Abstract
The only known volatile pheromone in Drosophila, 11-cis-vaccenyl acetate (cVA), mediates a variety of behaviors including aggregation, mate recognition, and sexual behavior. cVA is detected by a small set of olfactory neurons located in T1 trichoid sensilla on the antennae of males and females. Two components known to be required for cVA reception are the odorant receptor Or67d and the extracellular pheromone-binding protein LUSH. Using a genetic screen for cVA-insensitive mutants, we have identified a third component required for cVA reception: sensory neuron membrane protein (SNMP). SNMP is a homolog of CD36, a scavenger receptor important for lipoprotein binding and uptake of cholesterol and lipids in vertebrates. In humans, loss of CD36 is linked to a wide range of disorders including insulin resistance, dyslipidemia, and atherosclerosis, but how CD36 functions in lipid transport and signal transduction is poorly understood. We show that SNMP is required in pheromone-sensitive neurons for cVA sensitivity but is not required for sensitivity to general odorants. Using antiserum to SNMP infused directly into the sensillum lymph, we show that SNMP function is required on the dendrites of cVA-sensitive neurons; this finding is consistent with a direct role in cVA signal transduction. Therefore, pheromone perception in Drosophila should serve as an excellent model to elucidate the role of CD36 members in transmembrane signaling.
Keywords: CD36, olfaction, olfactory, sexual behavior, signal transduction
CVA (11-cis-vaccenyl acetate) mediates social behaviors in Drosophila, and its reception requires the odorant receptor Or67d and the extracellular pheromone-binding protein LUSH (1–4). Misexpression of Or67d receptors in trichoid neurons that are normally insensitive to pheromone confers cVA sensitivity but only if LUSH is present (3). However, Or67d and LUSH are not sufficient to confer cVA sensitivity to basiconic neurons (T.S.H. and D.P.S., unpublished work). This finding reveals that there are additional factors required for cVA sensitivity present in trichoid sensilla that are lacking in basiconic sensilla. Using a genetic screen, we set out to identify additional components important for cVA sensitivity. We screened ≈3,000 mutagenized third-chromosome lines selected for homozygous viability (5). We screened each mutant line for T1 electrophysiological responses to cVA using single sensillum electrophysiological recordings (2, 3, 6). We identified five complementation groups that were cVA-insensitive yet retained spontaneous activity in the pheromone-sensing neurons (the vains phenotype) (Fig. 1 and Table 1). The presence of spontaneous activity indicates that the neurons are present, are viable, and can sustain action potentials, thereby eliminating nonspecific mutants affecting development or general neuronal function. Of the five complementation groups recovered, two, Or67d and Or83b, affect genes previously implicated in cVA or general odorant detection, two remain unmapped, and the fifth encodes SNMP, a new cVA detection component.
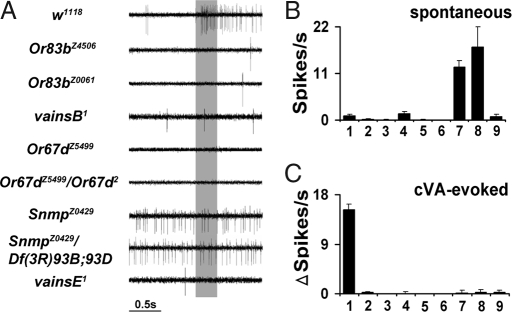
Fig. 1.
vains mutants are insensitive to cVA pheromone, in contrast to wild-type animals that show a strong response to cVA. Wild type is significantly different from the other genotypes (ANOVA; P < 0.001). (A) Single sensillum electrophysiological recordings from T1 sensilla from various genetic backgrounds. Wild type T1 sensilla (w1118) show robust responses to 1% cVA stimulation, but cVA stimulation fails to elicit responses above background from any of the vains mutants. The gray bar denotes cVA stimulus (300 ms). (B and C) Genotypes: 1, w1118; 2, Or83bZ4506; 3, Or83bZ0061; 4, vainsB1; 5, Or67dZ5499; 6, Or67dZ5499/Or67d2; 7, SnmpZ0429; 8, SnmpZ0429/ Df(3R)93B;93D; 9, vainsE1. (B) Quantitation of spontaneous activity in the same genotypes. Note the significantly increased spontaneous activity in SnmpZ0429 mutants and SnmpZ0429/Df(3R)93B;93D flies. Homozygous SnmpZ0429 and SnmpZ0429/Df(3R)93B;93D are not significantly different from each other, but both are significantly different from all other genotypes (ANOVA; P < 0.001). (C) cVA-evoked activity was quantified by measuring action potentials 1 sec after cVA stimulation and subtracting the number of action potentials 1 sec before stimulation to obtain a ΔSpikes value. Bar graphs represent mean responses ± SEM (n = 10–34).
Table 1.
The vains mutants
| Genotype | Phenotype | Gene affected |
|---|---|---|
| vainsA1 (Or83bZ4506) | cVA-insensitive | Or83b |
| vainsA2 (Or83bZ0061) | cVA-insensitive | Or83b |
| vainsB | cVA-insensitive | ND |
| vainsC (Or67dZ5499) | cVA-insensitive | Or67d |
| vainsD (SnmpZ0429) | cVA-insensitive, increased spontaneous activity | Snmp |
| vainsE | cVA-insensitive | ND |
ND, not determined.
We recovered two alleles of vainsA (vainsA1, Zuker Collection no.: Z4506, and vainsA2, Zuker Collection no.: Z0061). Fig. 1 shows that both mutants are defective for cVA sensitivity but also have striking defects in most olfactory responses. Deficiency mapping localized vainsA to the third chromosome at position 83 on the polytene map (7). A candidate gene in this interval, Or83b, encodes a coreceptor required to deliver odorant receptors to the dendrites (8). Mutants lacking Or83b are insensitive to most odorants due to lack of functional receptors exposed to the environment. Or83b mutants detect CO2 normally because this gas is detected by gustatory receptors Gr21a and Gr63a (9, 10), and gustatory receptors do not require Or83b for function (8). vainsA mutants, like previously reported Or83b mutants, have normal CO2 responses but lack responses to general odorants (Fig. 2A).
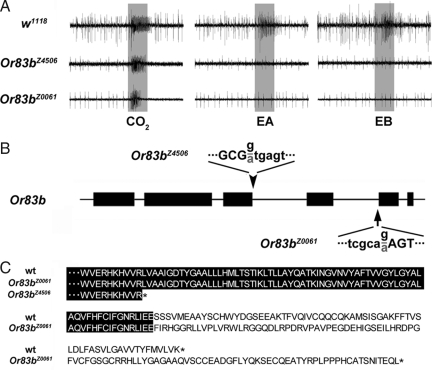
Fig. 2.
vainsA mutants are defective in Or83b expression. (A) The neurons in the large basiconic sensilla ab1 are defective for EA and EB responses in vainsA. CO2-sensitivity, mediated by the ab1c neuron that expresses gustatory receptors instead of odorant receptors, remains intact. Gray bar marks the odor stimulus (300 ms). (B) Or83b genomic locus. The black bars denote the six exons of Or83b separated by five introns. The downward arrowhead denotes the position of the point mutation that disrupts the splice donor sequence in vainsA 1(Or83bZ4506) at the start of intron 3 with the normal sequence (black letters) and the mutation (gray letter) below. Capital letters denote exon sequences, and lowercase letters are intron sequences. The upward arrow depicts the mutation in the splice acceptor site of intron 4 in vainsA2 (Or83bZ0061) that results in use of the AAG acceptor and deletion of two nucleotides and a resulting frame-shift mutant. (C) Alignment of C-terminal region of predicted Or83b polypeptides in wild type and the two Or83b mutants. For Or83bZ4506, intron 3 is included in the mature transcript, and the translation is prematurely terminated at amino acid 350. For Or83bZ0061, the frame shift causes the translated sequence to be altered after amino acid 418 and extended 37 aa past the normal stop codon in wild type Or83b.
We isolated DNA and RNA from vainsA1 and vainsA2 mutants and sequenced the genomic DNA and cDNAs encoding Or83b. Both vainsA alleles were found to contain lesions predicted to disrupt Or83b function (Fig. 2). vainsA1 mutants have a lesion in the splicing donor sequence GTGAGT at the start of intron 3 that is mutated to ATGAGT. Therefore, this intron is not recognized by the splicing machinery and is included in the mature transcript. Inclusion of this intron terminates the Or83b polypeptide prematurely at residue 350 (Fig. 2 B and C). vainsA2 mutants also have a single point mutation that produces a splicing defect. In this case, the mutants are defective in the splicing acceptor sequence, CAG, of intron 4, that is mutated from CAGAG to CAAAG. This mutation simultaneously creates a new splicing acceptor, AAG, two base pairs downstream that results in a 2-bp deletion in the mature message. Use of this novel acceptor results in a frame-shift mutation that encodes a polypeptide longer than wild type Or83b, which lacks the putative seventh transmembrane domain of the coreceptor (Fig. 2 B and C). To reflect the fact that vainsA mutants are new alleles of Or83b, we have renamed these mutants Or83bZ4506 and Or83bZ0061.
vainsC1 fails to complement Or67d2 null mutants (4), revealing that vainsC1 is defective for Or67d function (Fig. 1). Indeed, sequence analysis reveals that Or67d has a single-amino-acid substitution in vainsC1, C23W, which completely disrupts cVA signaling (Fig. 1). This mutation, near the N terminus, is predicted to be intracellular, so this mutation could disrupt the structural integrity of the receptor or its ability to activate downstream components. Henceforth, we refer to vainsC1 as Or67dZ5499.
vainsB1, vainsD1, and vainsE1 mutants complement lush and Or67d and thus represent previously uncharacterized sensitivity factors for cVA. vainsB and vainsE loci have not been mapped. However, we were able to map vainsD (see Fig. 1). vainsD1 T1 neurons are completely defective for cVA pheromone responses (Fig. 1) but are unique among the cVA detection mutants with respect to spontaneous activity. The T1 neurons from vainsD1 display increased basal activity (14–25 spikes per second compared with wild type at ≈1 spike per second). This phenotype is distinct from Or67d mutants and lush mutants which have almost no spontaneous neuronal activity present in the T1 neurons (2, 4).
To determine whether vainsD1 is required for olfactory responses in general, we surveyed the odor-evoked electrophysiological responses of large and small basiconic and non-T1 sensilla to a wide range of odorants (11, 12). Our results show that the basal activity and olfactory responses of basiconic neurons in vainsD1 mutants are indistinguishable from wild-type controls [supporting information (SI) Fig. S1]. Thus, vainsD1 is not an olfactory component mediating olfaction in a global manner but instead is selectively required for cVA activation of T1 neurons. Importantly, both Or67d and LUSH, the two factors known to be required for cVA detection, appear unaffected in the vainsD1 mutant background (Fig. S2).
We used deficiency mapping to localize the vainsD1 mutation. One deficiency, Df(3R)93B;93D, failed to complement vainsD1 (Fig. 1). We surveyed the known genes mapping to the 93B-93D interval for likely candidates. Notably, a strong candidate gene in this interval, Snmp (or CG7000), encodes a 551-aa homolog of SNMP, a moth protein expressed in pheromone-sensitive olfactory neuron dendrites (13–15). Moth SNMP is a 67-kDa polypeptide with similarity to members of the CD36 family of lipid binding proteins (15). In vertebrates, CD36 is an 88-kDa integral membrane protein receptor that mediates internalization of oxidized low-density lipoprotein by macrophages (16), formation of atherosclerotic plaques (17), and the import of long-chain fatty acids by adipose, heart, and other tissues (18, 19). In humans, loss of CD36 is linked to a wide range of disorders including insulin resistance, dyslipidemia, and atherosclerosis (17, 18, 20–22). CD36 molecules share a common domain structure with short intracellular domains at the N and C termini, two membrane spanning domains, and a large extracellular domain (19).
To examine whether Drosophila Snmp is defective in vainsD1 mutant animals, we determined its nucleotide sequence and compared it with parental controls (the isogenic stock used in the mutagenesis studies). Indeed, Snmp harbors a 5-bp deletion not present in parental controls that introduces a frame shift and a concomitant premature termination at residue 204, approximately halfway through the protein (Fig. 3 A and B). We surveyed Snmp mRNA to check global expression patterns and found abundant expression in antennae and heads lacking appendages (antennae and maxillary palps) and a lower expression level in the body (Fig. 3C). As expected, antiserum raised to the extracellular domain of the SNMP protein reveals that it is present in parental control flies and is clearly expressed in trichoid neurons and dendrites but is not detected in vainsD1 mutants (Fig. 3 D and E). To confirm that the vainsD1 (SnmpZ0429) phenotype results exclusively from the loss of the Snmp gene product, we expressed a wild type Snmp cDNA under control of the Or67d T1 neuron promoter or the lush nonneuronal supporting cell promoter in the SnmpZ0429 mutant background (Fig. 4). Expression of SNMP in the T1 neurons restored cVA sensitivity (Fig. 4 A and C), but cVA sensitivity was not restored when SNMP was expressed in the support cells with the lush promoter (Fig. 4 A and C). These findings provide direct evidence that cVA pheromone detection requires SNMP expression in T1 neurons and that this CD36 homolog has a specific role in pheromone detection in the antennae. Consistent with this finding, double mutants defective for both Snmp and lush have high spontaneous activity, indicating that SNMP functions downstream of LUSH in cVA signaling (Fig. 4 D–F).
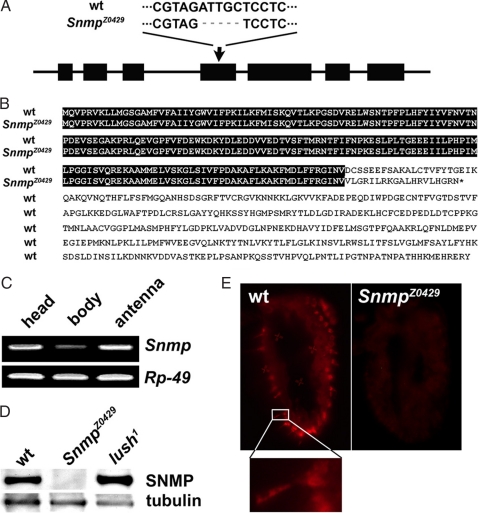
Fig. 3.
vainsD1 mutant is defective for SNMP. (A) Predicted gene structure of Snmp, composed of seven exons (solid bars). The arrow depicts the site of the lesion in vainsD1 (SnmpZ0429) and the five nucleotides deleted. (B) Alignment of deduced amino acid sequences from wild type (wt) and SnmpZ0429 mutant. SNMP is truncated in SnmpZ0429. (C) Snmp mRNA is widely expressed. The snmp-specific primers span intron regions to exclude potential genomic contamination of the cDNA. Predicted product sizes are 261 bp for Snmp and 141 bp for Rp-49. (D) Western blot of antennal extracts from wild type and SnmpZ0429 and lush1 mutants with antiserum against the entire putative extracellular domain of SNMP. Antitubulin monoclonal antibody was used to control for loading. (E) Immunofluorescent detection of SNMP protein in antennae sections in wild type and the SnmpZ0429 mutant. The magnification of wt shows expression of SNMP in a trichoid olfactory neuron cell body and dendrite. SNMP protein is not detected in the SnmpZ0429 mutant.
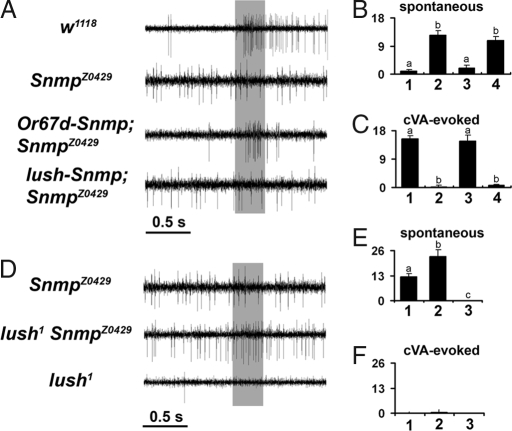
Fig. 4.
SNMP functions downstream of LUSH in cVA pheromone reception. (A) Representative recordings of T1 neurons from wild type, an SnmpZ0429 mutant, or SnmpZ0429 mutants rescued with neuronal-specific Snmp expression or with support-cell-specific Snmp expression. The gray bar denotes 1% cVA stimulus (300 ms). (B and C) Quantitation of spontaneous (B) and 1% cVA-evoked (C) activity in the different genotypes. Genotypes: 1, w1118; 2, SnmpZ0429; 3, Or67d-Snmp; SnmpZ0429; 4, lush-Snmp; SnmpZ0429. Bars represent mean response ± SEM (n = 11–34). Bars marked with the same letter are not significantly different from each other, but bars marked with different letters are significantly different (ANOVA; P < 0.001). (D) Representative traces of T1 sensilla recording from SnmpZ0429 mutants, SnmpZ0429, lush1 double mutants, or lush1 mutant flies. The gray bar denotes 1% cVA stimulus (300 ms). (E and F) Quantitation of mean spontaneous (E) and 1% cVA-evoked (F) activity. Genotypes: 1, SnmpZ0429; 2, lush1, SnmpZ0429; 3, lush1. Bars represent mean response ± SEM (n = 13–21). Bars labeled with different letters in E are significantly different (ANOVA; P < 0.001); there is no significant difference among the groups in F.
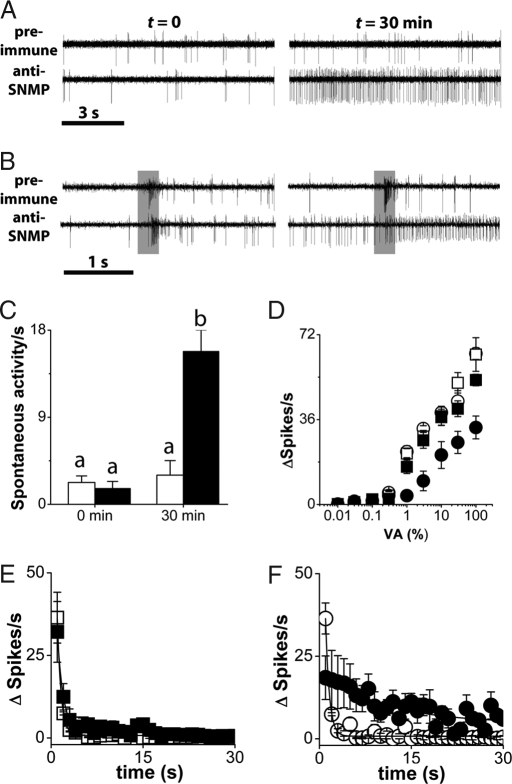
The rescue experiments prove that SNMP functions in T1 neurons but do not reveal whether SNMP directly mediates cVA detection or whether SNMP acts indirectly by mediating the expression or transport of another cVA sensitivity factor. If SNMP is required directly for cVA detection, we predict that SNMP function should be required on the surface of the T1 neuron dendrites. Therefore, we infused our antiserum to the extracellular domain of SNMP into the sensillum lymph of T1 sensilla from wild type flies through the recording pipette and monitored spontaneous activity and cVA sensitivity. Fig. 5 shows that initially the T1 neurons behave normally; but 30 min after immune serum is infused through the recording pipette, we observe striking effects on T1 behavior. First, spontaneous activity is dramatically increased, similar to what is observed in SnmpZ0429 mutants (Fig. 5 A and C). Second, dose–response analysis reveals that the cVA sensitivity is reduced ≈10-fold by the antibody treatment (Fig. 5D). Thus, disruption of SNMP function on the dendrites of T1 neurons phenocopies loss-of-function mutants in SNMP. Finally, we also observed an unexpected prolongation of cVA responses following treatment with anti-SNMP antiserum (Fig. 5 B and F). This finding suggests SNMP is also important for deactivation of cVA responses once initiated. Importantly, infusion of preimmune serum from the same animal at the same concentration had no effect on spontaneous activity, cVA sensitivity, or deactivation kinetics (Fig. 5). Essentially identical results were obtained with immune serum from two different animals (data not shown). These findings reveal that SNMP function is required on the dendritic surface where it is exposed to the sensillum lymph and support the view that SNMP functions directly in cVA signal transduction. Mutants in Snmp have been independently generated and analyzed by Benton et al. (23).
Fig. 5.
Antiserum to SNMP in the sensillum lymph phenocopies loss of Snmp. (A) Infusion of anti-SNMP immune serum (lower traces) but not preimmune serum increases the spontaneous activity of wild type T1 neurons. (Left) Traces were recorded immediately after introduction of the recording pipette containing the antiserum. (Right) Traces were recorded from the same sensillum 30 min later. (B) Anti-SNMP reduces cVA-evoked activity in T1 neurons. 3% cVA induces robust activity in neurons exposed to preimmune serum, and these responses are blunted by anti-SNMP serum. (C) Quantitation of increased spontaneous activity specific to immune serum. Open bars and filled bars represent preimmune serum and anti-SNMP infused into the T1 sensilla, respectively. Bars labeled with different letters are significantly different (ANOVA; P < 0.00001). (D) Dose–response analysis of neurons exposed to preimmune or anti-SNMP antiserum from the same animal. Preimmune (squares) and anti-SNMP (circles) at 0 min (open symbols) and 30 min (filled symbols) after penetration of the recording pipette. Each data point represents the mean ± SEM (n = 5–6). Above 1%, cVA-evoked activity in neurons exposed to anti-SNMP antiserum is significantly decreased (P = 0.0005) compared with preimmune serum. (E and F) Comparison of deactivation kinetics for preimmune and anti-SNMP antibody infused into the T1 sensilla. (E) cVA deactivation kinetics at electrode penetration (t = 0) for preimmune serum (open squares) and immune serum (filled squares). Deactivation time was constant for preimmune serum (0.68 sec ± 0.17) and for immune serum (0.73 sec ± 0.14). (F) cVA deactivation kinetics 30 min after diffusion of the antiserum through the recording pipette for preimmune serum (open circles) and immune serum (filled circles). Deactivation was constant for preimmune serum (0.69 sec ± 0.23) and for immune serum (13.68 sec ± 4.98). Net changes in spikes (ΔSpikes) were determined by subtraction of spike number before and after cVA delivery. Each data point represents average net change in spikes in 1-sec time bins.
Discussion
The results presented here, together with recent work (2, 3, 23), indicate that cVA perception in Drosophila requires supplemental factors not required for the detection of general food odorants. General food odorants are thought to activate odorant receptors through direct interactions with receptor proteins. Supporting this idea, Carlson and colleagues (24) have shown that misexpression of many Drosophila Ors in “empty” neurons (neurons lacking a functional odorant receptor) confers the odorant specificity profile of the misexpressed receptor. Thus, receptor expression is necessary and sufficient for neuronal activation by food odors. When Or67d was expressed in the empty neuron system, these workers detected responses to cVA in the absence of LUSH but only at concentrations that were orders of magnitude greater than the threshold sensitivity of wild type T1 neurons. Furthermore, these high cVA levels induced submaximal activation in the neurons (25). Other compounds with no ability to activate T1 neurons in vivo also activated Or67d under these conditions, suggesting that they may be nonspecific. Benton et al. (23) recently reported that Or67d alone failed to sensitize the empty neuron system to cVA. When Snmp was coexpressed with Or67d, high levels of cVA did elicit responses (23). However, flies with normal expression of Or67d but lacking LUSH or SNMP are electrophysiologically and behaviorally insensitive to cVA (2, 23). Thus, in vivo Or67d alone does not recapitulate the sensitivity or specificity to cVA observed in T1 neurons. LUSH and SNMP are members of a growing list of components in a unique signaling pathway used for pheromone perception but not for general odorants. It will be interesting to identify the genes affected in vainsB1 and vainsE1 mutants, both of which have normal responses to general odorants but are insensitive to cVA.
SNMP is a member of the CD36 family of lipoprotein binding proteins. CD36 knockout mice are defective for uptake of fatty acids into muscle and heart, and macrophages from these lines fail to take up oxidized cholesterol (18, 26–28). In Drosophila, other CD36 homologs are important for recognition and removal of dead cells (29) and bacteria (30), and absorption of vitamin A from the gut (31, 32) and transfer into the retina (33). In vertebrates, CD36 proteins function as receptors and signal transduction molecules. Binding to oxidized sterols triggers CD36 to interact with the nonreceptor tyrosine kinase lyn and MEKK2 which activate c-jun N-terminal kinase to mediate foam cell formation (34). SNMP clearly is required for pheromone signaling in Drosophila, and the signaling mechanisms downstream of Or67d are unknown. Whether SNMP signals through a tyrosine kinase pathway remains to be determined.
How does SNMP function in cVA signal transduction? lush1, SnmpZ0429 double mutants have high spontaneous activity as observed in SnmpZ0429 mutants, demonstrating that LUSH is upstream of SNMP in the cVA reception pathway. These genetic data are consistent with the finding that SNMP function is required in the T1 neurons, whereas LUSH is present outside the neurons (35). Based on the impaired cVA signaling and the increased spontaneous activity after treatment with antiserum to SNMP, we conclude that SNMP functions on the T1 neuron dendrites, consistent with a direct role in cVA signaling. Disruption of SNMP function, either genetically or with antiserum, results in increased spontaneous activity in T1 neurons. Thus, SNMP normally exerts an inhibitory influence on T1 activity in the absence of cVA. One model consistent with these data is that SNMP is an inhibitory subunit in a complex with Or67d. Such a role could also explain the abnormal deactivation kinetics we observed in the antibody experiments.
Detection of volatile pheromones is a specialized form of olfaction dedicated to perception of chemical cues with high biological information content delivered from other individuals of the same species. As such, pheromone detection is expected to be highly specific so that spurious environmental stimuli are not mistaken for biologically relevant pheromone cues. Our data support the idea that pheromone signaling is more specialized compared with general odor detection and requires additional factors including SNMP and LUSH. Future experiments will be required to elucidate the precise functional relationships among these factors.
Materials and Methods
Single Sensillum Recording and Odorants Preparation.
Extracellular electrophysiological recordings were carried out according to de Bruyne et al. (6). Flies (2–7 days old, males or females) were assayed under a constant stream of charcoal filtered air (36 ml/min, 22–25°C) to prevent any potential environmental odors from inducing activity during these studies. cVA, ethyl acetate (EA), and ethyl butyrate (EB) were diluted in paraffin oil (1% dilution for all cases in this report); 1 μl was applied to a filter paper and inserted in a Pasteur pipette; and air was passed over the filter and presented as the stimulus. The cVA-impregnated filters effectively evoke T1 neurons responses for over a year. Signals were amplified 1000×, fed into a computer via a 16-bit analog-to-digital converter, and analyzed offline with AUTOSPIKE software (USB-IDAC system; Syntech). The low cut-off filter setting was 200 Hz and the high cut-off setting was 3 kHz. Action potentials were recorded by inserting a glass electrode in the base of a sensillum. Data analysis was performed as reported by Xu et al. (2). Signals were recorded starting 10 sec before odorant stimulation. cVA-evoked action potentials were counted by subtracting the number of spikes 1 sec before cVA stimulation from the spike number 1 sec after cVA stimulation (ΔSpikes/sec). The recordings were performed from separate sensilla with a maximum of two sensilla recorded from any single fly. Deactivation time constants were calculated by using Origin 7.5 (OriginLab).
Genetic Screening Strategy.
The mutant lines of the Zuker EMS Collection (5) were screened by single sensillum recording using cVA stimulation. A minimum of one to three flies were tested for each line, and two to four T1 sensilla were tested for each animal. The lines with abnormal cVA response were retested in the next generation to confirm the cVA detection defect. The responses of non-T1 and basiconic sensilla from mutant lines with defective cVA response were recorded to study the effect of the mutant on global olfactory function.
Immunocytochemistry and Western Blotting.
The antiserum to SNMP was generated to the putative extracellular domain of SNMP (amino acids 42–456) expressed in E. coli and injected into rabbits as previously described (1). Immunocytochemistry was performed as previously described (1) with minor modifications. Briefly, heads were dissected with a razor blade and fixed in 4% paraformaldehyde for 4 h at 4°C and then incubated in 25% sucrose in 0.1 M NaPO4 overnight at 4°C. Fifteen-micrometer sections were collected on ProbeON Plus slides (Fisher Scientific), air-dried for 1 h, and then washed two times in 0.1 M NaPO4 and two times in 1× PBS. Slides were then blocked for 1 h in blocking buffer [3% normal goat serum, 100 mM Tris·HCl (pH 7.5), 150 mM NaCl], and incubated overnight at 4°C with a 1:500 dilution of anti-SNMP or anti-LUSH antibody (1) in blocking buffer. Slides were washed three times in TNT buffer [0.05% Tween 20, 100 mM Tris·HCl (pH 7.5), 150 mM NaCl] and incubated with a 1:500 dilution anti-rabbit IgG HRP-conjugated antibody (PerkinElmer) in blocking buffer. The slides were again washed three times in TNT buffer, and signals were amplified with the TSA Plus Cyanine 5 system (PerkinElmer), coverslipped with glycerol, and photographed. Western blot analysis was performed as previously reported (1). Between 20 and 25 antennae were dissected and homogenized for each lane, and anti-SNMP or anti-LUSH (1) serum was diluted 1:500 in blocking buffer. Antitubulin monoclonal antibody (1:400) was used for loading control.
Preparation and Delivery of Antiserum to Sensillum Lymph.
Preimmune and immune sera from the same animals were used for antiserum infusion experiments. Essentially identical results were obtained from two independent animals. Serum was diluted 1:100 in sensillum lymph buffer (36) resulting in a final protein concentration of 0.71 mg/ml for preimmune serum and 0.73 mg/ml for immune serum (Bradford assay; BioRad). Serum solutions were infused into T1 sensilla by passive diffusion through the recording electrodes, and responses were tested at t = 0 and 30 min.
Total RNA Isolation and RT-PCR.
Total RNA was purified by using TRIzol reagent as described by the manufacturer (Invitrogen). First-strand cDNA was synthesized from 1 μg of total RNA by using reverse transcriptase (Invitrogen) at 50°C for 1 h. PCR amplification was performed with Snmp-specific primers (5′-GTAGATTGCTCCTCGGAA-3′, 5′-CAGTGCCCACAAAGGTGTT-3′). Rp-49 was used as a PCR control with primer sets (5′-GCTTCAAGGGACAGTATCTG-3′, 5′-AAACGCGGTTCTGCATGAG-3′).
Generation of Transgenic Animals.
A cDNA-encoding SNMP-coding region was obtained by reverse transcription PCR amplification of RNA isolated from antennae from w1118 flies, and both strands were sequenced to confirm lack of PCR artifacts (University of Texas Southwestern Medical Center Sequencing Core Facility). The cDNA was cloned into pUAST (37), and transgenic animals were generated (38). SNMP was expressed in Or67d-expressing neurons by crossing the UAS-Snmp transgenic flies to flies expressing Gal4 under control of the Or67d promoter (39). The support cells expression was driven by Gal4 regulated by the lush promoter (1).
Insect Strains.
w1118 or bw;st, the parental line of the Zuker Collection (5), were used as wild type controls.
The detailed genotypes of the lines used are as follows:
vainsA1 (Or83bZ3–4506): + ; bw ; st Or83bZ3–4506
vainsA2 (Or83bZ3–0061): + ; bw ; st Or83bZ3–0061
vainsB1: + ; bw ; st vainsB1
vainsC1 (Or67dZ3–5499): + ; bw ; st Or67dZ3–5499
vainsD1 (SnmpZ3–0429): + ; bw ; st SnmpZ3–0429
vainsE1: + ; bw ; st vainsE1
Or67d2 knock-outs (4); w ; + ; Or67d2
Df(3R)93B;93D: Df(3R)e-R1, Ki1/TM3 Sb1 Ser1 (Bloomington Drosophila Stock Center, stock # Df3340)
w ; + ; lush1 (1)
w ; p[Or67d-GAL4 w+]; + (39)
w ; p[lush-GAL4 w+] ; +
Or67dZ5499/Or67d2: + ; bw/+ ; st Or67dZ5499/Or67d2
SnmpZ0429/Df3340: + ; bw/+ ; st SnmpZ0429/Df(3R)e-R1 Ki1
Or67d-Snmp;SnmpZ0429: w ; p[Or67d-GAL4 w+]/p[UAS-Snmp w+] ; SnmpZ0429
lush-Snmp;SnmpZ0429: w ;p[lush-GAL4 w+]/p[UAS-Snmp w+] ; SnmpZ0429
lush1, SnmpZ0429: w ; + ; lush1 SnmpZ0429
Supplementary Material
Acknowledgments.
We thank Charles Zuker for access to the Zuker Collection and Barry Dickson for Or67d2 mutant flies. This work was supported by National Institutes of Health Grant R01 DC02539.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803309105/DCSupplemental.
References
- 1.Kim M-S, Repp A, Smith DP. LUSH odorant binding protein mediates chemosensory responses to alcohols in Drosophila melanogaster. Genetics. 1998;150:711–721. doi: 10.1093/genetics/150.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu P-X, Atkinson R, Jones DNM, Smith DP. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron. 2005;45:193–200. doi: 10.1016/j.neuron.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 3.Ha TS, Smith DP. A pheromone receptor mediates 11-cis-vaccenyl acetate-induced responses in Drosophila. J Neurosci. 2006;26:8727–8733. doi: 10.1523/JNEUROSCI.0876-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurtovic A, Widmer A, Dickson BJ. A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature. 2007;446:542–546. doi: 10.1038/nature05672. [DOI] [PubMed] [Google Scholar]
- 5.Koundakjian EJ, Cowan DM, Hardy RW, Becker AH. The Zuker collection: A resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics. 2004;167:203–206. doi: 10.1534/genetics.167.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Bruyne M, Clyne P, Carlson JR. Odor coding in a model olfactory organ: The Drosophila maxillary palp. J Neurosci. 1999;19:4520–4532. doi: 10.1523/JNEUROSCI.19-11-04520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindsley DL, Zimm G. The Genome of Drosophila melanogaster. San Diego: Academic; 1992. [Google Scholar]
- 8.Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. 2007;445:86–90. doi: 10.1038/nature05466. [DOI] [PubMed] [Google Scholar]
- 10.Kwon JY, Dahanukar A, Weiss LA, Carlson JR. The molecular basis of CO2 reception in Drosophila. Proc Natl Acad Sci USA. 2007;104:3574–3578. doi: 10.1073/pnas.0700079104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobritsa AA, van der Goes van Naters W, Warr CG, Steinbrecht RA, Carlson JR. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37:827–841. doi: 10.1016/s0896-6273(03)00094-1. [DOI] [PubMed] [Google Scholar]
- 12.de Bruyne M, Foster K, Carlson JR. Odor coding in the Drosophila antenna. Neuron. 2001;30:537–552. doi: 10.1016/s0896-6273(01)00289-6. [DOI] [PubMed] [Google Scholar]
- 13.Rogers ME, Krieger J, Vogt RG. Antennal SNMPs (sensory neuron membrane proteins) of Lepidoptera define a unique family of invertebrate CD36-like proteins. J Neurobiol. 2001;49:47–61. doi: 10.1002/neu.1065. [DOI] [PubMed] [Google Scholar]
- 14.Rogers ME, Steinbrecht RA, Vogt RG. Expression of SNMP-1 in olfactory neurons and sensilla of male and female antennae of the silkmoth Antheraea polyphemus. Cell Tissue Res. 2001;303:433–446. doi: 10.1007/s004410000305. [DOI] [PubMed] [Google Scholar]
- 15.Rogers ME, Sun M, Lerner M, Vogt RG. Snmp-1, a novel membrane protein of olfactory neurons of the silk moth Antherea polyphemus with homology to the CD36 family of membrane proteins. J Biol Chem. 1997;272:14792–14799. doi: 10.1074/jbc.272.23.14792. [DOI] [PubMed] [Google Scholar]
- 16.Collot-Teixeira S, Martin J, McDermott-Roe C, Poston R, McGregor JL. CD36 and macrophages in atherosclerosis. Cardiovasc Res. 2007;75:468–477. doi: 10.1016/j.cardiores.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 17.Febbraio M, Guy E, Silverstein RL. Stem cell transplantation reveals that absence of macrophage CD36 is protective against atherosclerosis. Arterioscler Thromb Vasc Biol. 2004;24:2333–2338. doi: 10.1161/01.ATV.0000148007.06370.68. [DOI] [PubMed] [Google Scholar]
- 18.Coburn CT, et al. Defective uptake and utilization of long chain fatty acids in muscle and adipose tissues of CD36 knockout mice. J Biol Chem. 2000;275:32523–32529. doi: 10.1074/jbc.M003826200. [DOI] [PubMed] [Google Scholar]
- 19.Febbraio M, Silverstein RL. CD36: Implications in cardiovascular disease. Int J Biochem Cell Biol. 2007;39:2012–2030. doi: 10.1016/j.biocel.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pravenec M, Kurtz TW. Molecular genetics of experimental hypertension and the metabolic syndrome: From gene pathways to new therapies. Hypertension. 2007;49:941–952. doi: 10.1161/HYPERTENSIONAHA.107.086900. [DOI] [PubMed] [Google Scholar]
- 21.Miyaoka K, et al. CD36 deficiency associated with insulin resistance. Lancet. 2001;357:686–687. doi: 10.1016/s0140-6736(00)04138-6. [DOI] [PubMed] [Google Scholar]
- 22.Hirano K, et al. Pathophysiology of human genetic CD36 deficiency. Trends Cardiovasc Med. 2003;13:136–141. doi: 10.1016/s1050-1738(03)00026-4. [DOI] [PubMed] [Google Scholar]
- 23.Benton R, Vannice KS, Vosshall LB. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature. 2007;450:289–293. doi: 10.1038/nature06328. [DOI] [PubMed] [Google Scholar]
- 24.Hallem EA, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 25.van der Goes van Naters W, Carlson JR. Receptors and neurons for fly odors in Drosophila. Curr Biol. 2007;17:606–612. doi: 10.1016/j.cub.2007.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonen A, et al. A null mutation in skeletal muscle FAT/CD36 reveals its essential role in insulin- and AICAR-stimulated fatty acid metabolism. Am J Physiol. 2007;292:E1740–1749. doi: 10.1152/ajpendo.00579.2006. [DOI] [PubMed] [Google Scholar]
- 27.Thorne RF, Mhaidat NM, Ralston KJ, Burns GF. CD36 is a receptor for oxidized high density lipoprotein: Implications for the development of atherosclerosis. FEBS Lett. 2007;581:1227–1232. doi: 10.1016/j.febslet.2007.02.043. [DOI] [PubMed] [Google Scholar]
- 28.Guest CB, et al. Phagocytosis of cholesteryl ester is amplified in diabetic mouse macrophages and is largely mediated by CD36 and SR-A. PLoS ONE. 2007;2:e511. doi: 10.1371/journal.pone.0000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franc NC, Dimarcq JL, Lagueux M, Hoffmann J, Ezekowitz RA. Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity. 1996;4:431–443. doi: 10.1016/s1074-7613(00)80410-0. [DOI] [PubMed] [Google Scholar]
- 30.Philips JA, Rubin EJ, Perrimon N. Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science. 2005;309:1251–1253. doi: 10.1126/science.1116006. [DOI] [PubMed] [Google Scholar]
- 31.Kiefer C, Sumser E, Wernet MF, Von Lintig J. A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proc Natl Acad Sci USA. 2002;99:10581–10586. doi: 10.1073/pnas.162182899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu G, Yang J, Mitchell KA, O'Tousa JE. Drosophila ninaB and ninaD act outside of retina to produce rhodopsin chromophore. J Biol Chem. 2004;279:18608–18613. doi: 10.1074/jbc.M400323200. [DOI] [PubMed] [Google Scholar]
- 33.Wang T, Jiao Y, Montell C. Dissection of the pathway required for generation of vitamin A and for Drosophila phototransduction. J Cell Biol. 2007;177:305–316. doi: 10.1083/jcb.200610081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahaman SO, et al. A CD36-dependent signaling cascade is necessary for macrophage foam cell formation. Cell Metab. 2006;4:211–221. doi: 10.1016/j.cmet.2006.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shanbhag SR, Smith DP, Steinbrecht RA. Three odorant-binding proteins are co-expressed in the sensilla trichodea of Drosophila melanogaster. Arthropod Struct Dev. 2005;34:153–165. [Google Scholar]
- 36.Kaissling K-E, Thorson J. Insect olfactory sensilla: Structural, chemical, and electrical aspects of the functional organization. In: Sattelle DB, Hall LM, Hildebrand JG, editors. Receptors for Neurotransmitters, Hormones and Pheromones in Insects. Amsterdam: Elsevier; 1980. pp. 261–282. [Google Scholar]
- 37.Brand A, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 38.Karess RE, Rubin GM. Analysis of P transposable element functions in Drosophila. Cell. 1984;38:135–146. doi: 10.1016/0092-8674(84)90534-8. [DOI] [PubMed] [Google Scholar]
- 39.Couto A, Alenius M, Dickson BJ. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol. 2005;15:1535–1547. doi: 10.1016/j.cub.2005.07.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.