignments of >10% to all three parental species, suggesting that these individuals constitute a mixture of the genomes of all three parental species. In addition, one fish (146bw), classified by the AFLP likelihood triangle plot (Fig. 3) and phenotypic markers as a bluehead–white hybrid, had a flannelmouth sucker ND2 haplotype. Finally, all three of the fish we sequenced that had phenotypic and AFLP evidence for admixture of bluehead sucker and flannelmouth sucker (147bf, 104bf, and 16bf) had white sucker ND2 haplotypes (Fig. 2), suggesting that introduced white suckers and their hybrids provided the pathway for genetic exchange between the two native species.
Based on these lines of evidence, we conclude that the introduction of a nonnative catostomid species has eroded the genetic distinctiveness of two native species and could do the same in other locations in the Colorado River Basin. Flannelmouth suckers are in danger of extinction via hybridization because they hybridize readily with introduced white suckers, which have become pervasive throughout the range of flannelmouth suckers. Other studies have shown the potential for extinction caused by hybridization involving introduced and native taxa, such as rainbow trout and cutthroat trout (11) and gray ducks and mallard ducks (13). Such hybridization may represent a potentially common genetic consequence of introductions involving closely related species.
Hybridization may be a less imminent genetic threat to the bluehead sucker than to the flannelmouth sucker. Backcrossing of bluehead–white sucker hybrids appeared minimal, and hybrids between flannelmouth suckers and bluehead suckers were rare. That bluehead suckers were generally still genetically distinct was supported by the observation that those bluehead suckers from Muddy Creek judged to be phenotypically pure (white sucker present) were completely intermixed with bluehead suckers from the Ringdahl Reservoir (white sucker absent) in both the triangle likelihood plot (Fig. 3) and the neighbor-joining tree (Fig. 1). Furthermore, most of the bluehead–white hybrids formed a fairly distinct, intermediate grouping suggestive of hybridization rarely proceeding beyond the F1 generation (Figs. 1 and 3). The greater divergence between bluehead sucker and white sucker, compared with that between white sucker and flannelmouth sucker (Fig. 2), may partially explain why bluehead suckers show lower levels of introgressive hybridization with white suckers than do flannelmouth suckers (Figs. 1–4).
Nevertheless, muttsucker hybrids (e.g., 146bw, 147bf, and 16bf in Fig. 3), although infrequent, may represent an incipient genetic threat to the bluehead sucker through a breakdown of isolating mechanisms that previously separated flannelmouth suckers and bluehead suckers. Over longer time spans, such multiway hybrids may produce individuals more likely to hybridize with pure bluehead suckers. Thus, the introduced white sucker and its hybrids appear to be acting as a bridge that could eventually lead to a hybrid swarm involving all three species. Because the Utah sucker, and perhaps other species, are more closely related to the white sucker and flannelmouth sucker than to the bluehead sucker (Fig. 2), they may be even more susceptible to swamping by the muttsucker than is the bluehead sucker. Nolte et al. (5) showed that hybrid lineages can exhibit ecological adaptations that improve their ability to invade. In our study, hybridization also appears to play an important evolutionary role by facilitating the ability of an introduced species to hybridize sequentially with two native species, producing a complex, three-way admixture. Although our example involves an introduced species, faunal mixing can clearly occur naturally. In an aquatic context, the phenomenon of stream capture (26) can mix divergent fish faunas. In a terrestrial context, the Bering land bridge and Panamanian isthmus serve as examples of geological formations leading to faunal mixing that could well facilitate other cases of naturally occurring genetic bridging that still await detection.
Our findings have both conservation and evolutionary implications. From a conservation perspective, the introduced species now threatens the genetic integrity of not just one but two native species. Our results indicate that introductions of nonnative species may go beyond documented cases of single-species extinction by hybridization to a potentially cascading loss of genetic integrity in two or more native species that were previously reproductively isolated. From an evolutionary perspective, the genetic bridging suggests that we cannot assume that vertebrate evolution proceeds in a strictly bifurcating manner (Fig. 5). If hybridization has occurred in other vertebrate systems in a similar manner, then descendant species may have genomes inherited from three or more ancestral species and a consequently complex reticulate phylogeny that may span branches separated by (temporarily) uninvolved intermediate species. Concern for the impact of an introduced species led to this research, allowing us to glimpse a process, at or near its inception, that may be important in other, very different, contexts.
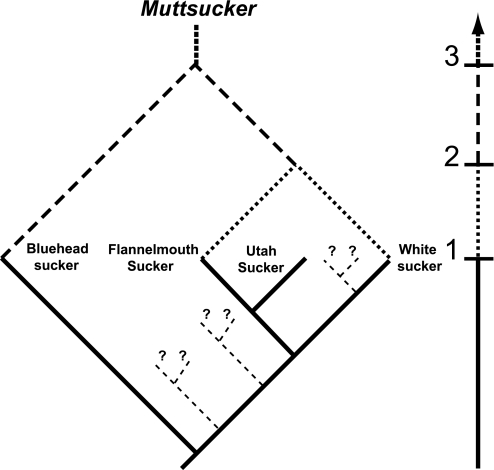
Fig. 5.
Hypothetical diagram for reticulate evolution among suckers (Catostomidae) in the drainages of the Colorado River. Before hybridization between flannelmouth sucker and white sucker (first bar on timeline at the right), little or no introgression occurred between native bluehead suckers and flannelmouth suckers. Once hybrid flannelmouth–white suckers appeared (second bar on timeline), hybridization with bluehead sucker became possible and may now proceed to produce more frequent hybrids (termed muttsuckers) with three ancestors (third bar on timeline). In the absence of the white sucker, direct hybridization between flannelmouth sucker and bluehead sucker might be absent or result in infertile F1 hybrids. Thus, flannelmouth–white hybridization may have produced the conditions necessary for introgression between the two native species and a consequently multireticulate phylogeny. The genetic bridging between the flannelmouth sucker and white sucker, and thence to bluehead sucker, extends across the intermediate Utah sucker. Dashed branch stubs (truncated for visual clarity) indicate the possibility that other, unstudied species may lie between the species studied and that they may also be at risk of being swamped by the multiorigin muttsucker.
Materials and Methods
Field Methods.
Samples of 137 individuals were collected from the Muddy Creek drainage of south-central Wyoming. At the time of sampling, all individual fish were classified as white sucker (n = 17), bluehead sucker (n = 24), or flannelmouth sucker (n = 38) after the criteria of Baxter and Stone (14) or as one of the three types of hybrids between the parental species (bluehead–flannelmouth, n = 6; bluehead–white, n = 25; flannelmouth–white, n = 27). Bluehead suckers, in particular, have a distinctive scraping mouth ridge that served as a morphological cue to possible hybridization with the other taxa. To provide a check on genotypic composition in the absence of hybridization, we included in all analyses 10 bluehead suckers from Ringdahl Reservoir in Wyoming, where white suckers do not occur, and 14 white suckers from the Laramie River near Laramie, WY, where neither bluehead suckers nor flannelmouth suckers occur.
Laboratory Techniques.
DNA was extracted from samples by using Qiagen DNeasy tissue kits beginning with 5 mg of tissue or a 1 × 1 mm fin clip. DNA extracts were visualized for quality on 1.5% agarose gels, and concentration was evaluated by using a 100-bp mass ladder (NEB).
The AFLP procedure was carried out as in Vos et al. (20) with slight modifications. Restriction digestion and adaptor-ligation were carried out simultaneously on 0.5 μg of genomic DNA by using the restriction endonucleases EcoRI and MseI (NEB). AFLP adaptor pairs were attached to digested fragments by using T4 DNA ligase (NEB). Restriction and ligation reactions were performed simultaneously in 11-μl volumes and incubated for 18 h at 38°C. After incubation, these reactions were diluted with 170 μl of 0.1× TE buffer (10 mM Tris/1 mM EDTA, pH 8.0). Preselective and selective primers were based on primer core sequences EcoRI 5′-GACTGCGTACCAATTC-3′ and MseI 5′-GATGAGTCCTGAGTAA-3′ (EcoRI and MseI hereafter). Preselective amplifications were run with 4 μl of the diluted restriction–ligation products, 15 μl of PCR core mix (Promega 10× reaction buffer, 1 mM MgCl2, 0.2 mM dNTPs, 1 unit of Promega Taq DNA polymerase), and 1 pmol of preselective primers, which consisted of the adaptor primer sequences with one additional nucleotide at the 3′ ends (EcoRI-A and MseI-C). Preselective PCR conditions were 20 times (94°C for 30 s, 56°C for 1 min, 72°C for 2 min) and a final extension at 60°C for 30 min. We diluted preselective amplification product with 170 μl of 0.1× TE.
Selective amplifications were run with 3 μl of diluted preselective amplification product, 15 μl of AFLP core mix, 1 pmol of selective MseI primer, and 1 pmol of the fluorescently labeled EcoRI-selective primer. Both EcoRI-selective and MseI-selective amplification primers had three extra nucleotides at the 3′ ends to reduce the number of fragments amplified to a manageable number. We used three selective primer combinations (EcoRI-ACT MseI-CTA, EcoRI-AAC MseI-CTT, and EcoRI-AAT MseI-CAT) to generate AFLP fragments. One microliter of each selective amplification product was run with 8.75 μl of formamide and 0.45 μl of GeneScan 500 ROX-labeled size standard (ABI) on an ABI 3130 capillary sequencer.
We amplified a 1,500-bp region of the mitochondrial ND2 by using the primers ND21500F (5′-TAAGCTTTCGGGCCCATACC-3′) and ND21500R (5′-GGCTCAGGCACCAAATACTA-3′) as described in ref. 19. PCRs were carried out in 20-μl volumes with 100 ng of DNA, 1× reaction buffer, 1 mM MgCl2, 0.2 mM dNTPs, 1 pmol of primers, and 1 unit of Taq DNA polymerase (Promega). DNA was denatured at 95°C for 2 min followed by 35 cycles of 95°C for 60 s, 58°C for 30 s, and 72°C for 90 s with a final 5-min extension at 72°C. The PCR product was cleaned up by using the enzymes exonuclease and shrimp alkaline phosphatase (NEB) with 1 unit of exonuclease 1 and 1 unit of shrimp alkaline phosphatase per 10 μl of product. These reactions were incubated at 37°C for 45 min and heated to 80°C for 15 min to deactivate the enzymes. Sequencing reactions were carried out from one direction by using the primer ND2459 F (5′-CACTGCAGCCGCTATAATC-3′) as in ref. 19 by using Big Dye sequencing reaction kits (ABI) and were visualized on an ABI 3100 capillary DNA sequencer. We sequenced this region for 12 white suckers, 11 bluehead suckers, 5 flannelmouth suckers, 6 bluehead–white sucker hybrids, 6 flannelmouth–white sucker hybrids, 3 bluehead–flannelmouth sucker hybrids, and 3 longnose suckers. All sequences have been submitted to the National Center for Biotechnology Information (NCBI). We also included haplotypes from 10 Utah Suckers deposited at the NCBI (GenBank accession nos. DQ360093, DQ360095, and DQ360099–DQ360106) (19).
Data Analysis.
AFLP fragment presence or absence in each lane file was analyzed by using the program Genemapper (ABI). We considered unambiguously discernible fragment sizes generated by each selective primer combination as dominant marker loci with two states, present (1) or absent (0). We limited analyses to fragment sizes between 70 and 400 bp, which resulted in a total of 467 unambiguously scoreable loci, recorded in a matrix that included all loci and all individuals. We calculated fragment frequency differentials as the difference in mean fragment frequencies over all loci between pairs of taxa (e.g., bluehead suckers vs. white suckers). Pairwise estimates of FST and Nei's genetic distance (D) among the three species and hybrid samples were obtained by using AFLP-SURV 1.0 (27).
We created a pairwise matrix of genetic distances among individuals with the restriction fragment approach of Nei and Li (21), as implemented in the RestDist routine of PHYLIP (ref. 28; available at http://evolution.genetics.washington.edu/phylip.html). One advantage of this distance measure is that it works directly from the 1 or 0 presence–absence form of the AFLP data, without the assumptions necessary when estimating allele frequencies from a dominant marker. From this distance matrix, we created an unrooted neighbor-joining tree with individuals as the operational taxonomic units by using the Neighbor routine of PHYLIP. We used TreeExplorer software (ref. 29; available at http://evolgen.biol.metro-u.ac.jp/TE/TE_man.html) to condense branch tips into wedges when individuals of the same type clustered.
Because of a lack of insertion and deletion events, sequences representing 393 base pairs of mitochondrial ND2 were easily aligned manually in BIOEDIT (ref. 30; available at www.mbio.ncsu.edu/BioEdit/bioedit.html). We estimated phylogenetic trees by using Bayesian methods implemented in MrBayes (31) and with parsimony by using PAUP (32). For Bayesian analyses, we used the general time reversible model of sequence evolution with among-site rate variation following a gamma distribution, enforced the molecular clock, and evaluated support at nodes with posterior probabilities generated in MrBayes. Support at the nodes for the parsimony analysis was evaluated with 1,000 bootstrap replicates (Fig. 2) in PAUP.
By using the fragment frequencies for each of the three sets of phenotypically judged parental species (white, flannelmouth, bluehead) we calculated likelihood as the natural logarithm of the product of the fragment frequencies across the 467 loci, in a manner analogous to the allelic assignment test of ref. 22. As a correction for zero-frequency fragments, we assumed a frequency of 0.005 (22). Each individual thus had a likelihood score for each of the three parental species. The resulting three-dimensional plot was then reduced to two dimensions by principal components analysis (PC1 and PC2 captured >98% of the variance). Thus, although the original data were discrete (0/1), the data used for the principal components analysis were continuous. We used the program STRUCTURE version 2.2.3 (ref. 24; available at http://pritch.bsd.uchicago.edu./structure.html), which implements the Bayesian approach of Pritchard et al. (23), to assess whether the sampled genotypes were substructured into multiple (K > 1) clusters or constitute a panmictic Hardy–Weinberg population (K = 1). Our primary purpose was to assess the degree of admixture among species. We used the method of Evanno et al. (25) to select the best-supported number of clusters. We used multiple runs with at least 10,000 burn-ins and 200,000 repetitions and without providing a priori information on population membership. We ran the best-supported cluster number (K = 7) with a burn-in of 50,000 and a run length of 500,000. All runs used the admixture model with correlated allele frequencies and all other settings at their default values.
Acknowledgments.
We thank A. Buerkle, D. Irwin, and three anonymous reviewers for comments that substantially improved the manuscript. M. Quist provided valuable assistance in the field. We also thank D. Bailey for kindly providing laboratory space and discussion at New Mexico State University during the early phases of this work. Funding came from the Bureau of Land Management to D.B.M., F.J.R., and W.A.H. and from an Environmental Protection Agency Greater Research Opportunities fellowship to T.L.P.
Footnotes
References
- 1.Arnold ML. Natural Hybridization and Evolution. Oxford: Oxford Univ Press; 1997. [Google Scholar]
- 2.Grant PR, Grant BR, Petren K. Hybridization in the recent past. Am Nat. 2005;166:56–67. doi: 10.1086/430331. [DOI] [PubMed] [Google Scholar]
- 3.Jorgensen S, Mauricio R. Hybridization as a source of evolutionary novelty: Leaf shape in a Hawaiian composite. Genetica (The Hague) 2005;123:171–179. doi: 10.1007/s10709-003-2740-2. [DOI] [PubMed] [Google Scholar]
- 4.Seehausen O. Hybridization and adaptive radiation. Trends Ecol Evol. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Nolte AW, Freyhoff J, Tautz D. When invaders meet locally adapted types: Rapid moulding of hybrid zones between sculpins (Cottus, Pisces) in the Rhine system. Mol Ecol. 2006;15:1983–1993. doi: 10.1111/j.1365-294X.2006.02906.x. [DOI] [PubMed] [Google Scholar]
- 6.Rhymer JM, Simberloff D. Extinction by hybridization and introgression. Annu Rev Ecol Syst. 1996;27:83–109. [Google Scholar]
- 7.Allendorf FW, Leary RF, Spruell P, Wenburg JK. The problems with hybrids: Setting conservation guidelines. Trends Ecol Evol. 2001;16:613–622. [Google Scholar]
- 8.Wolf DE, Takebayashi N, Rieseberg LH. Predicting the risk of extinction through hybridization. Conserv Biol. 2001;15:1039–1053. [Google Scholar]
- 9.Rahel FJ. Homogenization of freshwater faunas. Annu Rev Ecol Syst. 2002;33:291–315. [Google Scholar]
- 10.Olden JD, Poff NL, Douglas MR, Douglas ME, Fausch KD. Ecological and evolutionary consequences of biotic homogenization. Trends Ecol Evol. 2004;19:18–24. doi: 10.1016/j.tree.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Dowling TE, Childs MR. Impact of hybridization on a threatened trout of the Southwestern United States. Conserv Biol. 1992;6:355–364. [Google Scholar]
- 12.Utter FM, Allendorf FW. Phylogenetic relationships among species of Oncorhynchus: A consensus view. Conserv Biol. 1994;8:864–867. [Google Scholar]
- 13.Rhymer JM, Williams MJ, Braun MJ. Mitochondrial analysis of gene flow between New Zealand mallards (Anas platyrhynchos) and grey ducks (A. superciliosa) Auk. 1994;111:970–980. [Google Scholar]
- 14.Baxter GT, Stone MD. Fishes of Wyoming. Cheyenne: Wyoming Game and Fish Dept; 1995. pp. 128–131. [Google Scholar]
- 15.Utah Division of Wildlife Resources (UDWR) Range-wide Conservation Agreement and Strategy for Roundtail Chub Gila Robusta, Bluehead Sucker Catostomus discobolus, and Flannelmouth Sucker Catostomus latipinnis. 2006 (report to the Utah Dept of Natural Resources, Div of Wildlife Resources to Colorado River Fish and Wildlife Council, Salt Lake City), UDWR Publication No. 06-18. [Google Scholar]
- 16.Gompert Z, Fordyce JA, Forister ML, Shapiro AM, Nice C. Homoploid hybrid speciation in an extreme habitat. Science. 2006;314:1923–1925. doi: 10.1126/science.1135875. [DOI] [PubMed] [Google Scholar]
- 17.Linder CR, Rieseberg LH. Reconstructing patterns of reticulate evolution in plants. Am J Bot. 2004;91:1700–1708. [PMC free article] [PubMed] [Google Scholar]
- 18.Dowling TE, Secor CL. The role of hybridization and introgression in the diversification of animals. Annu Rev Ecol Syst. 1997;28:593–619. [Google Scholar]
- 19.Mock KE, et al. Rangewide molecular structuring in the Utah sucker (Catostomus ardens) Mol Ecol. 2006;15:2223–2238. doi: 10.1111/j.1365-294X.2006.02932.x. [DOI] [PubMed] [Google Scholar]
- 20.Vos P, et al. A new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paetkau D, Calvert W, Stirling I, Strobeck C. Microsatellite analysis of population structure in Canadian polar bears. Mol Ecol. 1995;4:347–356. doi: 10.1111/j.1365-294x.1995.tb00227.x. [DOI] [PubMed] [Google Scholar]
- 23.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pritchard JK. Pritchard Lab, Univ of Chicago; 2007. STRUCTURE, Software for Inferring Population Structure from Genetic Data. Version 2.2. [Google Scholar]
- 25.Evanno G, Regnaut S, Goudet J. Detecting the numbers of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 26.Matthews WJ. Patterns in Freshwater Fish Ecology. New York: Chapman and Hall; 1998. p. 178. [Google Scholar]
- 27.Vekemans X. Belgium: Laboratoire de Génétique et Ecologie Végétale, Université Libre de Bruxelles; 2002. AFLP-SURV, Software for Estimating Genetic Diversity and Population Genetic Structure from Population Samples. version 1.0. [Google Scholar]
- 28.Felsenstein J. Univ of Washington School of Medicine; 2007. PHYLIP, Phylogeny Inference Package. Version 3.6. [Google Scholar]
- 29.Tamura K. TreeExplorer Manual. Tokyo Metropolitan Univ; 2007. [Google Scholar]
- 30.Hall T. Carlsbad, CA: Ibis Biosciences; 2007. BioEdit, a Biological Sequence Alignment Editor. [Google Scholar]
- 31.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 32.Swofford DL. Sunderland, MA: Sinauer Associates; 2003. PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods) Version 4.0b 10. [Google Scholar]