Abstract
Calmodulin plays a vital role in mediating bidirectional synaptic plasticity by activating either calcium/calmodulin-dependent protein kinase II (CaMKII) or protein phosphatase 2B (PP2B) at different calcium concentrations. We propose an allosteric model for calmodulin activation, in which binding to calcium facilitates the transition between a low-affinity [tense (T)] and a high-affinity [relaxed (R)] state. The four calcium-binding sites are assumed to be nonidentical. The model is consistent with previously reported experimental data for calcium binding to calmodulin. It also accounts for known properties of calmodulin that have been difficult to model so far, including the activity of nonsaturated forms of calmodulin (we predict the existence of open conformations in the absence of calcium), an increase in calcium affinity once calmodulin is bound to a target, and the differential activation of CaMKII and PP2B depending on calcium concentration.
Keywords: allostery, synaptic plasticity, calcium binding, cooperativity, conformational transition
Activity-dependent changes in synaptic strength (1) have long been used as a paradigm to study learning and memory (reviewed in ref. 2). Calcium signaling is a key factor in both long-lasting increases [known as long-term potentiation (LTP)] and long-lasting decreases [long-term depression (LTD)] in synaptic strength. According to a model first proposed by Lisman (3), the coordinated activity of a pair of neurons leads to a large increase in calcium levels in the postsynaptic neuron and an increase in synaptic strength, whereas the activity of only one of the two neurons results in more moderate postsynaptic calcium levels and, consequently, a reduction in synaptic strength. Calcium entering the postsynaptic neuron through NMDA receptors and voltage-operated calcium channels and from the endoplasmic reticulum activates calmodulin. Activated calmodulin may bind to calcium/calmodulin kinase II (CaMKII) and increases its activity (4–6). Active CaMKII enhances the function of AMPA receptor channels by phosphorylating the GluR1 subunit (7). It also mediates an increase of AMPA receptor delivery to the postsynaptic membrane (8). These roles are consistent with reports implicating CaMKII in some forms of learning and memory (9). In contrast, lower amounts of calcium in the postsynaptic neuron will cause calmodulin to activate PP2B, leading to activation of protein phosphatase 1 and a subsequent reduction of CaMKII activity (reviewed in ref. 10). It remains to be explained, however, how calmodulin performs this dual function dependent on calcium levels.
Calmodulin is a ubiquitous regulatory protein that binds four calcium ions (11, 12). It is a single polypeptide chain of 148-aa residues (13) and can adopt two distinct conformations: in the absence of calcium, its EF hands typically adopt an inactive, compact (closed) form (14). When bound to four calcium ions, they are found in an open active form (15).
A variety of models for calmodulin activation and action have been used in the past. Each of these models reflects some properties of calmodulin and is reasonably applicable in contexts in which only these properties are relevant. However, none of these models can satisfactorily account for all of the observed properties of calmodulin such as cooperativity of calcium binding and different affinities for different calcium-binding sites (16), activation of targets by unsaturated calmodulin (17, 18), and increased affinity for calcium upon binding to targets (18–20). We propose an alternative model, based on a biophysical description of the conformational transitions. Originally applied to oligomeric proteins with symmetric identical subunits (21), this approach can also be adapted to a single polypeptide chain with multiple binding sites. The resulting generalized allosteric model of calmodulin can reconcile different properties of calmodulin, including differential activation of PP2B and CaMKII, residual activation of CaMKII at low calcium concentration, differences between the binding sites in terms of calcium affinity, and the existence of active and inactive conformations.
Allosteric Model of Calmodulin.
In our model, calmodulin can exist in two different states, the active open [relaxed (R)] state and the inactive closed [tense (T)] state. Each of these states can bind four calcium ions (Fig. 1). When no calcium is bound, the T state prevails, because its free energy is lower than that of the unbound R state. Consecutive binding of calcium ions, however, progressively stabilizes the R state until the free energy of the R state is lower than that of the T state, so the R state is favored.
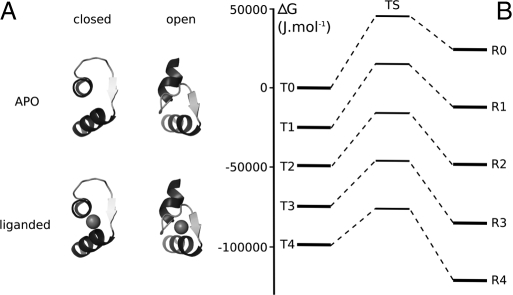
Fig. 1.
Thermodynamic model of calmodulin regulation by calcium. (A) Representative structures of a calmodulin EF hand modeled in this article. Residues 49–75 are shown. The closed apo structure comes from (14) [Protein Data Bank (PDB) ID code: 1CFD]. The closed calcium-liganded structure is inferred from (51) (PDB ID code: 2PQ3), using the position of Zn2+. The open structures come from (52) (PDB ID code: 3CLN). (B) Summarized free energy diagram for the different states of calmodulin. Energy levels (in joules per mole) were computed as in ref. 62. Each level of energy represents the average of all of the forms carrying the same number of calcium ions. Free energy differences between T state and corresponding R state relate to the allosteric isomerization constant. Between corresponding T and R states, a hypothetical transition state is depicted based on estimates of rate constants. Closed T state is shown on the left, open R state on the right, and the transition state in the middle.
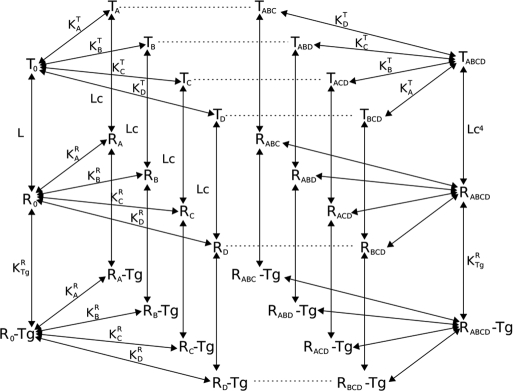
The four different binding sites are designated A, B, C, and D (A and B on the N-terminal domain, C and D on the C-terminal domain, with no sequential order being implied within the domains). Each of the states and each of the reactions is explicitly modeled, with distinct dissociation constants and R to T transition probabilities for each of the sites (Fig. 2). The constant L describes the equilibrium between both states when no calcium ion is bound: L = [T0]/[R0]. If L is very large, most of the protein exists in the tense state in the absence of calcium. If L is small (close to one), the R state is nearly as populated as the T state. The constants cA, cB, cC, and cD describe the ratio of dissociation constants for the R and T states for each site: ci = KiR/KiT. If c is one, both R and T states have the same affinity for calcium. The c values also indicate how much the equilibrium between T and R states changes upon calcium binding: the smaller c, the more the equilibrium shifts toward the R state.
Fig. 2.
Scheme of reactions used in the allosteric model of calmodulin. For clarity, only the first and fourth calcium-binding events are depicted in detail. Tg stands for target, which can be either CaMKII or PP2B.
The formula for fractional occupation of an allosteric protein in the absence of allosteric effectors (21) can be generalized to describe nonequivalent calcium-binding sites. In the case of four binding sites, the generalized expression is:
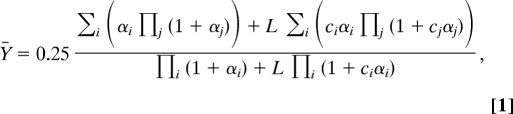 |
where i, j ∈ {A, B, C, D}, and j ≠ i. αi = [Ca2+]/KiR, where KiR denotes the dissociation constant for calcium to site i in the R state.
In a similar way, a general formulation for the fraction of protein in the R state, R̄, can be expressed as:
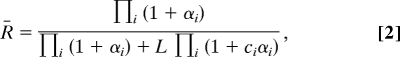 |
where i ∈ {A, B, C, D}. The model was further extended to include calmodulin binding to its targets. It was shown that autoinhibitor domains of CaMKII and PP2B bound to the same, open, conformation of calmodulin (22, 23). Therefore, we introduced the binding of both CaMKII and PP2B to calmodulin in the R state, independent of the number of bound calcium ions. Upon binding to its targets, the equilibrium of calmodulin shifts toward the R state, and the targets thus act as allosteric activators for calmodulin.
Parameter Determination.
The allosteric isomerization constant, L, the microscopic dissociation constants KiR for the R state for each binding site, and the ratios of R and T state affinity, ci, for each site, cannot be directly obtained from experimental literature. Experimental results do allow us, however, to constrain parameter space and obtain reasonable estimates. We used calcium-binding data in the presence and absence of an allosteric activator for mutant and wild-type forms of calmodulin.
We introduced the additional assumption that all four ci values are identical. This is based on the fact that both R and T states are symmetrical and on the idea that the free energy of transition is spread over the whole molecule.
To obtain an estimate of c and of the isomerization constant L, a reduced model of calmodulin was used. In this simple model, all four calcium-binding sites are equivalent, and one molecule of allosteric activator can bind to each molecule of calmodulin. In this case, the fractional occupation Ȳ of an allosteric protein in the presence of an allosteric activator can be described as (24):
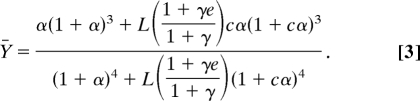 |
In this equation, α = [Ca2+]/KR, γ = [A]/KAR, and e = KAR/KAT; KR denotes the dissociation constant for calcium binding to the R state, L the allosteric equilibrium constant, [A] the concentration of allosteric activator, KAR the dissociation constant for binding of the allosteric activator to the R state, and KAT the dissociation constant for binding of the allosteric activator to the T state. The factor (1 + γ e)/(1 + γ) modulates the apparent isomerization constant as a function of activator concentration: if more target is present, the apparent value of L decreases. Peersen et al. (25) have measured calcium binding to calmodulin in the absence and presence of several target proteins, which act as allosteric effectors of calmodulin. By taking the calcium concentration at Ȳ = 1/2 from four different binding curves (absence of target, presence of skMLCK, PhK5, or CaATPase, respectively) and inserting them into the above equation, we obtained a system of four equations, which depend on L, c, and on e values for skeletal myosin light chain kinase, phosphorylase kinase, and Ca2+-ATPase, respectively. We minimized this system of equations by using the least-square function provided in Scilab (www.scilab.org). To avoid local minima, we ran 105 minimizations, each one with different initial values. By this approach we estimated L to be 20,670 and c to be 3.96 × 10−3. A full list of equations and details about the minimization process are given in supporting information (SI).
To determine the dissociation constants for all four binding sites, we used a similar approach, making use of the general formula for fractional occupancy presented above (Eq. 1). The formula can be simplified to reflect calmodulin binding to calcium under different experimental constraints. For instance, it can be reduced to the two N-terminal binding sites to describe recombinant versions of calmodulin in which the C-terminal binding sites are ablated. Using the available experimental literature, one thus obtains a system of four equations and corresponding data: the full equation for Ȳ (using data from ref. 26), two reduced equations for Ȳ with N- or C-terminal binding sites only, respectively (using data from experiments with recombinant calmodulin from ref. 18) and one reduced equation for Ȳ in which only the R state is considered (using data from ref. 25 in the presence of skMLCK, where it can be assumed that calmodulin exists mostly in the R state). There are thus four constraints that can be used to restrict parameter space. We used the calcium concentration at from the experimental literature to obtain eight equations that depend on KiR values. The eight equations are listed in SI.
To avoid the problem of local minima, we resorted to systematic sampling of parameter space and generated a script that tested all possible combinations of KiR values within a broad range. A detailed description of this can be found in SI. The resulting values are KAR = 8.32 × 10−6 M, KBR = 1.66 × 10−8 M, KCR = 1.74 × 10−5 M, and KDR = 1.45 × 10−8 M.
Kinetic Simulations.
Although Eq. 1 provides a general formula for calcium saturation in the case of four nonequivalent binding sites in the absence of allosteric effectors, the situation becomes less tractable in the presence of two competing allosteric activators (CaMKII and PP2B, in this case), especially at low calcium concentrations, where initial calcium concentration can differ significantly from free calcium concentration at steady state. Moreover, our intention was to provide an accurate model of calmodulin activation, which can serve as a basis for more complex models of signaling networks within the postsynaptic density (PSD). Both these concerns can be met by formulating the model as a system of reactions for kinetic simulations.
Every reaction was split into a separate forward and backward reaction. The full model comprises 352 reactions: 32 reactions describing calcium binding to the target-free T state, 32 reactions describing the corresponding dissociation events, 64 reactions describing calcium binding to and dissociation from the target-free R state, 32 reactions describing transitions between the R and T states, 32 reactions describing the binding and dissociation of CaMKII, another 32 reactions describing the binding and dissociation of PP2B, and 128 reactions describing calcium binding to and dissociation from CaMKII- or PP2B-bound calmodulin. A complete list of reactions can be found in SI.
The kon for calcium binding was assumed to be the same for all four binding sites and both states, because it can be assumed to be controlled only by calcium and calmodulin diffusion and size (random exploration). Different affinities were represented by different koff values.
Simulations were run by using the parameter-scan facility of the simulator COPASI (27).
Comparison of Calcium-Binding Curve to Experimental Results.
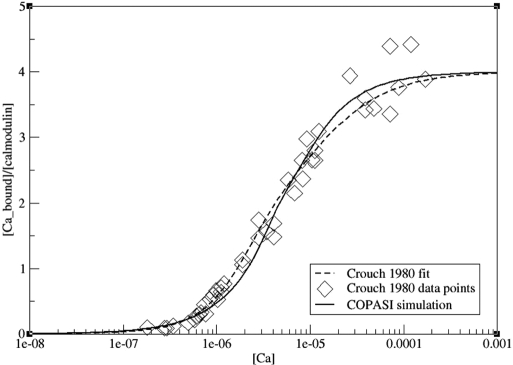
Our model reproduces experimental measurements of calmodulin binding to calcium. Fig. 3 compares the outcome of our simulation with results reported by Crouch and Klee (16). [For more figures comparing the simulation outcome with data reported by Peersen et al. (25) and Porumb (30), refer to SI.]
Fig. 3.
Comparison between simulation results and experimental results reported by Crouch and Klee (16). Moles of calcium bound per mole of calmodulin are shown as a function of calcium concentration. Diamonds, data points measured by Crouch and Klee (16); dashed line, curve used in Crouch and Klee to fit experimental data points; solid line, steady-state results of simulations at different initial calcium concentrations. Calmodulin concentration used was 2 × 10−7 M.
Apparent sequential association constants obtained by fitting a version of the Adair equation (43) to our model are listed in Table 1. A comparison to association constants reported in the experimental literature about calmodulin (16, 18, 28–30) shows that our association constants lie within the range of experimentally determined values.
Table 1.
Apparent sequential association constants (molar) for our model, and comparison to the models of Crouch and Klee (16), Porumb (30), and to other experimental reports (18, 28–30) and data reviews (28)
Activity of Various Forms of Nonsaturated Calmodulin.
By calculating the equilibrium constant for the transition between R and T states for calmodulin species that are bound to one or more calcium ions, one can see what fraction of calmodulin is active under steady-state conditions.
For instance, the relation between the R and T states for calmodulin with exactly two calcium ions bound is given by (R2/T2 = 1/(Lc2 ≈3). Similarly, without any calcium bound, there are more than 20,000× more calmodulin molecules in the T than in the R state. With one calcium ion bound, there are still ≈80× more calmodulin molecules in the T than in the R state. The equilibrium shifts toward the R state when two or more calcium ions are bound: with three calcium ions bound, there is ≈800× as much calmodulin in the R state as in the T state, and when fully saturated, there are nearly 200,000 calmodulin molecules in the R state for each calmodulin molecule in the T state. The shift of equilibrium from T to R state with three or more calcium ions bound can also be seen from the free energy diagram (see Fig. 1).
Altered Affinity for Calcium if Calmodulin Is Bound to a Target.
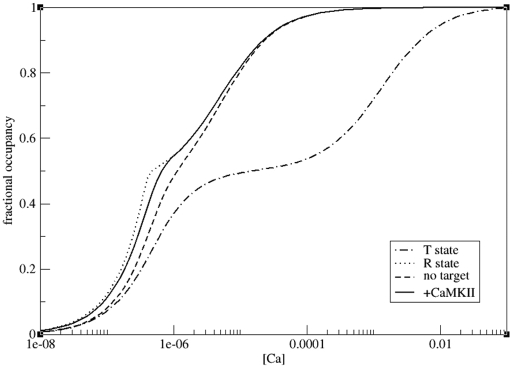
It has been reported (18–20) that the apparent affinity of calmodulin for calcium increases if calmodulin is bound to a target. To reproduce this effect, we simulated calcium binding to 2 × 10−7 M calmodulin at varying calcium concentrations in the absence of target and in the presence of CaMKII. We also performed simulations on two reduced models, representing only the R state or only the T state of calmodulin, respectively. Results are shown in Fig. 4. The fractional occupancy curve illustrates how, with increasing calcium concentration, the calmodulin population shifts from mostly T state to mostly R state, with the corresponding affinities for calcium. The presence of target further stabilizes the R state, resulting in the observed increase of affinity.
Fig. 4.
Increased affinity of calmodulin for calcium in the presence of a target protein. Upper dotted line, R state only; lower dotted/dashed line, T state only; dashed line, combined R and T states in the absence of target; solid line, combined R and T states in the presence of CaMKII. All lines are steady-state results of simulations at different initial calcium concentrations. Calmodulin concentration was 2 × 10−7 M.
Differential Activation of CaMKII and PP2B.
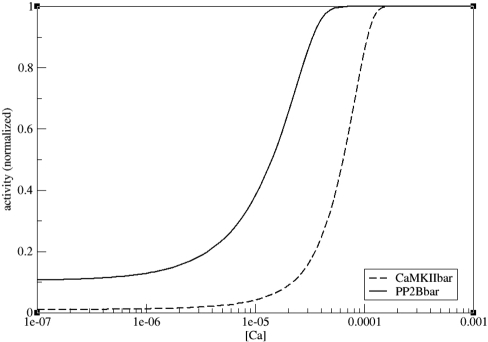
An intriguing feature of calmodulin is its ability to differentially activate PP2B or CaMKII, depending on calcium input. It is known that PP2B has a much higher affinity for calmodulin (31) than CaMKII (32). However, although PP2B has been detected in PSD fractions (reviewed in ref. 33), CaMKII is far more abundant and, in fact, constitutes a major PSD protein (34, 35). Together with our allosteric model, this information can explain the differential activation of PP2B at low calcium concentrations and of CaMKII at high calcium concentrations.
As Fig. 5 shows, we can reproduce a differential activation of PP2B and CaMKII in our model. At subsaturating calcium concentrations, PP2B becomes preferentially activated due to its high affinity for the R state. PP2B binding stabilizes the R state and thus increases the apparent affinity of calmodulin for its targets. At higher calcium concentrations, more calmodulin in the R state is available and can bind to CaMKII. Because of the abundance of CaMKII, there is more active CaMKII than PP2B in absolute terms.
Fig. 5.
Differential activation of PP2B and CaMKII at different calcium concentrations. Binding of both PP2B and CaMKII to calmodulin is shown normalized to maximum binding. Steady-state results of simulations at different initial concentrations of calcium are shown. Solid line, PP2B; dashed line, CaMKII. Calmodulin concentration was 3 × 10−5 M (see ref. 36).
Discussion
Induced-Fit Models vs. Thermal Equilibrium Models.
Both experimentalists (37) and theoreticians (38) have directly related calmodulin activity to calcium concentration, often by using a Hill function (39). This allows for some calmodulin activity even at low calcium concentrations. Furthermore, together with the facts that calmodulin affinity for PP2B (31) is higher than for CaMKII (32) and that CaMKII concentration in the PSD (40) is higher than PP2B concentration (41), it can explain why PP2B is preferentially activated at lower calcium concentrations. Models of this type do not allow for the possibility that the different binding sites might have different affinities for calcium. Recent experiments on recombinant calmodulin with site-directed mutations, however, suggest that the N-terminal binding sites have different affinities than the C-terminal binding sites (18). Finally, use of a Hill function to compute calmodulin activity as a function of calcium concentration is purely phenomenological and does not provide an explanation for how calcium binding relates to calmodulin activity.
Many investigators measuring calcium binding to calmodulin (16, 18, 28, 42) have used an Adair function (43), or a modified version thereof, to describe calmodulin binding to calcium. This approach allows for different microscopic affinities for the four binding events and thus for detailed models of calcium binding to calmodulin. These models do not, however, explain the transition of calmodulin between closed (“inactive”) and open (“active”) conformations.
Another way to model calmodulin action is by assuming a sequential mechanism, where calmodulin first binds four calcium ions and only then becomes activated (44, 45). These models are useful approximations of how calmodulin activates CaMKII, but they fail to account for important aspects of calmodulin function, all of which can be explained by our model.
A sequential model fails to explain why calmodulin can activate PP2B or CaMKII when bound to less than four calcium ions (17, 18). Some models assume that calmodulin can activate PP2B with two, three, or four calcium ions bound but requires four calcium ions bound to activate CaMKII (49). Although these models are decent phenomenological approximations for modeling the differential activation of CaMKII and PP2B, they do not provide a satisfactory mechanism by which calmodulin with three bound calcium ions activates PP2B, but not CaMKII. By postulating an equilibrium between inactive and active states that is shifted toward the active state by calcium, our model allows for calmodulin activity even when bound to less than four calcium ions. One can see the extent of this by looking at what fraction of subsaturated calmodulin becomes activated. As shown above, ≈75% of calmodulin molecules are active even when only two molecules of calcium are bound. This can explain experiments with mutant calmodulin showing that even when two of the calcium-binding sites are ablated, calmodulin can still activate CaMKII to a certain extent (18). It is also consistent with experimental data showing that conformational transitions exist in apo calmodulin (46), and that both apo calmodulin and calmodulin bound to only one calcium ion can exist in open and closed states (47). Finally, calmodulin open structures have been found where only one head was populated by calcium (48).
Because purely sequential models do not allow for subsaturated calmodulin binding to its target, they necessarily fail to account for the fact that the affinity of calmodulin for calcium increases once calmodulin is bound to a target (18–20). In our model, subsaturated calmodulin can bind to its targets. The apparent increase for calcium upon binding to targets arises from the fact that targets act as allosteric activators, drawing the equilibrium toward the high-affinity R state.
Note that a simple model based on thermal equilibrium has been proposed before (50), although based on sequential bindings of calcium. Furthermore, the author did not try to estimate parameters using experimental information or to validate the model.
Model Characteristics.
The association constants we determined from the simulation result by fitting it with an Adair-type equation (43) are the observable sequential association constants for the first, second, third, and fourth binding events. It is important to note, however, that our model does not assume a fixed order for calcium binding to the different binding sites. Rather, because the first calcium ion can bind to any one of four binding sites in either state, K1 is a combination of the microscopic KiR and KiT values for each of the sites in each of the states. The apparent sequential dissociation constants used in our model follow an Adair-type (43) framework as used, for instance, by Crouch and Klee (16) and Porumb (30). They are, in principle, Adair constants, except for a slight difference in nomenclature: K2 according to Adair (43) corresponds to K1 × K2 according to Crouch and Klee (16) and Porumb (30), and so on.
It is important to note that the conformation of calmodulin which we call the T state is not necessarily exactly identical to the reported apo structure (14) of calmodulin. Rather, the T state represents a collection of structures that may differ somewhat in the conformation of the calcium-binding sites, but whose overall structure resembles that of apo calmodulin. The existence of an ion-bound form that resembles the apo conformation has recently been established (51). Likewise, the R state is a collection of structures that resemble the reported open structure of active calmodulin (52, 53). Asymmetric forms of calmodulin with one lobe in an open state and one head in a closed state have been reported in the presence of some targets (54). However, the binding mechanism of these targets differs from that of CaMKII and PP2B, where both lobes bind to the target.
Lisman Hypothesis.
The model proposed here explains how different amounts of calcium can trigger the activation of PP2B or CaMKII and thus provides support for the Lisman hypothesis (3) on a molecular level. The question of how different frequencies of calcium signals lead to differential activation of PP2B or CaMKII is not addressed in the model. It has been suggested, however, that, at least under some conditions, high frequencies of calcium input result in high local concentrations of calcium, whereas low calcium frequencies result in moderate local calcium concentrations in the spine (44, 55, 56). In addition, calcium frequency has a direct impact on CaMKII, because of the requirement for two adjacent subunits to be active for autophosphorylation at threonine residue 286, which confers sustained activity (57–59). Other factors the model does not account for include variations in the subcellular localization of PP2B (reviewed in ref. 60) and CaMKII (61) and the inhibitory effect of PP2B on CaMKII (reviewed in ref. 10). The latter effect, if included, would increase the window of calcium concentrations at which PP2B is preferably activated, enhancing the distinction between PP2B and CaMKII activation. We believe, however, that our model provides a valid and useful biophysical basis on which to develop further models of synaptic plasticity mechanisms.
Supplementary Material
Acknowledgments.
We thank Julia Shifman (Hebrew University of Jerusalem) for sharing raw data on calcium binding to calmodulin in the presence and absence of CaMKII. We also thank Laurence Calzone, Stephen Martin, and Annalisa Pastore for helpful comments on the manuscript. S.J.E. was supported by an International Short Visit Fellowship from the Royal Society.
Note added in proof (64).
Experimental support for our equilibrium model of calmodulin function has been recently published by Gsponer et al.
Footnotes
The authors declare no conflict of interest.
Data deposition: The model reported in this paper has been deposited in the BioModels database, www.ebi.ac.uk/biomodels (accession code MODEL9885984404).
This article contains supporting information online at www.pnas.org/cgi/content/full/0804672105/DCSupplemental.
References
- 1.Bliss TV, Lomo T. Long-lasting potentiation of synaptic transmission in the dentate area of the anaesthetized rabbit following stimulation of the perforant path. J Physiol. 1973;232:331–356. doi: 10.1113/jphysiol.1973.sp010273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin SJ, Grimwood PD, Morris RG. Synaptic plasticity and memory: An evaluation of the hypothesis. Annu Rev Neurosci. 2000;23:649–711. doi: 10.1146/annurev.neuro.23.1.649. [DOI] [PubMed] [Google Scholar]
- 3.Lisman J. A mechanism for the Hebb and the anti-Hebb processes underlying learning and memory. Proc Natl Acad Sci USA. 1989;86:9574–9578. doi: 10.1073/pnas.86.23.9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulman H, Greengard P. Stimulation of brain membrane protein phosphorylation by calcium and an endogenous heat-stable protein. Nature. 1978;271:478–479. doi: 10.1038/271478a0. [DOI] [PubMed] [Google Scholar]
- 5.Schulman H, Greengard P. Ca2+-dependent protein phosphorylation system in membranes from various tissues, and its activation by “calcium-dependent regulator.”. Proc Natl Acad Sci USA. 1978;75:5432–5436. doi: 10.1073/pnas.75.11.5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lisman J, Schulman H, Cline H. The molecular basis of CaMKII function in synaptic and behavioural memory. Nat Rev Neurosci. 2002;3:175–190. doi: 10.1038/nrn753. [DOI] [PubMed] [Google Scholar]
- 7.Lee HK, Barbarosie M, Kameyama K, Bear MF, Huganir RL. Regulation of distinct AMPA receptor phosphorylation sites during bidirectional synaptic plasticity. Nature. 2000;405:955–959. doi: 10.1038/35016089. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi Y, et al. Driving AMPA receptors into synapses by LTP and CaMKII: Requirement for GluR1 and PDZ domain interaction. Science. 2000;287:2262–2267. doi: 10.1126/science.287.5461.2262. [DOI] [PubMed] [Google Scholar]
- 9.Silva AJ, Paylor R, Wehner JM, Tonegawa S. Impaired spatial learning in alpha-calcium-calmodulin kinase II mutant mice. Science. 1992;257:206–211. doi: 10.1126/science.1321493. [DOI] [PubMed] [Google Scholar]
- 10.Groth RD, Dunbar RL, Mermelstein PG. Calcineurin regulation of neuronal plasticity. Biochem Biophys Res Commun. 2003;311:1159–1171. doi: 10.1016/j.bbrc.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Lin YM, Liu YP, Cheung WY. Cyclic 3′:5′-nucleotide phosphodiesterase. Purification, characterization, and active form of the protein activator from bovine brain. J Biol Chem. 1974;249:4943–4954. [PubMed] [Google Scholar]
- 12.Xia Z, Storm DR. The role of calmodulin as a signal integrator for synaptic plasticity. Nat Rev Neurosci. 2005;6:267–276. doi: 10.1038/nrn1647. [DOI] [PubMed] [Google Scholar]
- 13.Watterson DM, Sharief F, Vanaman TC. The complete amino acid sequence of the Ca2+-dependent modulator protein (calmodulin) of bovine brain. J Biol Chem. 1980;255:962–975. [PubMed] [Google Scholar]
- 14.Kuboniwa H, et al. Solution structure of calcium-free calmodulin. Nat Struct Biol. 1995;2:768–776. doi: 10.1038/nsb0995-768. [DOI] [PubMed] [Google Scholar]
- 15.Babu YS, et al. Three-dimensional structure of calmodulin. Nature. 1985;315:37–40. doi: 10.1038/315037a0. [DOI] [PubMed] [Google Scholar]
- 16.Crouch TH, Klee CB. Positive cooperative binding of calcium to bovine brain calmodulin. Biochemistry. 1980;19:3692–3698. doi: 10.1021/bi00557a009. [DOI] [PubMed] [Google Scholar]
- 17.Kincaid RL, Vaughan M. Direct comparison of Ca2+ requirements for calmodulin interaction with and activation of protein phosphatase. Proc Natl Acad Sci USA. 1986;83:1193–1197. doi: 10.1073/pnas.83.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shifman JM, Choi MH, Mihalas S, Mayo SL, Kennedy MB. Ca2+/calmodulin-dependent protein kinase II (CaMKII) is activated by calmodulin with two bound calciums. Proc Natl Acad Sci USA. 2006;103:13968–13973. doi: 10.1073/pnas.0606433103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burger D, Stein EA, Cox JA. Free energy coupling in the interactions between Ca2+, calmodulin, and phosphorylase kinase. J Biol Chem. 1983;258:14733–14739. [PubMed] [Google Scholar]
- 20.Olwin BB, Edelman AM, Krebs EG, Storm DR. Quantitation of energy coupling between Ca2+, calmodulin, skeletal muscle myosin light chain kinase, and kinase substrates. J Biol Chem. 1984;259:10949–10955. [PubMed] [Google Scholar]
- 21.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 22.Meador WE, Means AR, Quiocho FA. Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science. 1992;257:1251–1255. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- 23.Ye Q, Li X, Wong A, Wei Q, Jia Z. Structure of calmodulin bound to a calcineurin peptide: A new way of making an old binding mode. Biochemistry. 2006;45:738–745. doi: 10.1021/bi0521801. [DOI] [PubMed] [Google Scholar]
- 24.Rubin MM, Changeux JP. On the nature of allosteric transitions: Implications of non-exclusive ligand binding. J Mol Biol. 1966;21:265–274. doi: 10.1016/0022-2836(66)90097-0. [DOI] [PubMed] [Google Scholar]
- 25.Peersen OB, Madsen TS, Falke JJ. Intermolecular tuning of calmodulin by target peptides and proteins: Differential effects on Ca2+ binding and implications for kinase activation. Protein Sci. 1997;6:794–807. doi: 10.1002/pro.5560060406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayley PM, Findlay WA, Martin SR. Target recognition by calmodulin: Dissecting the kinetics and affinity of interaction using short peptide sequences. Protein Sci. 1996;5:1215–1228. doi: 10.1002/pro.5560050701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoops S, et al. COPASI–a COmplex PAthway SImulator. Bioinformatics. 2006;22:3067–3074. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- 28.Burger D, Cox JA, Comte M, Stein EA. Conformational changes in calmodulin upon binding of calcium. Biochemistry. 1984;23:1966–1971. Sequential. [Google Scholar]
- 29.Haiech J, Klee CB, Demaille JG. Effects of cations on affinity of calmodulin for calcium: Ordered binding of calcium ions allows the specific activation of calmodulin-stimulated enzymes. Biochemistry. 1981;20:3890–3897. doi: 10.1021/bi00516a035. [DOI] [PubMed] [Google Scholar]
- 30.Porumb T. Determination of calcium-binding constants by flow dialysis. Anal Biochem. 1994;220:227–237. doi: 10.1006/abio.1994.1332. [DOI] [PubMed] [Google Scholar]
- 31.Quintana AR, Wang D, Forbes JE, Waxham MN. Kinetics of calmodulin binding to calcineurin. Biochem Biophys Res Commun. 2005;334:674–680. doi: 10.1016/j.bbrc.2005.06.152. [DOI] [PubMed] [Google Scholar]
- 32.Tzortzopoulos A, Török K. Mechanism of the T286A-mutant alphaCaMKII interactions with Ca2+/calmodulin and ATP. Biochemistry. 2004;43:6404–6414. doi: 10.1021/bi036224m. [DOI] [PubMed] [Google Scholar]
- 33.Collins MO, et al. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J Neurochem. 2006;97:16–23. doi: 10.1111/j.1471-4159.2005.03507.x. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, et al. Mass of the postsynaptic density and enumeration of three key molecules. Proc Natl Acad Sci USA. 2005;102:11551–11556. doi: 10.1073/pnas.0505359102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng D, et al. Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol Cell Proteomics. 2006;5:1158–1170. doi: 10.1074/mcp.D500009-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Kakiuchi S, et al. Quantitative determinations of calmodulin in the supernatant and particulate fractions of mammalian tissues. J Biochem. 1982;92:1041–1048. doi: 10.1093/oxfordjournals.jbchem.a134019. [DOI] [PubMed] [Google Scholar]
- 37.Pifl C, et al. Calmodulin X (Ca2+)4 is the active calmodulin-calcium species activating the calcium-, calmodulin-dependent protein kinase of cardiac sarcoplasmic reticulum in the regulation of the calcium pump. Biochim Biophys Acta. 1984;773:197–206. doi: 10.1016/0005-2736(84)90083-x. [DOI] [PubMed] [Google Scholar]
- 38.d'Alcantara P, Schiffmann S, Swillens S. Bidirectional synaptic plasticity as a consequence of interdependent Ca2+-controlled phosphorylation and dephosphorylation pathways. Eur J Neurosci. 2003;17:2521–2528. doi: 10.1046/j.1460-9568.2003.02693.x. [DOI] [PubMed] [Google Scholar]
- 39.Hill AV. The combinations of haemoglobin with oxygen and with carbon monoxide. Biochem J. 1913;5:471–480. doi: 10.1042/bj0070471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petersen JD, et al. Distribution of postsynaptic density (PSD)-95 and Ca2+/calmodulin-dependent protein kinase II at the PSD. J Neurosci. 2003;23:11270–11278. doi: 10.1523/JNEUROSCI.23-35-11270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goto S, Matsukado Y, Mihara Y, Inoue N, Miyamoto E. The distribution of calcineurin in rat brain by light and electron microscopic immunohistochemistry and enzyme-immunoassay. Brain Res. 1986;397:161–172. doi: 10.1016/0006-8993(86)91381-8. [DOI] [PubMed] [Google Scholar]
- 42.Mirzoeva S, et al. Analysis of the functional coupling between calmodulin's calcium binding and peptide recognition properties. Biochemistry. 1999;38:3936–3947. doi: 10.1021/bi9821263. [DOI] [PubMed] [Google Scholar]
- 43.Adair GS. The hemoglobin system. J Biol Chem. 1925;63:529–545. [Google Scholar]
- 44.Franks KM, Bartol TM, Sejnowski TJ. An M cell model of calcium dynamics and frequency-dependence of calmodulin activation in dendritic spines. Neurocomputing. 2001;38–40:9–16. [Google Scholar]
- 45.Naoki H, Sakamira Y, Ishii S. Local signaling with molecular diffusion as a decoder of Ca2+ signals in synaptic plasticity. Mol Syst Biol. 2005;1 doi: 10.1038/msb4100035. 2005.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tjandra N, Kuboniwa H, Ren H, Bax A. Rotational dynamics of calcium-free calmodulin studied by 15N-NMR relaxation measurements. Eur J Biochem. 1995;230:1014–1024. doi: 10.1111/j.1432-1033.1995.tb20650.x. [DOI] [PubMed] [Google Scholar]
- 47.Malmendal A, Evenäs J, Forsén S, Akke M. Structural dynamics in the C-terminal domain of calmodulin at low calcium levels. J Mol Biol. 1999;293:883–899. doi: 10.1006/jmbi.1999.3188. [DOI] [PubMed] [Google Scholar]
- 48.Schumacher MA, Rivard AF, Bächinger HP, Adelman JP. Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature. 2001;410:1120–1124. doi: 10.1038/35074145. [DOI] [PubMed] [Google Scholar]
- 49.Bhalla US, Iyengar R. Emergent properties of networks of biological signaling pathways. Science. 1999;283:381–387. doi: 10.1126/science.283.5400.381. [DOI] [PubMed] [Google Scholar]
- 50.Czerlinski GH. Allosteric competition in calmodulin. Physiol Chem Phys Med NMR. 1984;16:437–447. [Google Scholar]
- 51.Warren JT, Guo Q, Tang W-J. A 1.3-A structure of zinc-bound N-terminal domain of calmodulin elucidates potential early ion-binding step. J Mol Biol. 2007;374:517–527. doi: 10.1016/j.jmb.2007.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Babu YS, Bugg CE, Cook WJ. Structure of calmodulin refined at 2.2 A resolution. J Mol Biol. 1988;204:191–204. doi: 10.1016/0022-2836(88)90608-0. [DOI] [PubMed] [Google Scholar]
- 53.Fallon JL, Quiocho FA. A closed compact structure of native Ca(2+)-calmodulin. Structure. 2003;11:1303–1307. doi: 10.1016/j.str.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 54.Drum CL, et al. Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin. Nature. 2002;415:396–402. doi: 10.1038/415396a. [DOI] [PubMed] [Google Scholar]
- 55.Gamble E, Koch C. The dynamics of free calcium in dendritic spines in response to repetitive synaptic input. Science. 1987;236:1311–1315. doi: 10.1126/science.3495885. [DOI] [PubMed] [Google Scholar]
- 56.Bhalla US. Biochemical signaling networks decode temporal patterns of synaptic input. J Comput Neurosci. 2002;13:49–62. doi: 10.1023/a:1019644427655. [DOI] [PubMed] [Google Scholar]
- 57.Dosemeci A, Albers RW. A mechanism for synaptic frequency detection through autophosphorylation of CaM kinase II. Biophys J. 1996;70:2493–2501. doi: 10.1016/S0006-3495(96)79821-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Koninck P, Schulman H. Sensitivity of CaM kinase II to the frequency of Ca2+ oscillations. Science. 1998;279:227–230. doi: 10.1126/science.279.5348.227. [DOI] [PubMed] [Google Scholar]
- 59.Dupont G, Houart G, De Koninck P. Sensitivity of CaM kinase II to the frequency of Ca2+ oscillations: A simple model. Cell Calcium. 2003;34:485–497. doi: 10.1016/s0143-4160(03)00152-0. [DOI] [PubMed] [Google Scholar]
- 60.Colbran RJ. Protein phosphatases and calcium/calmodulin-dependent protein kinase II-dependent synaptic plasticity. J Neurosci. 2004;24:8404–8409. doi: 10.1523/JNEUROSCI.3602-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bayer KU, et al. Transition from reversible to persistent binding of CaMKII to postsynaptic sites and NR2B. J Neurosci. 2006;26:1164–1174. doi: 10.1523/JNEUROSCI.3116-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Edelstein SJ, Schaad O, Henry E, Bertrand D, Changeux JP. A kinetic mechanism for nicotinic acetylcholine receptors based on multiple allosteric transitions. Biol Cybernet. 1996;75:361–379. doi: 10.1007/s004220050302. [DOI] [PubMed] [Google Scholar]
- 63.Keller CH, Olwin BB, LaPorte DC, Storm DR. Determination of the free-energy coupling for binding of calcium ions and troponin I to calmodulin. Biochemistry. 1982;21:156–162. doi: 10.1021/bi00530a027. [DOI] [PubMed] [Google Scholar]
- 64.Gsponer J, et al. A coupled equilibrium shift mechanism in calmodulin-mediated signal transduction. Structure (London) 2008;16:736–746. doi: 10.1016/j.str.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.