Abstract
Many DNA-interacting proteins diffuse on DNA to perform their biochemical functions. Processivity factors diffuse on DNA to permit unimpeded elongation by their associated DNA polymerases, but little is known regarding their rates and mechanisms of diffusion. The processivity factor of herpes simplex virus DNA polymerase, UL42, unlike “sliding clamp” processivity factors that normally form rings around DNA, binds DNA directly and tightly as a monomer, but can still diffuse on DNA. To investigate the mechanism of UL42 diffusion on DNA, we examined the effects of salt concentration on diffusion coefficient. Ensemble studies, employing electrophoretic mobility shift assays on relatively short DNAs, showed that off-rates of UL42 from DNA depended on DNA length at higher but not lower salt concentrations, consistent with the diffusion coefficient being salt-dependent. Direct assays of the motion of single fluorescently labeled UL42 molecules along DNA revealed increased diffusion at higher salt concentrations. Remarkably, the diffusion coefficients observed in these assays were ≈104-fold higher than those calculated from ensemble experiments. Discrepancies between the single-molecule and ensemble results were resolved by the observation, in single-molecule experiments, that UL42 releases relatively slowly from the ends of DNA in a salt-dependent manner. The results indicate that UL42 “hops” rather than “slides,” i.e., it microscopically dissociates from and reassociates with DNA as it diffuses rather than remaining so intimately associated with DNA that cation condensation on the phosphate backbone does not affect its motion. These findings may be relevant to mechanisms of other processivity factors and DNA-binding proteins.
Keywords: herpes simplex virus, linear diffusion
DNA polymerases are central to DNA replication. Most replicative DNA polymerases include accessory subunits that promote replication of long stretches of DNA without dissociating from the template. The best known of these processivity factors are the “sliding clamps” (reviewed in ref. 1), which include polymerase subunits of bacteria, eukaryotes, and archaea that form multimeric rings around DNA with the aid of ATP-dependent clamp-loaders. These rings then tether their cognate catalytic subunits to DNA, permitting processive DNA synthesis.
A variety of cellular and viral polymerases include processivity subunits that do not use ATP or other proteins for loading onto DNA. Of these, herpes simplex virus (HSV) UL42 is one of the best characterized. This protein's structure resembles that of a monomer of the sliding clamp proliferating cell nuclear antigen (2), yet UL42 binds directly to DNA as a monomer with relatively high affinity (apparent dissociation constant (Kd) in the nanomolar range) (3–5). This direct binding of DNA by UL42 tethers the catalytic subunit of HSV DNA polymerase (Pol) to DNA, thereby enabling processivity (3, 5–7).
An important attribute of processivity factors is their ability to diffuse on DNA. Such diffusion permits tethering of the catalytic subunit without impeding translocation of the enzyme. The loose topological association of sliding clamps with DNA, as opposed to direct binding, is widely thought to permit diffusion [although a recent study has shown direct binding to DNA by the E. coli sliding clamp (8)]. Indeed, biochemical studies have shown that sliding clamps can diffuse on DNA (9, 10); however, to our knowledge, their rates and mechanisms of diffusion have not been identified. Similarly, HSV UL42, despite its tight binding to DNA, does not slow the elongation of the viral polymerase (5) and does diffuse on DNA (11). Its diffusion coefficient (D) on DNA has been estimated to be ≈10 bp2/s from ensemble studies measuring the dependence of the off-rate of UL42 from DNA of different lengths (7, 11). This value is consistent with the rate of HSV polymerase translocation (5, 7). However, as noted in ref. 11, this estimate was based on an indirect analysis that relied on several assumptions.
Berg et al. (12) have defined two major mechanisms of diffusion on DNA, three-dimensional and one-dimensional. In three-dimensional diffusion, proteins dissociate from DNA and rebind a distance away on the same DNA or on another DNA in a manner that is not positionally correlated; i.e., not adjacent to or near the starting point of diffusion. The folding of DNA in solution increases the likelihood of three-dimensional diffusion (12). By contrast, in one-dimensional or linear diffusion, which is the mechanism that is relevant to processivity factor function, the protein samples sites on the DNA in a positionally correlated manner. Berg et al. describe two mechanisms of one-dimensional diffusion, “sliding” and “hopping,” and provide an experimental test to distinguish these mechanisms (12). Sliding is defined as motion along the contour length of the DNA via transfer of bound protein between linearly contiguous sites, implying a helical path of movement. Hopping is defined as microscopic dissociation of the protein from DNA to a point where the protein becomes free to move but can quickly and with high probability reassociate with the same or a nearby site. In effect, the protein remains macroscopically bound but can test nearby binding sites through repeated microscopic dissociations. Microscopic dissociations can be defined as ones in which the protein is removed just far enough from DNA to permit recondensation of cations onto the phosphate backbone of DNA. Thus, operationally, diffusion mediated by hopping is accelerated by higher salt concentrations, whereas sliding is unaffected.
Compared with our knowledge of processivity factors, we know much more about the mechanisms and rates of diffusion on DNA of other DNA binding proteins. By the criteria described above, certain of these proteins have been shown to slide on DNA by either indirect measurements or by direct single-molecule observations (e.g., refs. 13–15). Other proteins have been shown to exhibit salt-dependent diffusion (e.g., ref. 16), but it has been difficult to distinguish between hopping and three-dimensional diffusion events, especially on longer DNAs in solution (17, 18). In at least some cases, three-dimensional diffusion has been shown to dominate at lengths >50 bp, even at low ionic strengths (19). However, to our knowledge, no examples of direct observation of hopping at the single molecule level have been reported. Moreover, despite the terminology sliding clamps, to our knowledge, no processivity factor has been examined for its mechanism of diffusion
To address the mechanism of one-dimensional diffusion by UL42, we examined its diffusion in the presence of different salt concentrations, first in ensemble studies employing relatively short DNAs, and then, to permit direct measurements in the absence of three-dimensional diffusion, using single-molecule methods on long-stretched DNAs.
Results
Effects of Salt Concentration on Diffusion Calculated from Ensemble Measurements.
Multiple assays have shown that UL42 diffuses on DNA (7, 11). In one assay, the observed dissociation rate of the protein from DNA [koff(observed)] decreased with increasing DNA length, implying that UL42 not only dissociates from internal sites on DNA, but also can diffuse to and dissociate from the ends of DNA (7, 11). koff(observed) is the sum of the dissociation rate from internal sites on the DNA [koff(internal)], which is independent of DNA length, and the dissociation rate via the ends of DNA [koff(ends)], which depends on the time required for UL42 to diffuse to the DNA ends and therefore depends on the length of the DNA. An equation has been derived that relates the length of DNA, b, and the half-life of the protein on DNA, t½, that permits calculation of the diffusion coefficient (D) and koff(internal): t½ = ln2/((12D/b2) + koff(internal)) (7). To measure t½, UL42 was incubated with a radiolabeled DNA of defined length under conditions where only one UL42 was bound to each DNA and then mixed with a vast excess of unlabeled DNA to prevent rebinding to the labeled template. Then, at various times, the amount of bound probe was measured using an electrophoretic mobility shift assay (EMSA).
To test the effect of salt concentration on UL42 diffusion on DNA, we conducted these assays using three different concentrations of NaCl in the binding reaction. The UL42 preparation was a C-terminal truncation mutant that retains all known biochemical and biological activities of the full-length protein (20–22). In contrast to the studies in refs. 7 and 11, we omitted MgCl2 from the binding buffer, because divalent salts can complicate analysis of cation effects (23). The resulting plots of t½ versus DNA length are presented in Fig. 1. A clear dependence of half-life on DNA length was observed at 50 mM NaCl (Fig. 1C), permitting a calculation of D = 12 ± 3 bp2/s [supporting information (SI) Table S1], slightly lower than the value calculated from data obtained at 75 mM NaCl and 3 mM MgCl2 (7). However, at 10 mM and 25 mM NaCl, no dependence of D on DNA length could be ascertained (Fig. 1 A and B). This result differs from previous observations at 10 mM NaCl and 3 mM MgCl2 [ref. 11 and G.K.-M. and D.H.C., unpublished observations), which can be attributed to the lack of MgCl2 in the present assay. Using the equation above, we calculated from these data that D is <1 bp2/s at 10 mM and 25 mM NaCl, and thus at least 10 times lower than D at 50 mM NaCl, implying a very strong dependence of diffusion on salt concentration. In contrast, only a <2-fold difference in koff(internal) between 25 and 50 mM NaCl was calculated from these assays (Table S1). In these ensemble experiments, the strong dependence of D on NaCl concentration was consistent with UL42 diffusion on DNA by a hopping mechanism.
Fig. 1.
Effect of salt concentration on DNA length dependence of UL42 dissociation from DNA. Half-lives of UL42-DNA complexes were measured by EMSAs at 10 mM (A), 25 mM (B), and 50 mM (C) NaCl. Data were fitted using the equation t½ = ln 2/((12D/b2) + koff(internal)), relating the length of DNA, b, and the half-life of the protein on DNA, t½, to permit calculation of the diffusion coefficient (D) and koff(internal) (7) (continuous line). In A and C, data for krelease(ends) (Table 1) from single-molecule experiments at 10 mM (A) and 50 mM (C) NaCl were fitted using the equation t½ = ln2·(2krelease(ends)/b + koff(internal))−1, where the koff(internal) values were derived from either single-molecule studies (dotted line) or EMSA studies (dashed line).
Single-Molecule Assay of UL42 Diffusion.
The EMSA experiments described above had several limitations: Diffusion was not observed or measured directly; three-dimensional diffusion (in addition to hopping) was possible; only a narrow range of lengths of relatively short DNAs could be tested; the gel electrophoresis step could artifactually affect dissociation and reassociation; and several assumptions, including UL42 dissociating from DNA as soon as it encounters an end, were required to calculate D. Therefore, we turned to an assay that would permit direct observation of single molecules of UL42 diffusing on long DNA molecules. In this assay, one end of bacteriophage lambda DNA (48.5 kb) was biotinylated and bound to a streptavidin-polyethylene glycol coated glass slide. The DNA was stretched by slowly flowing buffer across the slide, thereby preventing the possibility of three-dimensional diffusion. Movie S1 shows dye-stained DNA being stretched in this fashion.
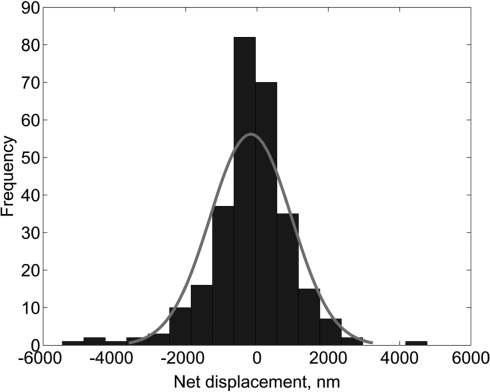
To visualize UL42, we labeled the protein using the dye Cy3B, which covalently modifies cysteine residues on the protein. These residues lie distant from the basic surface of UL42 that mediates DNA-binding (2, 7, 24), and the Cy3B-labeled protein bound and dissociated from DNA in EMSAs in a manner indistinguishable from unlabeled protein (Fig. S1). To measure diffusion, UL42 was incubated with the lambda DNA attached to the slides, the DNA was stretched by flow, images were collected by video fluorescence microscopy, and the motion of single molecules of UL42 on the DNA was analyzed to determine D (see Movie S2 and SI Materials and Methods). We first asked whether UL42 diffused in both directions on DNA despite the flow of buffer in one direction. The net displacements of 132 UL42 molecules from their initial locations were measured and found to be symmetric around the starting point (Fig. 2). Similar results were obtained by using lambda DNA molecules that were stretched and then attached at both ends on slides such that the external buffer flow could be omitted (graph in Fig. S2 and Movie S3). Thus, the flow did not measurably affect the bidirectional diffusion of UL42.
Fig. 2.
UL42 diffuses bidirectionally on DNA. Histogram showing net displacement of UL42 molecules on lambda DNA measured by single-molecule assays in the presence of buffer flow. Curve represents Gaussian fit.
Increasing Salt Concentrations Increase Diffusion of UL42.
We then used the single-molecule assay described above to measure the diffusion of UL42 in 10 mM, 50 mM, and 100 mM concentrations of NaCl. At each salt concentration, we studied the motion of 65–150 molecules of UL42. As shown in Fig. 3A, when the time that each UL42 molecule remained on DNA was plotted vs. time, the data fit an exponential decay, consistent with a single rate constant controlling the dissociation of the protein from an internal site on the DNA. From the exponential fits, we determined the mean binding lifetimes (equivalent to t½/ln2) of UL42 molecules on lambda DNA (Fig. 3B). As expected from our EMSA experiments (Fig. 1 and Table S1) and from measurements of equilibrium binding of UL42 to DNA in various salts (24), increasing NaCl decreased the binding lifetimes of UL42 molecules on DNA. The mean binding lifetimes obtained at 10 mM and 50 mM NaCl were 2- to 3-fold lower than those obtained for the longest DNAs used in the EMSA studies (Table S1). We think it likely that the longer lifetimes observed in the EMSAs are due to artifactual reassociation of UL42 onto DNA during gel electrophoresis.
Fig. 3.
Effect of salt concentration on UL42 binding lifetimes and diffusion coefficients measured by single-molecule experiments. (A) Distribution of binding times of UL42 molecules on lambda DNA at 10 mM, 50 mM, and 100 mM NaCl. Data were fitted to single-exponential decay curves. (B) Mean binding lifetimes of UL42 on lambda DNA determined from curve fit in A (open circle, left axis) and mean diffusion coefficients (D) of UL42 on lambda DNA (open square, right axis) at three different salt concentrations. The error bar for the highest D value indicates standard deviation, which was ≈10% of the mean for each of the three D values. (C) Histograms showing distributions of diffusion coefficients of UL42 on lambda DNA at 10 mM, 50 mM, and 100 mM NaCl. Lines represent smoothed envelope curves. The asterisk in Lower indicates that the frequency shown actually represents the frequency for all D values greater than or equal to the indicated D.
We then analyzed the diffusion coefficients of UL42 molecules on DNA at the different salt concentrations. As expected, there was heterogeneity among different UL42 molecules, but, at each salt concentration, the distribution of diffusion coefficients could be fit to a Gaussian curve. As salt increased, the distributions moved toward higher D values, particularly between 50 mM and 100 mM NaCl (Fig. 3C). Mean D values also increased (Fig. 3B). Indeed, the overall salt dependence of diffusion between 10 mM and 100 mM NaCl was similar to that for binding to DNA (Fig. 3B). Thus, single-molecule diffusion of UL42 on DNA increased with increasing NaCl, fulfilling the operational definition for hopping provided by Berg et al. (12).
Salt-Dependent Effects on UL42 Release From Ends Explain Major Discrepancies Between Ensemble and Single-Molecule Measurements of UL42 Diffusion.
The UL42 diffusion coefficients that were directly measured in the single-molecule assays (Fig. 3) differed dramatically from those calculated from the ensemble EMSA experiments (Fig. 1 and Table S1): Diffusion was readily detected at 10 mM NaCl in the single-molecule assays, whereas none was detected in the EMSA experiments. Remarkably, D values were ≈104 higher in the single-molecule assays than in the EMSA experiments.
One possible explanation for these major discrepancies was that UL42 does not immediately dissociate from DNA upon reaching a DNA end, as we had assumed in our calculations from the EMSA experiments. A lag in release from DNA ends would effectively decrease koff(ends) and thus koff(observed) and D. Therefore, we devised a single molecule assay to test whether UL42 released rapidly or slowly from the ends of DNA. Because too few UL42 molecules could be observed releasing from ends in our standard single-molecule assay, we modified the assay so that UL42 could be pushed to the end of lambda DNA using flow. Because flow does not ordinarily affect the diffusion of UL42 on DNA (Fig. 2), we increased the cross-sectional area of UL42 by coupling it to a quantum dot (see Materials and Methods). The hydrodynamic radius of the quantum dot-coupled protein was estimated to be ≈12.5 nm, a ≈6-fold increase compared with the ≈2 nm radius of the protein alone. The diffusion of quantum dot-coupled UL42 was bidirectional on doubly tethered DNA in the absence of flow, like that of uncoupled UL42 (Movie S4). We then incubated the quantum dot-coupled protein with lambda DNA in the absence of flow and recorded the positions of fluorescent signal (Fig. 4A). Almost immediately upon initiation of buffer flow, the quantum dot-coupled proteins were observed to translocate by ≈16 μm from the starting point, corresponding to the length of lambda DNA. Strikingly, however, the quantum dots did not immediately disappear into the flow (Fig. 4B and Movie S5). Similar results were obtained for blunt and staggered DNA ends (data not shown).
Fig. 4.
Release of UL42 from the ends of DNA. (A and B) Images of UL42 proteins conjugated to fluorescent quantum dots and bound to lambda DNA in the absence (A) or presence (B) of buffer flow. (Scale bar: 5 μm.) (C) Change in fluorescence intensity at the end of lambda DNA over time at 20 mM NaCl. Flow was started at t = 10 s. Data were fitted to a single-exponential decay curve.
We then measured the intensities of fluorescence at the positions corresponding to the ends of lambda DNA at 10 mM, 20 mM, and 50 mM NaCl. The fluorescence intensity decreased with time (20 mM data shown in Fig. 4C), and the data could be fit to a single-exponential decay. The decay is presumably due to a sequential series of events in which the rate limiting step is the release of individual UL42 molecules from the ends of DNAs. This permitted calculation of mean binding lifetimes on DNA ends, which, in turn, permitted calculation of the rates at which UL42 was released from the ends [krelease(ends)] at the different NaCl concentrations. As summarized in Table 1, UL42 released from the ends of DNA in a highly salt-dependent manner, with rates ≈103-fold higher at 50 mM NaCl than at 10 mM NaCl. At 10 mM NaCl, the mean binding lifetime on DNA ends was even higher than the mean binding lifetime on internal sites on DNA (compare Table 1 and Fig. 3B), providing an explanation for the lack of length dependence of dissociation at this salt concentration observed in Fig. 1A.
Table 1.
Salt-dependent release of UL42 from DNA ends
| [NaCl] | Mean binding lifetime on ends, s | krelease(ends), s−1 |
|---|---|---|
| 10 mM | 480 | 0.001 |
| 20 mM | 15 | 0.048 |
| 50 mM | 0.14–0.49 | 1.4–5.0 |
Based on these observations, we revisited the equation used to calculate D from EMSA studies. That calculation assumed that UL42 diffuses to and dissociates from DNA ends immediately upon reaching them. However, given the slow release from DNA ends and the high D values that we directly observed in single-molecule experiments, it is much more likely that UL42 rapidly diffuses back and forth between the ends of short DNAs and that the probability of UL42 release from DNA ends is a function of the frequency of ends relative to the length of the DNA (b), which is 2/b. Additionally, at high values of D, krelease(ends) becomes rate-limiting. These considerations yield the following equation: t½ = ln2·(2krelease(ends)/b + koff(internal))−1. Applying this equation and the values for krelease(ends) and koff(internal) derived from single-molecule experiments to DNA lengths between 1 and 600 bp yielded the dotted curves in Fig. 1 A and C. These curves mirrored both the lack of length dependence seen in the EMSA experiments at 10 mM NaCl (Fig. 1A) and the positive length dependence at 50 mM NaCl (Fig. 1C), but the predicted t½ values were lower than those observed. However, as noted above, the binding lifetimes of UL42 on DNA in the EMSA experiments were higher than those in the single-molecule studies, likely because of artifactual reassociation of UL42 with DNA during gel electrophoresis. If one inserts koff(internal) values derived from the half-lives observed in the EMSA studies, then the derived dashed curves in Fig. 1 provide an excellent fit for the 10 mM NaCl EMSA data (Fig. 1A) and are within 2-fold of the 50 mM EMSA data (Fig. 1C). Thus, the major discrepancies between the results of the EMSAs and the single-molecule assays were explained.
Discussion
In this study, we found that the diffusion of HSV UL42 on DNA increased with increasing NaCl concentration in both ensemble studies of the dissociation of UL42 from different lengths of DNA and, more directly, single-molecule studies of UL42 motion on stretched DNA. The latter studies, in particular, removed the possibility of three-dimensional diffusion. Thus, by the criteria developed by Berg et al. (12), UL42 hops as it diffuses one-dimensionally on DNA. We cannot exclude the possibility that UL42 can also undergo three-dimensional diffusion in solution; it is likely based on studies of other hopping proteins (19). Three-dimensional diffusion is not likely, however, in the context of UL42's function as a processivity factor, bound to the catalytic subunit of HSV DNA polymerase.
We also found a remarkable discrepancy between the values for UL42 diffusion calculated from the ensemble data and those measured directly in the single-molecule studies. This discrepancy was resolved by our finding that UL42 releases slowly from ends of DNA in a salt-dependent manner, which further emphasizes the value of single-molecule assays. Below, we discuss the relevance of these findings to how UL42 interacts with DNA, how UL42 and certain other processivity factors function, and how other proteins, including other processivity factors, diffuse on DNA.
How Does UL42's Interaction with DNA Result in Hopping?
In considering this question, it is useful to compare HSV UL42 to lac repressor, a protein that diffuses by sliding (14). As noted by Winter et al. (14), the binding of lac repressor to nonoperator sequences is ideal for sliding, because its binding to this DNA is entirely due to electrostatic interactions and thus, diffusion occurs on an isopotential surface (14, 25). As the protein moves and displaces counterions from the phosphate backbone, the same number of counterions bind where the protein has left. Electrostatic interactions are also important for UL42 binding to DNA (7, 24). However, despite UL42's binding to DNA with higher affinity than lac repressor, fewer electrostatic interactions are involved in DNA binding by UL42 than by lac repressor (24, 26). Indeed, when plots of log(apparent Kd) of UL42 for DNA vs. log([NaCl]) (24) are extrapolated to the standard state [log(1 M salt) = 0], the analysis indicates that non-electrostatic interactions must also be important for equilibrium binding and most likely help govern dissociation of UL42 from internal sites on DNA. Thus, we envision that UL42 diffuses relatively slowly because of both electrostatic and nonelectrostatic interactions in low salt, and upon condensation of counterions by higher salt concentrations, UL42 becomes more mobile and can diffuse without macroscopic dissociation. A crystal structure of UL42 bound to DNA would be invaluable for understanding the details of the interactions that govern this process.
Winter et al. (14) have suggested that, in general, hopping should be slower than sliding, because hopping depends upon dissociation followed by reassociation to a different site. Furthermore, the rate of sliding depends on the hydrodynamic radius of the protein because of the required rotational movement of the protein on the phosphate backbone, with larger proteins expected to slide more slowly (13, 15). UL42 and hOgg1 are similar in size, and thus their hydrodynamic radii are expected to be similar, but the maximal mean D that we observed for UL42 (1.8 × 105 bp2/s at 100 mM NaCl) is substantially less than the corresponding value for hOgg1 (5 × 106 bp2/s) (13), consistent with hopping by UL42.
Implications of Hopping for UL42 Function.
The ability of UL42 to diffuse on DNA should be crucial for its function as a processivity factor, because such diffusion is expected to permit elongation by HSV DNA polymerase. Our earlier calculations of D from ensemble experiments, assuming 1-bp steps by UL42, provided rates of diffusion that were similar to the median rates of translocation achieved by HSV polymerase under similar conditions (7). Our current measurements of D from single-molecule assays are several orders of magnitude higher, making it even more likely that UL42 does not apply meaningful resistance to DNA elongation by HSV polymerase. This possibility could be tested directly by measuring the force required to move UL42 unidirectionally on DNA and the force generated by the catalytic subunit, Pol.
When UL42 is functioning within the HSV DNA polymerase holoenzyme, it almost certainly moves in 1-bp increments along a helical path; i.e., its step size and path of motion should be dictated by Pol. However, there is a considerable excess of UL42 over Pol in HSV-infected cells, and it is likely that much UL42 is bound to DNA and diffusing on it, unencumbered by Pol. The function of this excess UL42 is not known. Interestingly, diffusion of the T4 bacteriophage processivity subunit plays a role in stimulation of late gene expression (10). Similarly, there is evidence that processivity factors of other herpesviruses have functions other than DNA replication, including stimulation of gene expression (27, 28). Thus, how UL42 diffuses on DNA may well be relevant to these other functions. Hopping, by definition, can entail step sizes >1 bp. Such step sizes raise the possibility of non-helical paths of motion, e.g., translation parallel to the DNA axis (29, 30). Such paths of diffusion could be important for moving past proteins bound to one face of a DNA helix.
Slow Release of UL42 from DNA Ends.
Despite our awareness of the possibility that UL42 might remain on DNA upon encountering an end, we were nevertheless surprised to find that this was the case—and by just how slow the release from DNA ends was, particularly at 10 mM NaCl, where the mean binding lifetimes were greater on ends than on long DNAs. Notably, UL42 can resume diffusion on DNA once the flow force that pushes it to the end is relaxed (data not shown). Slow release from ends presumably reflects both the large energy difference between UL42 bound to DNA and free UL42, as shown by the low apparent Kd of UL42 for DNA, and a high kinetic barrier to dissociation. Interestingly, release from ends is much more strongly salt-dependent than is dissociation from internal sites. This suggests in turn that the barrier to release from ends depends more strongly on electrostatic interactions than does the barrier to dissociation from internal sites. Given the recent evidence that a sliding clamp interacts with DNA via electrostatic interactions (8), we wonder whether sliding-clamp processivity factors could also exhibit slow release from ends. Although the rate of release from DNA ends is likely to be even higher at intracellular salt concentrations, it is still tempting to wonder whether binding to ends by UL42 (or other processivity factors) plays a physiological role. In this light, it is interesting that many models of lagging-strand DNA replication include a step in which processivity factors are deposited at primer-template junctions before the arrival of the polymerase catalytic subunit (e.g., ref. 31). This leads to the speculation that UL42 might dwell on primer-template junctions during lagging-strand replication, or that UL42 binding to DNA ends might play some other role.
Do Other Processivity Factors Slide or Hop?
Although UL42 differs substantially from sliding clamps in its mechanism of DNA interaction, both types of processivity factors share structural folds and positive charge on residues that interact with DNA (2, 8, 24). The term sliding clamp was first used to describe the action of bacteriophage T4 accessory subunits in promoting polymerase activities (32, 33) rather than to describe its mechanism of diffusion. To our knowledge, the rates and mechanisms of diffusion of sliding-clamp processivity factors or other processivity factors have not been defined. Single-molecule assays such as those described here could help determine whether these other processivity factors slide or whether, like UL42, they hop.
Materials and Methods
Protein Expression and Purification.
UL42ΔC340, the N-terminal 340 residues of UL42, which retains the biochemical and biological activities of full-length UL42 (20–22), and MBP-UL42ΔC340, which has maltose binding protein fused to the N terminus, were expressed and purified as described in refs. 7, 20, and 24, except that 2 mM TCEP was used instead of DTT as a reducing reagent in the buffer used for the heparin column.
EMSAs.
EMSAs were performed and analyzed as described in ref. 7, except that no MgCl2 was used in the binding buffer. For experiments in which no length dependence of UL42 dissociation could be ascertained, the average of all of the half-lives of UL42-DNA complexes was used to calculate koff(internal).
Single-Molecule Assays.
λ DNA with a single biotin on one end or with biotins on each end was prepared by annealing and ligating biotinylated oligonucleotides (Integrated DNA Technologies) to the 12-base overhangs of the λ DNA. UL42ΔC340 was fluorescently labeled by reacting its surface-exposed cysteines with Cy3B Mono Maleimide (Amersham Biosciences). For observation of the dissociation of the proteins from DNA ends, MBP-UL42ΔC340 was labeled with fluorescein-5-maleimide (Invitrogen) and coupled to anti-fluorescein-coated quantum dots (QD565; Molecular Devices). Biotinylated DNA was coupled to the streptavidin-coated surface of a flow cell and stretched by introducing a laminar buffer flow. For DNA biotinylated on one end, the buffer flow had to be maintained to keep the DNA stretched; for DNA biotinylated on both ends, the flow could be stopped after the DNA had coupled to the surface on each end. An Olympus IX-71 inverted microscope with a 60x TIRF objective (N.A. = 1.45) was used to direct the excitation light of the 520-nm line of an Ar/Kr laser (Coherent; I70 Spectrum) to the sample and to image the emission onto an EM-CCD camera (iXon, Andor Technologies). Image-analysis software (MetaMorph; Molecular Devices) was used to process the images and track the position of the fluorescent proteins along the DNA as a function of time. For more details on protein labeling, DNA preparation, imaging, and particle-tracking, see SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank John Randell, Patrick Yacono, Paul Blainey, and Oleg Tsodikov for thoughtful discussions and Satoshi Habuchi for help with the DNA, slides, and microscope. This work was supported by National Institutes of Health Grants R01 AI019838 (to D.M.C.), R01 GM077248 (to A.M.v.O.), R01 HL032854 (to D.E.G.), and T32 ES00715522 (to R.M.).
Note Added in Proof.
After this paper was approved, Laurence et al. reported measurements of D for the E. coli sliding clamp (34).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802676105/DCSupplemental.
References
- 1.Johnson A, O'Donnell M. Cellular DNA replicases: Components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 2.Zuccola HJ, Filman DJ, Coen DM, Hogle JM. The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C-terminus of its cognate polymerase. Mol Cell. 2000;5:267–278. doi: 10.1016/s1097-2765(00)80422-0. [DOI] [PubMed] [Google Scholar]
- 3.Gottlieb J, Challberg MD. Interaction of herpes simplex virus type 1 DNA polymerase and the UL42 accessory protein with a model primer template. J Virol. 1994;68:4937–4945. doi: 10.1128/jvi.68.8.4937-4945.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Randell JCW, Coen DM. The herpes simplex virus processivity factor, UL42, binds DNA as a monomer. J Mol Biol. 2004;335:409–413. doi: 10.1016/j.jmb.2003.10.064. [DOI] [PubMed] [Google Scholar]
- 5.Weisshart K, Chow CS, Coen DM. The herpes simplex virus processivity factor, UL42, imparts increased DNA-binding specificity on viral DNA polymerase and decreased dissociation from primer-template without reducing elongation rate. J Virol. 1999;73:55–66. doi: 10.1128/jvi.73.1.55-66.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow CS, Coen DM. Mutations that specifically impair the DNA binding activity of the herpes simplex virus protein UL42. J Virol. 1995;69:6965–6971. doi: 10.1128/jvi.69.11.6965-6971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Randell JCW, et al. Effects of substitutions of arginine residues on the basic surface of herpes simplex virus UL42 support a role for DNA binding in processive DNA synthesis. J Virol. 2005;79:12025–12034. doi: 10.1128/JVI.79.18.12025-12034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Georgescu RE, et al. Structure of a sliding clamp on DNA. Cell. 2008;132:43–54. doi: 10.1016/j.cell.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stukenberg PT, Studwell-Vaughn PS, O'Donnell M. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J Biol Chem. 1991;266:11328–11334. [PubMed] [Google Scholar]
- 10.Tinker RL, Kassavetis GA, Geiduschek EP. Detecting the ability of viral, bacterial and eukaryotic replication proteins to track along DNA. EMBO J. 1994;13:5330–5337. doi: 10.1002/j.1460-2075.1994.tb06867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Randell JC, Coen DM. Linear diffusion on DNA despite high-affinity binding by a DNA polymerase processivity factor. Mol Cell. 2001;8:911–920. doi: 10.1016/s1097-2765(01)00355-0. [DOI] [PubMed] [Google Scholar]
- 12.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. I. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 13.Blainey PC, et al. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc Natl Acad Sci USA. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winter RB, Berg OG, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids 3. The Escherichia coli lac repressor-operator interaction: Kinetic measurements and conclusions. Biochemistry. 1981;20:6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 15.Gorman J, et al. Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2-Msh6. Mol Cell. 2007;28:359–370. doi: 10.1016/j.molcel.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeltsch A, Pingoud A. Kinetic characterization of linear diffusion of the restriction endonuclease EcoRV on DNA. Biochemistry. 1998;37:2160–2169. doi: 10.1021/bi9719206. [DOI] [PubMed] [Google Scholar]
- 17.Stanford NP, Szczelkun MD, Marko JF, Halford SE. One- and three-dimensional pathways for proteins to reach specific DNA sites. EMBO J. 2000;19:6546–6557. doi: 10.1093/emboj/19.23.6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halford SE, Szczelkun MD. How to get from A to B: Strategies for analysing protein motion on DNA. Eur Biophys J. 2002;31:257–267. doi: 10.1007/s00249-002-0224-4. [DOI] [PubMed] [Google Scholar]
- 19.Gowers DM, Wilson GG, Halford SE. Measurements of the contributions of 1D and 3D pathways to the translocation of a protein along DNA. Proc Natl Acad Sci USA. 2005;102:15883–15888. doi: 10.1073/pnas.0505378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bridges KG, Chow CS, Coen DM. Identification of crucial hydrogen-bonding residues for the interaction of herpes simplex virus DNA polymerase subunits via peptide display, mutational, and calorimetric approaches. J Virol. 2001;75:4990–4998. doi: 10.1128/JVI.75.11.4990-4998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Digard P, Chow CS, Pirrit L, Coen DM. Functional analysis of the herpes simplex virus UL42 protein. J Virol. 1993;67:1159–1168. doi: 10.1128/jvi.67.3.1159-1168.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao M, DiTusa SF, Cordingley MG. The C-terminal third of UL42, a HSV-1 DNA replication protein, is dispensable for viral growth. Virology. 1993;194:647–653. doi: 10.1006/viro.1993.1304. [DOI] [PubMed] [Google Scholar]
- 23.Lohman TM. Kinetics of protein-nucleic acid interactions: Use of salt effects to probe mechanisms of interaction. CRC Crit Rev Biochem. 1986;19:191–245. doi: 10.3109/10409238609084656. [DOI] [PubMed] [Google Scholar]
- 24.Komazin-Meredith G, et al. The positively charged surface of herpes simplex virus UL42 mediates DNA binding. J Biol Chem. 2008;283:6154–6161. doi: 10.1074/jbc.M708691200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalodimos CG, et al. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 26.deHaseth PL, Lohman TM, Record MT., Jr Nonspecific interaction of lac repressor with DNA: An association reaction driven by counterion release. Biochemistry. 1977;16:4783–4790. doi: 10.1021/bi00641a004. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Q, et al. Functional and physical interactions between the Epstein-Barr virus (EBV) proteins BZLF1 and BMRF1: Effects on EBV transcription and lytic replication. J Virol. 1996;70:5131–5142. doi: 10.1128/jvi.70.8.5131-5142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isomura H, et al. The late promoter of the human cytomegalovirus viral DNA polymerase processivity factor has an impact on delayed early and late viral gene products but not on viral DNA synthesis. J Virol. 2007;81:6197–6206. doi: 10.1128/JVI.00089-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kampmann M. Obstacle bypass in protein motion along DNA by two-dimensional rather than one-dimensional sliding. J Biol Chem. 2004;279:38715–38720. doi: 10.1074/jbc.M404504200. [DOI] [PubMed] [Google Scholar]
- 30.Shimamoto N. One-dimensional diffusion of proteins along DNA: Its biological significance and chemical significance revealed by single-molecule measurements. J Biol Chem. 1999;274:15293–15296. doi: 10.1074/jbc.274.22.15293. [DOI] [PubMed] [Google Scholar]
- 31.Kelly TJ, Stillman B. In: DNA replication and human disease. DePamphilis ML, editor. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 2006. pp. 1–29. [Google Scholar]
- 32.Venkatesan M, Nossal NG. Bacteriophage T4 gene 44/62 and gene 45 polymerase accessory proteins stimulate hydrolysis of duplex DNA by T4 DNA polymerase. J Biol Chem. 1982;257:12435–12443. [PubMed] [Google Scholar]
- 33.Huang C-C, Hearst JE, Alberts BM. Two types of replication proteins increase the rate at which T4 DNA polymerase traverses the helical regions in a single-stranded DNA template. J Biol Chem. 1981;256:4087–4094. [PubMed] [Google Scholar]
- 34.Laurence TA, et al. Motion of a DNA sliding clamp observed by single molecule fluorescence spectroscopy. J. Biol Chem. 2008 doi: 10.1074/jbc.M800174200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.