Abstract
The eukaryotic cell division cycle is characterized by a sequence of orderly and highly regulated events resulting in the duplication and separation of all cellular material into two newly formed daughter cells. Protein phosphorylation by cyclin-dependent kinases (CDKs) drives this cycle. To gain further insight into how phosphorylation regulates the cell cycle, we sought to identify proteins whose phosphorylation is cell cycle regulated. Using stable isotope labeling along with a two-step strategy for phosphopeptide enrichment and high mass accuracy mass spectrometry, we examined protein phosphorylation in a human cell line arrested in the G1 and mitotic phases of the cell cycle. We report the identification of >14,000 different phosphorylation events, more than half of which, to our knowledge, have not been described in the literature, along with relative quantitative data for the majority of these sites. We observed >1,000 proteins with increased phosphorylation in mitosis including many known cell cycle regulators. The majority of sites on regulated phosphopeptides lie in [S/T]P motifs, the minimum required sequence for CDKs, suggesting that many of the proteins may be CDK substrates. Analysis of non-proline site-containing phosphopeptides identified two unique motifs that suggest there are at least two undiscovered mitotic kinases.
Keywords: cell cycle, cyclin-dependent kinase, mass spectrometry, proteomics
Execution of the eukaryotic cell division cycle results in the coordinated replication and separation of cellular material into two newly formed daughter cells. The process is precisely regulated to ensure that each step completes faithfully before the next begins (1). Errors at any point can be catastrophic to the cell and in humans lead to numerous disease states, including cancer (2). Elucidating the events and regulatory mechanisms of the cell cycle is of wide interest and has been intensely studied.
Seminal experiments identified the M-phase promoting factor (MPF), so called because of its ability to induce mitosis (3, 4). MPF has since been shown to be a cyclin-dependent kinase (CDK) complex, Cdc2–CyclinB (5). During mitosis, cells undergo dramatic changes: transcription and translation cease, chromosomes condense, the nuclear envelope breaks down, the nucleolus dissolves, and the mitotic spindle forms. Phosphorylation by CDKs and other mitotic kinases is instrumental in regulating these processes (6).
Enormous effort has been devoted to identifying cell cycle kinase substrates and understanding the consequences of their phosphorylation. The rate of progress, however, has been limited by the difficulty of proteome-wide application of traditional methods for detecting phosphorylation, such as the use of radioactive phosphate, phospho-specific antibodies, and gel-mobility shifts (7). The emergence of mass spectrometry as a tool for biological analysis and the refinement of techniques for enriching and detecting phosphopeptides have made it possible to identify thousands of phosphorylation sites from a single sample (8–10). With the recent development of quantitative methods, it is possible to monitor in vivo phosphorylation changes for thousands of proteins (10, 11).
In this study, we used several recent refinements developed in our lab and elsewhere, along with metabolic labeling, to survey relative levels of protein phosphorylation in unfractionated lysates from HeLa cells arrested in opposing phases of the cell cycle. We identified >14,000 unique sites of phosphorylation on 3,682 different proteins. Importantly, we measured the relative abundances of peptides harboring the majority of the sites. The data reveal a massive wave of phosphorylation during mitosis, predominantly within the cyclin-dependent kinase target motif [pS/pT]-P, suggesting that most of the sites may be direct targets of CDKs. Among the regulated phosphorylation sites are many canonical cell cycle regulated events and many unreported sites on known cell cycle regulators. Functional analysis shows a strong bias for proteins involved in critical cell cycle processes, suggesting that many of the unknown targets may themselves be regulators of the cell cycle.
Results
We grew asynchronous HeLa cells in media containing 13C615N2-lysine and 13C615N4-arginine (“heavy”). Cells were mixed with either G1 or M-phase arrested cells grown in conventional media (“light”) and lysed in 8 M urea (Fig. 1). The mixed whole-cell extracts were directly proteolyzed with trypsin. The resultant peptides were desalted by solid-phase extraction and then separated by strong cation exchange (SCX) chromatography. Twelve fractions were collected, split equally, and enriched for phosphopeptides by iron immobilized metal ion affinity chromatography (12, 13) (IMAC) or with TiO2 resin (14, 15). IMAC and TiO2 eluates from the same SCX fractions were pooled. Twelve samples from each experiment were analyzed in duplicate for a total of 48 90-min LC-MS/MS runs. Data collection was performed with a hybrid Orbitrap mass spectrometer operating in a dual detection mode. Intact peptide ions were detected in the Orbitrap by high-resolution survey scans, and the 10 most abundant ions from each survey scan were selected for MS/MS fragmentation and analysis in the linear ion trap, resulting in >325,000 MS/MS spectra. We used the SEQUEST algorithm (16) with a concatenated target-decoy database (17) of human proteins to match spectra to peptide sequences. Matches were filtered with common parameters (mass deviation, XCorr, dCn′), using decoy matches as a guide (17). The probability of correct phosphorylation site assignment based on the observation of phosphorylation-specific fragment ions was assessed by using the Ascore algorithm (9) for every site on every phosphopeptide [supporting information (SI) Table S1].
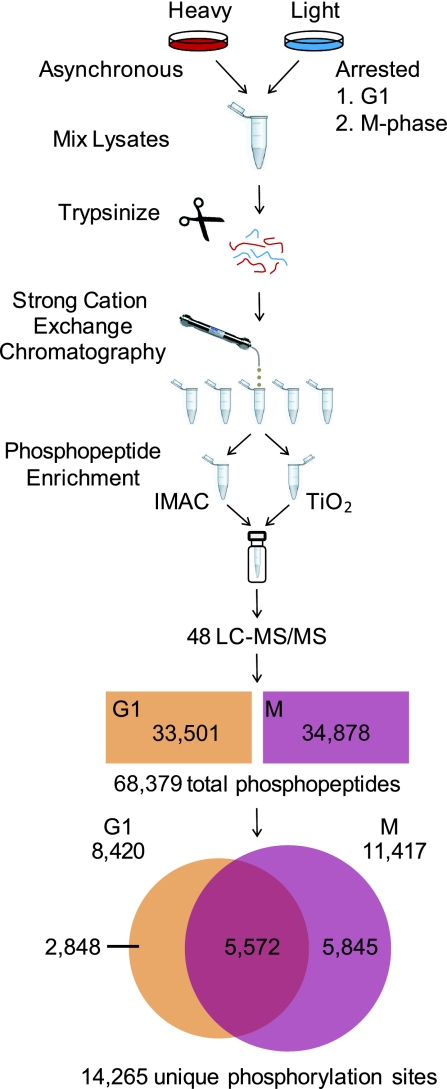
Fig. 1.
Sample preparation and data analysis for quantitative cell cycle phosphoproteome profiling. Asynchronous HeLa cells were cultured in media containing 13C615N2-lysine and 13C615N4-arginine, lysed, and mixed 1:1 by cell number with lysates from double-thymidine or nocodazole arrested cells cultured in standard media and digested with trypsin in solution. Peptides from each experiment were subjected to strong-cation exchange chromatography and the eluates were collected in 12 fractions for each run. Each fraction was split and enriched for phosphopeptides with IMAC and TiO2. Enriched eluates were recombined by fraction and analyzed in duplicate on a hybrid linear ion trap–orbitrap, mass spectrometer. MS/MS spectra were searched by using SEQUEST and filtered to a 1% false-discovery rate before further automated analysis to determine phosphorylation site localization and perform quantification. The filtered dataset contained 68,379 phosphopeptides with a false discovery rate of 0.3%. These peptides contain 14,265 unique phosphorylation sites.
The final dataset contained 68,379 phosphopeptides with an estimated false-discovery rate of 0.3% (200 total reverse hits) from 3,682 different proteins. This included 33,501 phosphopeptides (2,731 proteins) from double-thymidine arrested (G1) cells and 34,878 phosphopeptides (3,181 proteins) from nocodazole arrested (M) cells. These data correspond to at least 14,265 different phosphorylation events (Table S1). Among the identified sites are many canonical cell cycle phosphorylations. These include S-22 and S-392 of Lmna (18) in M-phase required for nuclear envelope breakdown, inhibitory sites T-14 and Y-15 of Cdc2 in G1 (1), the Cdc2 activating site T-167 in mitosis (1), activating sites on the Cdc2 activating phosphatase Cdc25c (1), and inhibitory sites on the Cdc2 inhibiting kinase Pkmyt1 (1) in mitosis.
By adding IMAC and TiO2 phosphopeptide enrichment steps after SCX chromatography, we increased by nearly an order of magnitude the number of sites identified from nocodazole treated HeLa cell lysates in previous work (9) (Fig. S1). We observed 75% of the sites found in that study along with nearly 13,000 more. Phosphoproteomic analysis by Olsen et al. (10) identified 6,600 sites in epidermal growth factor-treated HeLa cells. Our data include 3,545 common sites, 53% of which were in the previous study (Fig. S1). The PhosphoSite database (19) (www.phosphosite.org) is a curated collection of phosphorylation sites with >13,000 human sites from the literature. Our data contain 4,051 (31%) of these sites. Of the sites in our dataset localized with high confidence (12,297; P < 0.05), 6,764 have not, to the best of our knowledge, been reported in the literature.
To further benchmark the breadth of our dataset, we compared our results to those from three directed analyses, using purified components (Fig. S1 and Table S2). A study of taxol stabilized mitotic spindles purified from HeLa cells (20) identified 312 sites from 72 spindle proteins. Without enriching for spindle proteins and considering only localized sites found in M-phase cells, we identified 577 sites from 93 known spindle proteins, including 165 (53%) of the previously reported sites. We also identified 32 sites on the nine proteins of the Nup107–160 complex, including 6 of 12 sites identified after affinity purification (21) and 47 sites from proteins of the anaphase promoting complex, including 27 of 43 sites identified from purified APC (22). We compiled additional site lists for proteins of the cohesin complex, prereplicative complex, and proteins localizing to the nuclear envelope (Table S2).
Although double-thymidine and nocodazole arrest do not perfectly mimic physiological cell cycle states, they have been shown to faithfully reproduce many cell cycle regulatory events and have provided invaluable insights into cell cycle biology. Asynchronous cells were chosen as a common reference sample to allow the detection of large changes in mitotic phosphorylation and provide a means of comparison between the two arrested states. Relative abundances of coeluting light and heavy peptide pairs from arrested and asynchronous cells respectively, were calculated automatically, using the software program Vista (23) (Fig. S2). We measured abundance ratios for 52,934 phosphopeptides, including 11,243 nonredundant peptides from 3,049 different proteins (Table S3). Duplicate analyses of each sample allowed us to assess the reproducibility of the abundance ratios for peptides detected in both trials (Fig. S3). For measurements made with S/N ≥ 3, 91% (G1) and 89% (M) of all abundance ratios fell within 20% of the mean of replicate measurements.
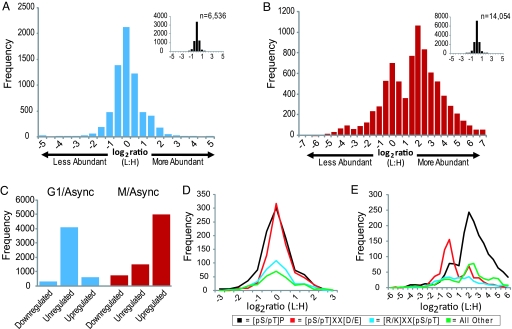
Light-to-heavy ratios >1 indicate increased abundance in the arrested cells. The log2 transformation of this ratio provides a convenient measure of relative abundance (Fig. 2). Based on the distribution of unphosphorylated peptide ratios, we consider phosphorylated peptides whose relative abundance changed ≥2.5-fold as regulated and those that changed <1.5-fold as unregulated. In G1-arrested cells, several hundred phosphopeptides changed in abundance (Fig. 2A and Table S3). In contrast, thousands of phosphopeptides were up-regulated in mitotic cells (Fig. 2 B and C).
Fig. 2.
Phosphopeptide abundance distributions. (A and B) Log2-transformed light:heavy (arrested:asynchronous) ratios for all quantified phosphopeptides from G1 (A) and M (B) phase arrested cells. Bins are 0.5 units wide; e.g., the ‘0′ bin stretches from −0.25 to +0.25. (Insets) Shown is the distribution of unphosphorylated peptides in each experiment. (C) Peptides with ≥2.5-fold changes were deemed regulated and those with ≤1.5-fold changes unregulated. (D and E) Log2 phosphopeptide abundance distributions for peptides with different phosphorylation motifs are shown for G1 phase cells (D) and for mitotic cells (E). Phosphorylation sites were classified into 1 of 3 motifs, [pS/pT]-P, [pS/pT]-X-X-[D/E], or [K/R]-X-X-[pS/pT]. Sites lacking these motifs were grouped into “other.” Only peptides containing a single motif class were included in the analysis.
To control for changes in protein abundance levels that could lead to apparent changes in phosphorylation abundance levels, we analyzed a large pool of unphosphorylated peptides derived from the M-phase SCX fractions before phosphopeptide enrichment (Fig. 2B). Using the median abundance ratios from the unphosphorylated peptides for each protein (see SI Materials and Methods), we obtained quantitative measurements for 2,884 proteins in the M-phase samples (Table 1 and Table S4). Most protein ratios did not change; 2,327 proteins (81%) changed <1.5-fold and only 182 (6.3%) changed ≥2.5-fold. We identified 1,944 M-phase proteins with regulated phosphopeptides and measured protein abundances for 842 (43%) of them. Ninety-six proteins (11%) changed ≥2.5-fold. We conclude that the vast majority of phosphorylation changes cannot be explained by changing protein levels. One surprising finding was that many of the proteins with the greatest abundance changes, such as Aurora Kinase B, Polo Kinase 1, and Cyclin B1, are known to be degraded by the APC (24–26) (Table 1 and Table S4), suggesting that others might also be substrates.
Table 1.
Examples of increased M-phase protein abundance
| Protein | Description | Fold change |
|---|---|---|
| Cyr61 | Cysteine-rich, angiogenic inducer, 61 | 18.2 |
| Fam91a1 | Family with sequence similarity 91, member A1 | 16.3 |
| Ube2s | Ubiquitin-conjugating enzyme E2S | 14.4 |
| Scfd1 | Sec1 family domain containing 1 | 13.1 |
| Ccnb1 | Cyclin B1 | 12.9 |
| Anln | Anillin, actin binding protein | 12.3 |
| Cdca8 | Cell division cycle associated 8 | 11.8 |
| Aurka | Aurora kinase A (STK6) | 11.3 |
| Tpx2 | TPX2, microtubule-associated, homolog | 10.9 |
| Chmp4a | Chromatin modifying protein 4A | 10.7 |
| Kif20a | Kinesin family member 20A | 9.9 |
| Plk1 | Polo-like kinase 1 (Drosophila) | 9.7 |
| Fosl1 | FOS-like antigen 1 | 9.2 |
| Kif11 | Kinesin family member 11 (Eg5) | 8.3 |
| Aurkb | Aurora kinase B | 8.2 |
| Apoc3 | Apolipoprotein C-III | 8.1 |
| Tbc1d15 | TBC1 domain family, member 15 | 8.0 |
| Pttg1 | Pituitary tumor-transforming 1 (securin) | 7.5 |
| Cenpf | Centromere protein F, 350/400ka (mitosin) | 7.2 |
| Top2a | Topoisomerase (DNA) II alpha 170 kDa | 7.2 |
| Nusap1 | Nucleolar and spindle-associated protein 1 | 7.1 |
Underlined proteins are known APC substrates. See Table S4 for the complete dataset with references.
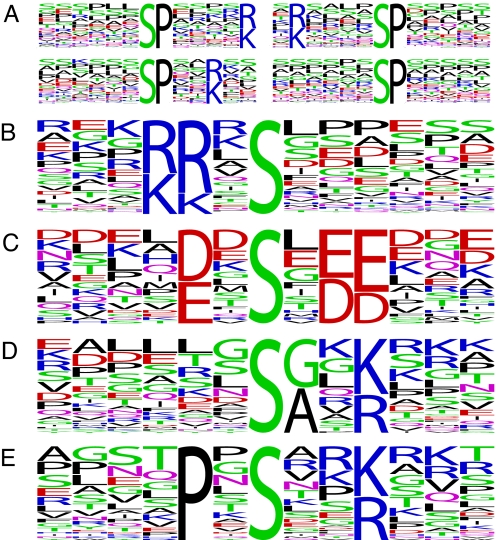
Cyclin-dependent kinases require a proline at the amino acid immediately carboxyl-terminal to the phospho-acceptor residue (27). The distributions of the log2 abundance ratios for peptides that contained phosphorylation sites of three broad motif classifications, proline-directed ([pS/pT]-P), acidophilic ([pS/pT]-X-X-[D/E]), and basophilic ([K/R]-X-X-[pS/pT]) show that most of the changing phosphopeptides contain [pS/pT]-P sites (Fig. 2). However, there were significant, although modest, numbers of mitotically regulated acidophilic and basophilic site containing phosphopeptides and a sizable number lacking a classic motif. The same analysis of the G1 phosphorylation sites revealed a nearly uniform distribution for all motifs. Motif discovery analysis of the up-regulated mitotic sites yielded many pS-P motifs (Fig. 3) and two motifs similar to those phosphorylated by Aurora kinase A, [K/R]-[K/R]-X-pS [with a bias for L at +1, consensus = [K/N/R]-R-X-[pS/pT]-Φ (28)] and Polo-like kinase 1, [D/E]-X-pS-X-[D/E]-[D/E] [with a bias for L at +1, consensus = [D/E]-X-[pS/pT]-Φ-X-[D/E] (29)] (Fig. 3). Based on these consensus sequences, we compiled a list of regulated candidate substrates of Aurora kinase A and Polo kinase 1 from our data (Table S5). Among these are known substrates Tacc3 (30) and Dlg7 (Hurp) (31). In addition, we identified two motifs, pS-[A/G]-X-[K/R] and P-X-pS-X-X-[K/R], that account for a significant portion of the regulated phosphopeptides lacking a classic motif. Proteins with regulated phosphopeptides harboring these motifs include the ribonuclease Dicer1 and the Wnk1 kinase (Table S5).
Fig. 3.
Substrate motif discovery. We extracted phospho-serine motifs from mitotically regulated peptides using Motif-X (9). Only sites with Ascore ≥ 13 and median abundance ratio (L:H) ≥ 4 (n = 2,949) were included. (A) More than half of these sites, 1,670, lie in pS-P motifs. (B and C) We identified two motifs similar to those for the mitotic kinases (B) Aurora kinase A and (C) Polo-like kinase 1, whose consensus substrate sequences are [K/N/R]-R-X-[pS/pT]-Φ and [D/E]-X-[pS/pT]-Φ-X-[D/E], respectively, where Φ denotes any hydrophobic residue. Note the bias for leucine in the +1 position for both. (D and E) We also found two notable motifs that included a basic residue at +3 but that lacked the +1 proline.
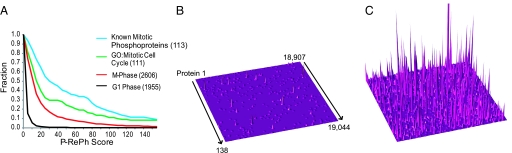
To facilitate protein-level analysis, we developed a simple metric, the regulated phosphorylation (RePh) score. The RePh score is the median abundance ratio from multiple observations of the same phosphorylation site. For each protein, we recorded both a Max RePh, representing the highest value for any site, and a P-RePh score, representing the sum of individual site RePh scores (Table S3). To evaluate the utility of RePh scores, we examined the scores assigned to two sets of mitotic proteins, 153 known mitotically up-regulated phosphoproteins identified in the literature (Table S6) and 273 proteins associated with the gene ontology (GO) term “mitotic cell cycle” [GO:0000278 (32)]. We found phosphorylation sites in our M-phase data from 113 of the mitotic phosphoproteins and at least one regulated phosphopeptide from all but seven of these; 111 of the GO-associated genes were found, 91 of them with a regulated phosphopeptide (Table S6). Known mitotic proteins scored consistently higher than those in the full set and known mitotic phosphoproteins even higher (Fig. 4A). The distributions of P-RePh scores (Fig. 4 B and C) show strikingly different protein phosphorylation landscapes for G1 and M-phase cells. Only 42 G1 proteins received a P-RePh score >10 compared with 947 proteins from M-phase cells.
Fig. 4.
Protein regulated phosphorylation (P-RePh) scores. The P-RePh score is a cumulative assessment of regulated phosphorylation sites assigned to each protein in the dataset. (A) The fraction of M-phase P-RePh scores above a given threshold was plotted for all G1 (black trace) and M-phase (red trace) phosphoproteins along with those for proteins annotated to GO:0000278 (mitotic cell cycle) (green trace) and known mitotic phosphoproteins identified from the literature (blue trace). (B and C) Entire proteome topographical plots of up-regulated phosphorylation in G1 (B) and M-phase (C) cells. Each protein in the human proteome is represented as a single point on a continuous 138 × 138 plane. P-RePh scores are represented on the z axis. Plots are on the same scale with a maximum P-RePh of 500. The clipped peak in the mitotic plot corresponds to Ki-67 (P-RePh = 1,103).
High scoring proteins were submitted to GO analysis, using DAVID (33) to identify statistically overrepresented term associations (Table S7). We found well known mitotic biological processes, such as mitotic sister chromatid segregation and chromosome condensation, and the broader terms M-phase and mitosis. Cellular component GO analysis also revealed expected mitotic categories, such as kinetochore, spindle, and nuclear envelope. Although few proteins were up-regulated in G1 cells, we found GO terms related to the DNA damage response. This is not surprising as double thymidine arrest mildly activates the DNA damage response (S. Elledge, data not shown). Many of the most highly regulated G1 phosphorylation sites, such as those in Smc1 and Utp14a, occur in [pS/pT]-Q motifs targeted by the DNA damage kinases ATM and ATR.
We also performed additional directed analyses of candidate protein classes and complexes that may be preferentially regulated by phosphorylation. These sets include many previously undescribed regulated sites on known cell cycle proteins including elements of the APC, prereplicative complex, and cohesin complex; kinases; and nuclear envelope proteins (Table S8).
Discussion
Because of its intimate ties to human pathologies, cell cycle biology is an intense area of study with a constantly growing cast of players and an increasingly complex regulatory network. The importance of phosphorylation in the cell cycle and, specifically, as cells pass through mitosis has long been recognized. Attempts to identify mitotically phosphorylated proteins have met with modest success. Monoclonal antibodies raised against mitotic extracts revealed 16 phosphorylated protein bands present only in mitotic cell extracts (34); a later screen for phosphorylation of in vitro-translated Xenopus laevis cDNAs in mitotic egg extracts identified 20 proteins (35); and traditional single-gene analyses have identified >150 phosphoproteins (see Table S6 for references). In this study, using mass spectrometry, we identified >1,000 mitotic phosphoproteins whose phosphorylation levels vary in the cell cycle; measured the relative abundances of the phosphorylated peptides in M-phase and G1 arrested cells; and, in most cases, identified the precise sites of phosphorylation. This work provides a sizable list of suspects in the hunt for players in phosphorylation-dependent cell cycle regulation and represents major progress toward a mechanistic understanding of how phosphorylation regulates the cell-division cycle.
The most striking findings were (i) the large number of proteins that became heavily phosphorylated in mitotic cells and (ii) how many of them were multiply phosphorylated. More than 1,000 proteins were phosphorylated more than four times; 280 proteins were phosphorylated on ≥10 sites; and the highest scoring protein, Ki-67, was phosphorylated on >100 mitotically up-regulated sites. Multiple phosphorylation can act in signal integration, create signaling thresholds, or generate dynamic behaviors, such as ultrasensitivity (36). There may be complex roles for phosphorylation that are not easily dissected by individual site analysis. The addition of multiple phosphates confers physicochemical changes that can affect protein conformation, interactions, subcellular localization, and stability. In the case of NFAT1, the dephosphorylation of 13 sites promotes a conformational change that affects its localization (37). Multiply phosphorylated proteins in our dataset may undergo similar regulation.
Genetic and biochemical experiments have identified a number of mitotic kinases (6). We detected >50 kinases with mitotically up-regulated phosphorylation sites (Table S7), including a site in the glycine-rich loop of the Cdc2 regulator Pkmyt1. This site is homologous to Cdc2 threonine-14, an inhibitory site phosphorylated by Pkmyt1 and Wee1 (1). It may also inhibit Pkmyt1 kinase activity. We also identified 18 kinases with mitotically down-regulated sites (Table S8). Strikingly, we found phosphopeptides with sites in the activation loop of three Map kinases, Mapk1 (Erk2), Mapk3 (Erk1), Mapk14 (p38 Mapkα), and an upstream kinase, Map2k2 (Mek2) that were down-regulated 20-, 35-, 11-, and 5-fold, respectively. This is consistent with an earlier report (38).
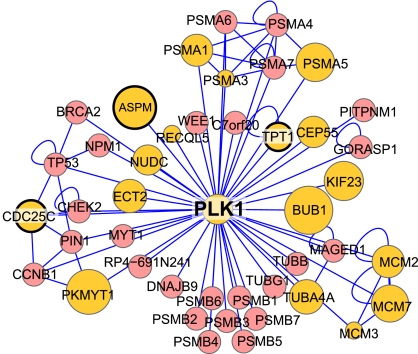
Identifying kinase-substrate pairs continues to be a major barrier to understanding cell signaling networks. Our data provide an opportunity for the discovery of substrate motifs that provide clues to the identity of the kinases. In addition to proline-containing CDK-like sites, we found motifs similar to those for the mitotic kinases AurkA and Plk1 and numerous candidate substrates for each (Table S5). We also identified two motifs bearing a basic residue at the +3 position but lacking a +1 proline. One of these requires a small amino acid, either alanine or glycine at the +1 position. These could be unconventional CDK sites that require a small residue to overcome the need for proline. Alternatively, they may be phosphorylated by unknown mitotic kinases. Peptides with these motifs could be used in a biochemical assay to purify and identify the kinases. Protein–protein interactions can also inform kinase–substrate relationships. By mapping the mitotic P-RePh scores onto the human protein-interaction network, we identified a large number of mitotically phosphorylated proteins that directly interact with the major mitotic kinases Plk1, Cdc2, Aurka, and Aurkb (Fig. 5 and Fig. S4). For Plk1 and AurkA, a number of these interacting proteins (Aspm1, Cdc25c, and Tpt1 for Plk1; Pml and Tpx2 for AurkA) also appear in the regulated candidate substrates list (Fig. 5, Fig. S4, and Table S5).
Fig. 5.
Mitotic phosphorylation and the human protein interaction network. Shown is the first-neighbor human protein interaction network (41) for Polo-like kinase 1. Yellow nodes were found phosphorylated in our mitotic dataset. The size of each node corresponds to its mitotic P-RePh score, larger nodes are more heavily phosphorylated in mitosis. Nodes with a heavy black border also contain regulated candidate Plk1 sites. Additional networks for Cdc2, AurkA, and AurkB appear in Fig. S4.
Functional analysis of our mitotic phosphoprotein data identified familiar processes and structures central to cell cycle biology, including proteins localizing to the spindle and nuclear envelope and functioning in chromosome condensation and chromatid segregation. Many of the proteins were not known to undergo cell cycle regulated changes in phosphorylation.
In this study, our phosphopeptide enrichment strategy reduced interference from many of the most abundant cellular proteins. This was demonstrated by the detection of 1,883 phosphoproteins that were not identified by unphosphorylated peptides. The depth of our analysis is further underscored by the identification of more than two-thirds of known mitotic phosphoproteins. We identified thousands of cell cycle regulated events, substantially broadening an already crowded landscape. Although these data imply provocative new functional connections and regulatory mechanisms, only more directed experimentation can reveal the true nature and importance of these relationships. It is our hope that this work will aid in the formulation of concrete hypotheses about the regulation of cell cycle events and serve as a launching pad for future biological investigations.
Experimental Procedures
Sample Preparation and Mass Spectrometry.
Standard cell culture techniques were used. SCX chromatography, IMAC, and TiO2 enrichment were performed as described (8, 39). LC-MS/MS was performed on a LTQ Orbitrap hybrid mass spectrometer (ThermoFisher). Samples were resuspended in 5% acetonitrile/5% formic acid, loaded onto a fused silica microcapillary C18 column, separated by using a 47-min linear gradient of 6–27% acetonitrile, and introduced into the mass spectrometer. One full MS scan per cycle was acquired at high mass accuracy in the orbitrap, followed by 10 data-dependent MS/MS spectra in the linear ion trap from the 10 most abundant ions. Further details appear in SI Materials and Methods.
Database Searching and Filtering and Phosphorylation Site Assignment.
MS/MS spectra were searched via the SEQUEST algorithm against a composite database containing the human IPI protein database and its reversed complement. Parameters allowed for up to three missed cleavages, a mass tolerance of ±100 ppm, a static modification of 57.02146 Da on cysteine and dynamic phosphorylation on serine, threonine, and tyrosine, methionine oxidation; 10.00827 Da on arginine; and 8.01420 Da on lysine. Search results were filtered to include <1% matches to reverse sequences by restricting the mass tolerance window and setting thresholds for Xcorr and dCn′. We used the Ascore algorithm (9) to assign phosphorylation site localization. For further details, see SI Materials and Methods.
Protein Abundance Measurements, RePh Scores, and Motif Analysis.
Unphosphorylated peptides found in the final sample along with those from an unfractionated pool for each sample were searched and quantified as described for phosphopeptides (see SI Materials and Methods). For the M-phase arrested cells, we also examined SCX fractions before any phosphopeptide enrichment step. Relative protein levels were determined from the median log2-transformed ratios for all unique peptides in each protein. RePh scores were calculated independently for G1 and M-phase phosphopeptides. The score for a given site or combination of sites on a peptide is the median abundance ratio of all peptides harboring that unique combination of sites with scores capped at 100. Protein RePh scores (P-RePh) are the sum of all individual RePh scores for each protein. Phosphorylation motifs were identified with the Motif-X algorithm (40). Further details appear in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank members of the S.P.G. laboratory for discussion and critical comments. This work was supported by a grant from the National Institutes of Health (to S.J.E.) and in part by National Institutes of Health Grants HG3456 and GM67945 (to S.P.G.) and a postdoctoral fellowship from the Spanish Ministry of Education and Science (to J.V.). S.J.E. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805139105/DCSupplemental.
References
- 1.Morgan DO. Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 2.Massague J. G1 cell cycle control and cancer. Nature. 2004;432:298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 3.Rao PN, Johnson RT. Mammalian cell fusion: Studies on the regulation of DNA synthesis and mitosis. Nature. 1970;225:159–164. doi: 10.1038/225159a0. [DOI] [PubMed] [Google Scholar]
- 4.Masui Y, Markert CL. Cytoplasmic control of nuclear behavior during meiotic maturation of frog oocytes. J Exp Zool. 1971;177:129–145. doi: 10.1002/jez.1401770202. [DOI] [PubMed] [Google Scholar]
- 5.Lohka MJ, Hayes MK, Maller JL. Purification of maturation-promoting factor, an intracellular regulator of early mitotic events. Proc Natl Acad Sci USA. 1988;85:3009–3013. doi: 10.1073/pnas.85.9.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 7.Johnson SA, Hunter T. Kinomics: Methods for deciphering the kinome. Nat Methods. 2005;2:17–25. doi: 10.1038/nmeth731. [DOI] [PubMed] [Google Scholar]
- 8.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci USA. 2007;104:1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 10.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 11.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 12.Andersson L, Porath J. Isolation of phosphoproteins by immobilized metal (Fe3+) affinity chromatography. Anal Biochem. 1986;154:250–254. doi: 10.1016/0003-2697(86)90523-3. [DOI] [PubMed] [Google Scholar]
- 13.Ficarro SB, et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 14.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Pinkse MW, Uitto PM, Hilhorst MJ, Ooms B, Heck AJ. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal Chem. 2004;76:3935–3943. doi: 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- 16.Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 17.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 18.Ward GE, Kirschner MW. Identification of cell cycle-regulated phosphorylation sites on nuclear lamin C. Cell. 1990;61:561–577. doi: 10.1016/0092-8674(90)90469-u. [DOI] [PubMed] [Google Scholar]
- 19.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 20.Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci USA. 2006;103:5391–5396. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glavy JS, et al. Cell-cycle-dependent phosphorylation of the nuclear pore Nup107–160 subcomplex. Proc Natl Acad Sci USA. 2007;104:3811–3816. doi: 10.1073/pnas.0700058104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kraft C, et al. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 2003;22:6598–6609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakalarski CE, et al. The impact of peptide abundance and dynamic range on proteome-scale quantitative analyses. J Proteome Res. 2008 doi: 10.1021/pr800333e. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura M, Matsuda Y, Yoshioka T, Sumi N, Okano Y. Identification and characterization of STK12/Aik2: A human gene related to aurora of Drosophila and yeast IPL1. Cytogenet Cell Genet. 1998;82:147–152. doi: 10.1159/000015089. [DOI] [PubMed] [Google Scholar]
- 25.Golsteyn RM, et al. Cell cycle analysis and chromosomal localization of human Plk1, a putative homologue of the mitotic kinases Drosophila polo and Saccharomyces cerevisiae Cdc5. J Cell Sci. 1994;107(Pt 6):1509–1517. doi: 10.1242/jcs.107.6.1509. [DOI] [PubMed] [Google Scholar]
- 26.Pines J, Hunter T. Isolation of a human cyclin cDNA: Evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2. Cell. 1989;58:833–846. doi: 10.1016/0092-8674(89)90936-7. [DOI] [PubMed] [Google Scholar]
- 27.Songyang Z, et al. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 28.Ferrari S, et al. Aurora-A site specificity: A study with synthetic peptide substrates. Biochem J. 2005;390:293–302. doi: 10.1042/BJ20050343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakajima H, Toyoshima-Morimoto F, Taniguchi E, Nishida E. Identification of a consensus motif for Plk (Polo-like kinase) phosphorylation reveals Myt1 as a Plk1 substrate. J Biol Chem. 2003;278:25277–25280. doi: 10.1074/jbc.C300126200. [DOI] [PubMed] [Google Scholar]
- 30.Peset I, et al. Function and regulation of Maskin, a TACC family protein, in microtubule growth during mitosis. J Cell Biol. 2005;170:1057–1066. doi: 10.1083/jcb.200504037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu CT, et al. Phosphorylation and stabilization of HURP by Aurora-A: Implication of HURP as a transforming target of Aurora-A. Mol Cell Biol. 2005;25:5789–5800. doi: 10.1128/MCB.25.14.5789-5800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashburner M, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dennis G, Jr, et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 34.Davis FM, Tsao TY, Fowler SK, Rao PN. Monoclonal antibodies to mitotic cells. Proc Natl Acad Sci USA. 1983;80:2926–2930. doi: 10.1073/pnas.80.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stukenberg PT, et al. Systematic identification of mitotic phosphoproteins. Curr Biol. 1997;7:338–348. doi: 10.1016/s0960-9822(06)00157-6. [DOI] [PubMed] [Google Scholar]
- 36.Cohen P. The regulation of protein function by multisite phosphorylation—a 25 year update. Trends Biochem Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 37.Okamura H, et al. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol Cell. 2000;6:539–550. doi: 10.1016/s1097-2765(00)00053-8. [DOI] [PubMed] [Google Scholar]
- 38.Tamemoto H, et al. Biphasic activation of two mitogen-activated protein kinases during the cell cycle in mammalian cells. J Biol Chem. 1992;267:20293–20297. [PubMed] [Google Scholar]
- 39.Wilson-Grady JT, Villen J, Gygi SP. Phosphoproteome analysis of fission yeast. J Proteome Res. 2008;7:1088–1097. doi: 10.1021/pr7006335. [DOI] [PubMed] [Google Scholar]
- 40.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale datasets. Nat Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 41.Mishra GR, et al. Human protein reference database—2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.