Abstract
During translation, usually only one in ≈400 misincorporations affects the function of a nascent protein, because only chemically similar near-cognate amino acids are misincorporated in place of the cognate one. The deleterious misincorporation of a chemically dissimilar noncognate amino acid during the selection process is precluded by the presence of a tRNA at the ribosomal E-site. However, the selection of first aminoacyl-tRNA, directly after initiation, occurs without an occupied E-site, i.e., when only the P-site is filled with the initiator tRNA and thus should be highly error-prone. Here, we show how bacterial ribosomes have solved this accuracy problem: In the absence of a Shine–Dalgarno (SD) sequence, the first decoding step at the A-site after initiation is extremely error-prone, even resulting in the significant incorporation of noncognate amino acids. In contrast, when a SD sequence is present, the incorporation of noncognate amino acids is not observed. This is precisely the effect that the presence of a cognate tRNA at the E-site has during the elongation phase. These findings suggest that during the initiation phase, the SD interaction functionally compensates for the lack of codon–anticodon interaction at the E-site by reducing the misincorporation of near-cognate amino acids and prevents noncognate misincorporation.
Keywords: E-site, translational errors
The binding of aminoacyl-tRNAs to the ribosome is dictated by the complementarity between the anticodon of the tRNA and the codon of the mRNA. To ensure the high fidelity of translation, the correct stereochemistry of the mRNA-tRNA codon–anticodon interaction is monitored by components of the small ribosomal subunit in a process known as decoding (reviewed in ref. 1). During decoding, the first and second nucleotide positions (in terms of the codon) of the mRNA-tRNA duplex are closely monitored, whereas interaction at the third or wobble position is less strictly recognized. Consistently, the misincorporation of the wrong amino acids into polypeptide chains usually occurs through the binding of near-cognate aminoacyl-tRNAs, i.e., those tRNAs carrying an anticodon similar to that of the cognate aminoacyl-tRNA, rather than noncognate aminoacyl-tRNAs, which carry dissimilar anticodons.
Nascent polypeptide chains are surprisingly tolerant to misincorporation, with only one in ≈400 misincorporations being deleterious for the protein's activity (reviewed in ref. 2). The reason for this is that usually near-cognate aminoacyl-tRNAs are selected instead of the cognate aminoacyl-tRNA, and the genetic code lexicon is organized in such a way that near-cognate tRNAs bear amino acids that are chemically similar to those carried by the cognate tRNA. For example, the misincorporation of an aspartate (codon: GAU/C) by near-cognate Asp-tRNA, instead of glutamate (GAA/G) by the cognate Glu-tRNA, both incorporate acidic amino acids. The middle and, in most cases, the first position of a codon are almost never misread, even under error-inducing conditions such as high magnesium or the presence of aminoglycosides, and are thus considered noncognate (for review and references see ref. 3). The terms cognate, near-cognate, and noncognate are also defined functionally, namely, the misincorporation of near-cognate amino acids in vivo and in vitro require higher GTP consumption than for cognate, whereas noncognate amino acids are never incorporated, and no GTP is consumed (4, 5), or if incorporation is observed, the rate is greatly reduced (6, 7).
Aminoacyl-tRNAs can, however, occupy the A-site without being subjected to the decoding process. For example, the tRNA moiety of Ala-tmRNA does not even have an anticodon but still binds efficiently to the A-site in complex with EF-Tu·GTP and the SmpB protein (8). Another example is when the ribosome has an empty E-site, i.e., a peptidyl-tRNA occupies the P-site whereas the A- and E-sites are free. In such a situation, even a noncognate aminoacyl-tRNA can enter the A-site leading to an incorporation of the noncognate amino acid into the nascent peptide chain (this article) (5). During elongation, when a peptidyl-tRNA resides at the P-site after translocation, the E-site is tightly occupied by a deacylated tRNA (9, 10). The E-tRNA is released through an active mechanism, whereby interaction of a ternary complex aminoacyl-tRNA·EF-Tu·GTP at the A-site is coupled to the release of the E-tRNA (11, 12). E-tRNA release follows the decoding step but occurs before accommodation of the aminoacyl-tRNA into the A-site (13).
The presence of an E-tRNA has been shown to be important for maintaining the reading frame both in vivo (14, 15) and in vitro (10) but also, as mentioned above, for preventing the selection of noncognate aminoacyl-tRNAs (5). In the latter experiment, Geigenmüller et al. (5) demonstrated that when the E-site was unoccupied, the noncognate acidic Asp (codon GAC/U) could be misincorporated in place of the cognate aromatic hydrophobic Phe (codon UUU/C), however, no misincorporation of Asp was observed when the E-site was occupied. Because tRNA near-cognate to the E-site codon could not prevent the incorporation of a noncognate amino acid, the conclusion was that codon–anticodon interaction at the E-site is required to prevent misincorporation at the A-site. This is consistent with the genetic (14, 15), biochemical (12, 16, 17), and structural (18) evidence demonstrating the likelihood of codon–anticodon interaction at the E-site.
So how does the presence of an E-tRNA influence decoding at the A-site? An occupied E-site dramatically increases (almost 3-fold) the activation energy barrier for A-site occupation, namely from ≈40 kJ/mol to ≈115 kJ/mol, in a physiological buffer with 3–6 mM Mg2+ and polyamines (19). These findings were incorporated into the allosteric three-site model (4, 9, 12) stating that the A- and E-sites are reciprocally linked, such that occupation of the E-site induces a low-affinity A-site, and vice versa. This model explains why, in native polysomes from both eukaryotes and bacteria, precisely two tRNAs per ribosome are observed (20, 21).
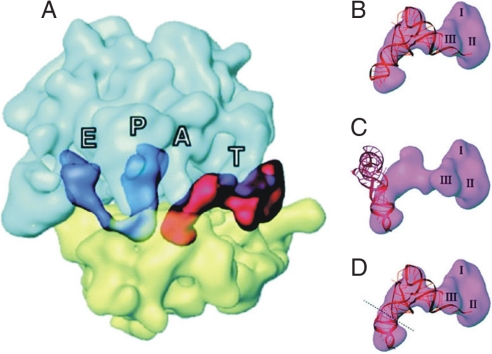
The next question is how the low-affinity A-site excludes the selection of noncognate aminoacyl-tRNAs? One possibility is that the low-affinity A-site restricts the binding of the ternary complex (EF-Tu·GTP·aminoacyl-tRNA) with the ribosome to only the interaction between the A-site codon and the anticodon of the tRNA, until successful decoding is completed. In this model, contacts outside of the codon–anticodon interaction would not contribute to the selection precision because they are common to all ternary complexes, regardless of cognate or noncognate, and therefore would allow even noncognate aminoacyl-tRNAs to interfere with the selection process, as well as leading to the occasional misincorporation before the decoding potential of the codon–anticodon interaction has been exploited (22). Indeed, cryoelectron microscopic (cryo-EM) studies reveal that during A-site decoding, the incoming ternary complex aminoacyl-tRNA·EF-Tu·GTP binds in an initial A/T state, where codon–anticodon interaction is checked in the decoding center of the A-site before the aminoacyl-tRNA fully moves into the classic A-site (23–25). Interestingly, the anticodon loop is kinked relative to the anticodon stem by ≈40° to allow decoding, whereas simultaneously preventing interaction of the tRNA outside the anticodon loop with the A-site (Fig. 1, A-D). However, EF-Tu interaction with the ribosome visualized in these complexes probably reflects a state after the decoding process has been completed, and therefore it is unclear whether EF-Tu interacts with the ribosome before or during the selection process. We assume that EF-Tu contacts the ribosome only after the decoding process (22), but we note that this point remains controversial (6, 7).
Fig. 1.
The ternary complex interacting with the decoding center of the A-site as seen by using cryo-EM. (A) The ribosome position of the ternary complex during the decoding process (A/T-site). (B–D) Fitting the aminoacyl-tRNA within the ribosomal-bound ternary complex. To satisfactorily fit the crystal structure of a tRNA into the corresponding cryo-EM density requires the introduction of a kink ≈40° in the anticodon stem, just below the anticodon stem of the aminoacyl-tRNA. (According to ref. 23, modified.)
If an occupied E-site is important for translational fidelity by reducing near-cognate or preventing noncognate misincorporation at the A-site, as explained by the allosteric three-site model, this raises the question as to how accuracy is maintained when the first aminoacyl-tRNA binds to the A-site directly after the initiation phase. This is a unique situation, in which ribosomes contain only one tRNA, namely an initiator-tRNA bound at the P-site, referred to as a Pi state. Therefore, directly after initiation, the binding of ternary complex to the ribosome and decoding at the A-site occurs with an empty E-site, and according to the allosteric three-site model, should be error-prone. This would be surprising because there is a strong codon bias at the second position for GCN codons in highly expressed genes (26), i.e., the codon directly after the start codon, and this position has been shown to have a strong influence on the efficiency of translation initiation (27). Indeed, stable cognate codon–anticodon interaction at this position has been proposed to be important for preventing premature peptidyl-tRNA drop-off (26). Furthermore, the first few N-terminal amino acids modulate the stability of proteins as well as providing determinants for the cleavage of the N-terminal formyl-methionine residue from nascent peptide (28, 29). Collectively, this suggests that accurate decoding at the second position is important for gene expression, and therefore bacteria must have developed a mechanism to ensure accurate decoding at the A-site in the absence of an E-tRNA.
Here, we demonstrate that Pi state ribosomes, lacking an E-tRNA, are indeed error-prone and misincorporate both near- and noncognate-tRNAs when the mRNA has no Shine–Dalgarno (SD) sequence. However, the presence of a SD sequence leads to a reduction in the misincorporation of near-cognate amino acids and abolishes misincorporation of noncognate amino acids, analogous to the presence of E-tRNA. These experiments reveal that the SD sequence functionally compensates for the absence of E-tRNA in conferring accuracy to the first decoding step of translation elongation.
Results
The E-tRNA Is Important for the Accurate Selection of Aminoacyl-tRNA at the A-Site.
Previous experiments demonstrating that a cognate deacylated tRNA at the E-site is important for the fidelity of aminoacyl-tRNA selection at the A-site, were performed by using ribosomes programmed with poly(U) mRNA (5). However, with poly(U) mRNA, the binding of the deacylated tRNAPhe is ambiguous because both the A- and E-site codon display a Phe (UUU) codon. When a heteropolymeric mRNA displaying different codons at A- and P-sites is used, this ambiguity is resolved because noncognate deacylated tRNA cannot bind to the E-site under the experimental conditions used (17).
Under steady-state conditions, such as that seen during poly(U)-dependent poly(Phe) synthesis, the misincorporation of the near-cognate amino acid Leu is <0.1% (Leu versus Leu plus Phe incorporation) by using our polyamine buffer system (3, 30). We analyzed the error of near-cognate dipeptide formation in the presence of a heteropolymeric mRNA with ribosome·P-tRNA complexes, containing either a free (Pi) or an occupied E-site (posttranslocational state (POST) with tRNAs at P- and E-sites). A moderate reduction of the near-cognate error by a factor of ≈2 on E-site occupation was observed in agreement with previous experiments (data not shown) (5).
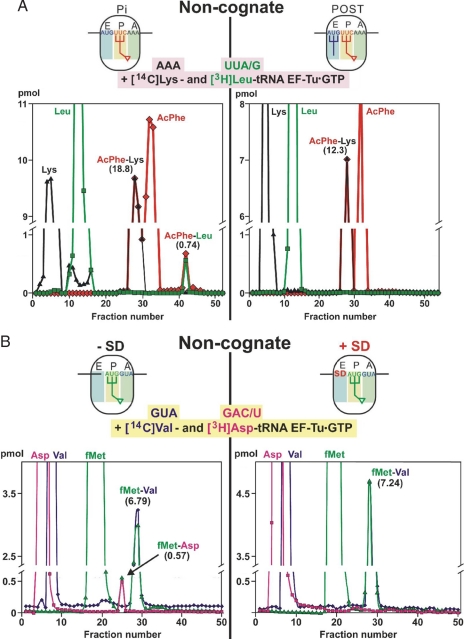
A more important case deals with the influence of the E-tRNA on the misincorporation of noncognate amino acid, a process much more detrimental to protein synthesis than the incorporation of a near-cognate one. Therefore, the formation of noncognate amino acid-containing dipeptides was analyzed in the presence of a heteropolymeric mRNA with and without a tRNA at the E-site. Both Pi and POST state ribosomes were prepared by using the heteropolymeric MFK-mRNA (AUG-UUC-AAA encoding for Met-Phe-Lys), such that Ac[14C]Phe-tRNA is positioned at the P-site, and the POST state, formed via translocation, contains an additional deacylated [32P]tRNA fMet bound to the E-site. A stoichiometric mixture of ternary complexes containing cognate Lys-tRNA (codon AAA) and noncognate Leu-tRNAs (codons UUA/G) were then added, and the formation of cognate and noncognate dipeptides was assessed via HPLC in one and the same experiment. Supporting information (SI) Fig. S1 and Fig. 2 demonstrate the excellent resolution of the HPLC system to separate the free amino acids from the dipeptides, such as AcPhe-Leu and fMet-Val. At least three experiments were performed for each experimental design, and representative examples are shown in Fig. 2.
Fig. 2.
Noncognate misincorporation levels. (A) The influence of the E-tRNA: HPLC analysis of dipeptides formed by the addition of a stoichiometric mixture of ternary complexes containing cognate [14C]Lys-tRNA and noncognate [3H]Leu-tRNA to either (i) Pi-state ribosomes (Left) containing AcPhe-tRNA at the P-site or (ii) POST-state ribosomes (Right) carrying AcPhe-tRNA at the P-site and deacylated [32P]tRNAfMet at the E-site, generated via EF-G-dependent translocation. The codons are given above the amino acids, and the specific activity of [3H]Leu-tRNALeu was very high (10,120 dpm/pmol), enabling even very low-level misincorporation of [3H]Leu-tRNALeu to be detected, i.e., the resolution limit was 0.03 pmol for noncognate AcPhe-Leu dipeptide. Puromycin reaction demonstrated a high specificity of >90% for the Pi and POST translocational complexes. (B) The effects of SD on the selection of noncognate aminoacyl-tRNA in the presence of MVF-mRNA. After filling the P-site with f[3H]Met-tRNA (1,670 dpm/pmol), a mixture of ternary complexes was added containing cognate [14C]Val-tRNA (codon GUA; 540 dpm/pmol) and noncognate [3H]Asp-tRNA (GAC/U; 21,750 dpm/pmol). The resolution limit of the noncognate dipeptide fMet-Asp was 0.06 pmol in this HPLC analysis. In the absence of the SD sequence, an error of 7.7% was observed (Left), whereas in its presence, a significant amount of noncognate fMet-Asp is not observed (Right). Each experiment was performed at least three times, a representative run is shown. For further details see Materials and Methods.
In the absence of E-tRNA, the formation of the noncognate AcPhe-Leu dipeptide represented 3.8% of the total dipeptide formed (Pi state in Fig. 2A Left), whereas with a filled E-site (POST), no traces of noncognate dipeptide were observed within the resolution limitations of the experiment (Fig. 2A Right and Table S1). This demonstrates that E-site occupation is essential for preventing the misincorporation of noncognate amino acids and is in agreement with the results by using the poly(U) mRNA, where the misincorporation of Asp (codon GAC/U) at the A-site codon UUU was monitored (5).
The SD Sequence Confers Accuracy to the First Decoding Step After Translation Initiation.
We next wanted to determine the accuracy of the first decoding step after the translation initiation, in particular to ascertain whether or not the SD sequence can functionally compensate for the absence of an E-tRNA and reduce the level of misincorporation of near-cognate aminoacyl-tRNAs at the A-site codon. To this end, we programmed the ribosome by using the MA-mRNA (AUG-GCU encoding Met-Ala) with a GGAGG SD sequence (+SD) and without (−SD), separated by a spacer of seven nucleotides upstream from the AUG codon. Initiator fMet-tRNA was bound enzymatically to the AUG start codon at the P-site, thus placing the GCU codon cognate for Ala-tRNA at the A-site. Mixtures of ternary complexes containing constant amounts of cognate [14C]Ala-tRNA (anticodon 3′-CGU) but increasing amounts of the near-cognate [3H]Thr-tRNA (anticodon 3′-UGU) were then added, and the binding of the respective tRNAs was assessed via nitrocellulose filtration. Fig. 3 shows that at all ratios of cognate:near-cognate aminoacyl-tRNAs, the binding of the near-cognate Thr-tRNA was significantly lower when the SD sequence was present in the mRNA (+SD). For example, at equimolar ratios of cognate and near-cognate tRNAs, the binding of the near-cognate Thr-tRNA was approximately two times lower with the +SD-mRNA compared with the −SD-mRNA. This moderate effect on reducing misincorporation of near-cognate tRNAs is similar to that observed when deacylated tRNA is present at the E-site (see Table 1) (5).
Fig. 3.
The effects of SD sequence on the selection of near-cognate aminoacyl-tRNA in the presence of MA-mRNA. 70S ribosomes were programmed with mRNAs with or without an SD sequence. After binding enzymatically f[3H]Met-tRNA (15 dpm/pmol) to the P-site, a mixture of ternary complexes were offered containing cognate [14C]Ala-tRNA (codon GCU; 360 dpm/pmol) and various amounts of near-cognate [3H]Thr-tRNA (ACU; 8,640 dpm/pmol). The molar ratio Ala-tRNA:70S = 1:1 was kept constant, where Thr-tRNA was 1, 5, 10, and 15.
Table 1.
Comparison of the improvements of the accuracy of aminoacyl-tRNA selection for A-site occupation in the presence of E-tRNA or a SD sequence
| Category | E-site factor, % | SD factor, % |
|---|---|---|
| Improved accuracy (cognate versus near-cognate) | Free/occupied E-site <2.0 (ref. 5) | No SD/SD7.3/3.8 = 1.9(Fig. 3) |
| Improved accuracy (cognate versus noncognate) | Free/occupied E-site 3.8/<0.5 = >7.6 (Fig. 2A) | No SD/SD7.7/<0.8 = >9.6(Fig. 2B) |
E-site factor (SD factor) is the ratio of the error observed with a free and occupied E-site (without and with SD sequence). The factors given for improved accuracy for the discrimination against noncognate amninoacyl-tRNAs are minimal values, because no significant error above the resolution limit could be measured in the presence of an occupied E-site or an SD sequence. Therefore, the resolution limits of 0.03 and 0.06 pmol for noncognate dipeptides, respectively, were taken as misincorporation values (see legend of Fig. 2B).
To monitor the influence of the SD sequence on the misincorporation of noncognate aminoacyl-tRNAs during the first decoding step, ribosomes were programmed with either the MVF-mRNA (−SD) or the MVF-mRNA (+SD), containing a GGAGGU SD sequence 5 nt upstream of the AUG start codon (see Materials and Methods). In both cases, the P-site was filled with the initiator fMet-tRNA, and then a stoichiometric mixture of ternary complexes containing cognate Val-tRNA (codon GUA) and noncognate Asp-tRNA (GAC/U) was added. In the absence of the SD sequence (Fig. 2B Left), the fraction of the noncognate fMet-Asp dipeptide (0.57 pmol) constituted 7.7% of the total (6.79 pmol + 0.57 pmol), whereas in the presence of the SD sequence (Fig. 2B Right), no formation of the fMet-Asp dipeptide was observed above the background. Because the resolution limit was 0.03 pmol, the 0.57 pmol value of the noncognate dipeptide could be determined with high precision. This striking effect of the SD sequence to abolish the detrimental noncognate misincorporation during the first decoding step at the A-site parallels the corresponding effects of the E-tRNA during translation elongation (see Table 1 for comparison) (5).
Discussion
Occupation of the E-site with a cognate deacylated tRNA has three important, and probably mutually related, consequences for the A-site: (i) it induces a low-affinity A-site (11, 12), (ii) it increases the activation energy for A-site occupation dramatically, from ≈40 to 115 kJ/mol (19), and (iii) it prevents the incorporation of noncognate amino acids, i.e., those most likely to affect protein structure and function, and less importantly reduces the incorporation of near-cognate ones. Thus occupation of the E-site makes a significant overall contribution to the accuracy of translation (Fig. 2A and ref. 5). In vivo studies have also demonstrated that the weakening of E-tRNA binding, or the absence of codon–anticodon interaction at the E-site, induces errors and frameshifting (refs. 31 and 32, respectively). The induction of a low-affinity A-site by the presence of E-tRNA has been demonstrated to occur on ribosomes from all three domains of life (for review see ref. 33), with the most impressive example being observed with yeast 80S ribosomes, where an occupied E-site prevents the binding of the cognate ternary complex aminoacyl-tRNA·EF1A·GTP, unless the EF3 together with ATP is present and opens the E-site (12). This view was recently supported by a cryo-EM analysis of a yeast EF3–80S ribosome complex revealing how EF3 opens the E-site (34).
Here, we have demonstrated that the presence of a SD sequence located in the 5′ untranslated region of an mRNA can functionally compensate for the lack of a cognate tRNA at the E-site, a situation that occurs directly after the initiation phase of translation. We show that the SD sequence confers similar beneficial effects as an E-tRNA, in terms of accuracy during the selection of ternary complexes aminoacyl-tRNA·EF-Tu·GTP at the decoding center: The selection of the near-cognate aminoacyl-tRNA is moderately improved by a factor of two (Fig. 3), but, most significantly, the misincorporation of detrimental noncognate amino acids is abolished (Fig. 2B and Table 1). This feature provides a likely explanation as to why only 1 in 400 misincorporations have deleterious consequences for folding, stability, and/or function of the protein (2), because, as mentioned, misincorporation of near-cognate aminoacyl-tRNA substitutes chemically similar amino acids, whereas misincorporation of noncognate aminoacyl-tRNA introduces chemically unrelated amino acids.
Recent x-ray crystallography studies have visualized the interaction of the SD sequence with the anti-SD sequence located in the 3′-end of the 16S rRNA on the ribosome (18, 35–37). These studies reveal that the SD helix sits in a pocket located between the head and platform of the 30S subunit, adjacent to but not directly in the E-site. The SD–anti-SD interaction probably reduces the time necessary for mRNA-ribosome programming, because it helps to guide the mRNA from an initial stand-by site into a position whereby the AUG start codon is correctly positioned in the presence of initiator tRNA (36, 38, 39). Interestingly, the conformation of the mRNA in the E-site appears to be influenced by the state of the ribosome. In the initiation state, the mRNA is considered to be in a structurally constrained conformation, such that codon–anticodon interaction would not be possible in the E-site (18, 37). However, after initiation, the whole SD helix rotates on the ribosome toward the E-site, which leads to conformational relaxation in the mRNA, such that the A-form helix is adopted by the E-codon of the mRNA that now allows codon–anticodon interaction at the E-site (18). The recent observation that the SD helix appears to fix the orientation of head of the 30S subunit (35) might provide the first structural hint as to how the SD helix (or E-tRNA) influences A-site accuracy, however further work using both in vitro and in vivo experimental systems will be required to fully elucidate this mechanism.
A recent analysis of 162 completely sequenced prokaryotic genomes (with 141 of bacterial origin), revealed that an astonishingly large percent (46%) of mRNAs do not contain a SD sequence, with the corresponding value for Escherichia coli mRNA being 39% (40). However, it is well documented that a SD sequence is preferentially found in highly expressed genes (41), suggesting that accuracy during the first decoding step may be more important for high expression, although it is unclear exactly why. In addition to the SD sequence, an optimal spacer length of ≈6 nt between SD and the initiator AUG codon, an A/U-rich enhancer upstream of the SD sequence and the absence of strong secondary structures around the SD sequence are most important determinants for high expression (for review and references see ref. 42).
We do not know whether or how the eukaryotic 80S ribosomes overcome the accuracy problem during the first decoding step, because their mRNAs do not contain SD sequences. Eukaryotic translation systems require ≈12 initiation factors, some of which are composed of several different subunits (for reviews see refs. 43, 44), whereas bacterial systems use only 3 monomeric initiation factors. Thus, we can only speculate that the more complicated system required for the formation of both the 40S and subsequent 80S initiation complexes solves the accuracy problem of the first aminoacyl-tRNA selection.
Finally, archaeal mRNAs often contain an identifiable SD, but their set of initiation factors is similar to, although somewhat simpler than, that in eukaryotes (45). Curiously, archaea contain a number of leaderless mRNAs, i.e., mRNAs that have no 5′ untranslated region (therefore no SD sequence), and start directly with an AUG start codon. Based on the findings presented here, we would predict that leaderless mRNAs are error-prone at the step of forming the initial dipeptide. Whether the corresponding proteins can tolerate an increased error at the N-terminal or whether another mechanism operates remains unknown. At least in bacteria, only a fraction (<0.1%) of mRNAs are leaderless and do not comprise mRNAs of essential genes (46), therefore the accuracy problem might not pose a significant problem toward cell viability in these cases.
In summary, we have demonstrated here that the SD sequence, in addition to its canonical function related to mRNA positioning, has a second important function. This is seen in the fact that the SD–anti-SD interaction can functionally replace the E-tRNA to confer accurate decoding of the codon after the AUG. Specifically, the SD sequence reduces near-cognate misincorporation and precludes the selection of noncognate aminoacyl-tRNAs, thereby protecting the cell from amino acids substitutions detrimental to protein folding, stability, and function. Because noncognate aminoacyl-tRNAs in the cell are in >5- to 10-fold excess over cognate aminoacyl-tRNAs, it is clear that the absence of the beneficial effects of cognate E-tRNA and SD–anti-SD interactions would practically exclude the synthesis of a protein of a length ≈400 aa, with an undisturbed structure and function.
Materials and Methods
Translational Components.
Sources for materials, various tRNAs, isolation procedures for EF-Tu and EF-G with C-terminal His-tags, charged tRNAs, and reassociated 70S ribosomes were described (10). MFK-mRNA, encoding Met-Phe-Lys, sequence GGG(A4G)3AAAAUGUUCAAAAG(A4G)2AAAU (47), and MVF-mRNA sequence GGG(A4G)3AAAAUGGUAUUC(A4G)3AAAU, encoding Met-Val-Phe, were prepared accordingly (48). The MVF-mRNAs without and with a SD sequence, used in the experiments shown in Fig. 2B, had the sequences GGGAA(GA4)CACAUAUGGUAUUCAAA(GA4)5UGGACUCAGA-GCUACGGAAAUAUUCG and GGGAA(GA4)GGAGGUCACAUAUGGU-AUUCAAA(GA4)5UGGACUCAGAGCUACGGAAAUAUUCG (coding sequence in bold and italics and SD sequence underlined).
mRNA encoding the sequence for Met-Ala, in the presence of a SD sequence (+SD MA-mRNA) GGC6GGAGGC4CCCAUGGCUUCUC16A, and in its absence (−SD MA-mRNA) GGC15CCCAUGGCUUCUC16A, were cloned between the BstNI and BamHI restriction sites of plasmid pET7 from New England BioLab, under control of a T7 promotor. Plasmids were linearized with BamHI for run-off transcription by T7 RNA polymerase. All mRNAs were purified via denaturing 15% polyacrylamide gel electrophoresis. Initiation factors IF1, IF2, and IF3 were prepared as described (49).
Effects of E-Site Occupation on Accuracy of Aminoacyl-tRNA Selection.
MFK mRNA, Pi complex (E-site free).
220 pmols of reassociated 70S ribosomes were incubated in the presence of MFK mRNA (molar ratio mRNA:70S = 6:1) and N-Ac[14C]Phe-tRNA (1,139 dpm/pmol; molar ratio to ribosomes 2:1) for P-site binding in the following buffer: 20 mM Hepes (pH 7.6) at 0°C, 4.5 mM Mg(acetate)2, 150 mM NH4(acetate), 4 mM 2-mercaptoethanol, 2 mM spermidine, and 0.05 mM spermine (H20M4.5N150SH4Sd2Spm0.05); total volume 660 μl. After an incubation for 30 min at 37°C, the P-site location was determined via puromycin reaction in a 30-μl aliquot, 95–100% of the bound AcPhe-tRNA was present at the P-site.
MFK mRNA, POST complex (E-site occupied).
The first incubation was conducted in the same way as described above except that [32P]tRNAfMet (specific activity 323–5,000 dpm/pmol exactly determined for each experiment; molar ratio tRNA:70S = 2:1) was used to block the P-site. Ac[14C]Phe-tRNA was added in a second step (molar ratio to ribosomes 2:1), and an incubation followed for 30 min at 37°C. After an EF-G-dependent translocation, a puromycin reaction and a filtration control revealed that 80% of the AcPhe-tRNA was at the P-site, each with a [32P]tRNAfMet at the E-site.
Addition of a mixture of cognate and near-cognate aminoacyl-tRNAs to the A-site and dipeptide analysis.
Both Pi and POST complexes were centrifuged through a 10% sucrose cushion at 44,000 rpm for 18 h at 4°C (TLA-100.3 rotor Beckman) to remove nonbound tRNAs and EF-G, if present. Pelleted Pi and POST complexes were resuspended in binding buffer (H20M4.5N150SH4Sd2Spm0.05), and tRNA binding was controlled by nitrocellulose filtration, demonstrating that none of the tRNAs were lost from the ribosomes. Aliquots of both Pi- and POST-translocational complexes containing 80–90 pmol of ribosomes were mixed with 126 pmol of EF-Tu, 504 pmol of EF-Ts, 400 μM of GTP, and a mixture of cognate [14C]Lys-tRNALys (208 dpm/pmol) and noncognate [3H]Leu-tRNALeu (10,120 dpm/pmol) in a 1.5 molar ratio per ribosome of each tRNA. Enzymatic A-site binding was allowed for 5 min at 37°C. The binding of aminoacyl-tRNAs was controlled by nitrocellulose filtration; the POST-case occupation of the A-site led to a quantitative release of the E-tRNA, as observed (50). Aliquots containing ≈60–80 pmol of 70S ribosomes were subjected to phenolization, and the dipeptides formed were analyzed via HPLC as described (10). Ac[14C]Phe and Ac[14C]Phe-[14C]Lys can be easily distinguished in the HPLC analysis.
Effects of the SD Sequence on the Accuracy of Aminoacyl-tRNA Selection at the A-Site.
Four pmol of ribosomes were incubated at 37°C for 20 min with 40 pmol of MA messenger (with and without a SD sequence), 6.4 pmol of f[3H]Met-tRNAfMet (15 dpm/pmol, just enough to recognize binding), a mixture of three initiation factors (10 pmol each), and 0.5 mM GTP, under the ionic conditions of H20M6N150SH4Spd2Spm0.05. Ternary complexes were formed in 10 μl containing 4 pmol cognate [14C]Ala-tRNAAla (360 dpm/pmol), 4, 20, 40, or 60 pmol of near-cognate [3H]Thr-tRNAThr (8,640 dpm/pmol), 0.5 mM GTP, and two molar excess of EF-Tu per tRNA, under the same ionic conditions, incubated at 37°C for 5 min and added to the ribosome mixture, followed by an incubation of at 37°C for 5 min. The bound tRNAs were assessed via nitrocellulose filtration, and the percentage of error was calculated from the bound aminoacyl-tRNAs per ribosome: [near-cognate Thr-tRNAThr/(cognate Ala-tRNA + near-cognate Thr-tRNA)] × 100.
In the case of MVF mRNA without or with the SD sequence, the f[3H]Met-tRNAfMet was first bound to P-site. The cognate A-site tRNA was [14C]Val-tRNA and noncognate tRNA was [3H]Asp-tRNA. Enzymatic binding was achieved by adding these two aminoacyl-tRNAs, each >1.5-fold over ribosomes, with 1.5-fold EF-Tu per aminoacyl-tRNA and incubating for 5 min at 37°C. The remaining conditions and steps were as described above.
Supplementary Material
Acknowledgments.
We thank Drs. Roland Krause and Oliver Vesper for help and discussions, and Edda Einfeldt for technical assistance. The work was supported by Ministry of Science and Technology of China Project 973 Grant 2006CB910903 (to Y.Q.) and National Natural Science Foundation of China Grant 30770436 (to Y.Q.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801974105/DCSupplemental.
References
- 1.Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 2.Kurland CG, et al. In: The Ribosome- Structure, Function, and Evolution. Dahlberg A, et al., editors. Washington, DC: Am Soc Microbiol; 1990. pp. 513–526. [Google Scholar]
- 3.Szaflarski W, et al. New features of the ribosome and ribosomal inhibitors: Non-enzymatic recycling, misreading and back-translocation. J Mol Biol. 2008;380:193–205. doi: 10.1016/j.jmb.2008.04.060. [DOI] [PubMed] [Google Scholar]
- 4.Nierhaus KH. The allosteric three-site model for the ribosomal elongation cycle: features and future. Biochemistry. 1990;29:4997–5008. doi: 10.1021/bi00473a001. [DOI] [PubMed] [Google Scholar]
- 5.Geigenmüller U, Nierhaus KH. Significance of the third tRNA binding site, the E Site, on E. coli ribosomes for the accuracy of translation: An occupied E site prevents the binding of noncognate aminoacyl-transfer RNA to the A site. EMBO J. 1990;9:4527–4533. doi: 10.1002/j.1460-2075.1990.tb07904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cochella L, Green R. Fidelity in protein synthesis. Curr Biol. 2005;15:R536–R540. doi: 10.1016/j.cub.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 7.Daviter T, Gromadski KB, Rodnina MV. The ribosome's response to codon–anticodon mismatches. Biochimie. 2006;88:1001–1011. doi: 10.1016/j.biochi.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 8.Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 9.Rheinberger H-J, Nierhaus KH. Testing an alternative model for the ribosomal peptide elongation cycle. Proc Natl Acad Sci USA. 1983;80:4213–4217. doi: 10.1073/pnas.80.14.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marquez V, et al. Maintaining the ribosomal reading frame: The influence of the E site during translational regulation of release factor 2. Cell. 2004;118:45–55. doi: 10.1016/j.cell.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 11.Rheinberger H-J, Nierhaus KH. Allosteric interactions between the ribosomal transfer RNA-binding sites A and E. J Biol Chem. 1986;261:9133–9139. [PubMed] [Google Scholar]
- 12.Triana-Alonso FJ, Chakraburtty K, Nierhaus KH. The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. J Biol Chem. 1995;270:20473–20478. doi: 10.1074/jbc.270.35.20473. [DOI] [PubMed] [Google Scholar]
- 13.Dinos G, Kalpaxis DL, Wilson DN, Nierhaus KH. Deacylated tRNA is released from the E site on A site occupation but before GTP is hydrolyzed by EF-Tu. Nucleic Acids Res. 2005;33:5291–5296. doi: 10.1093/nar/gki833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leger M, Dulude D, Steinberg SV, Brakier-Gingras L. The three transfer RNAs occupying the A, P and E sites on the ribosome are involved in viral programmed -1 ribosomal frameshift. Nucleic Acids Res. 2007;35:5581–5592. doi: 10.1093/nar/gkm578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanders CL, Curran JF. Genetic analysis of the E site during RF2 programmed frameshifting. RNA. 2007;13:1483–1491. doi: 10.1261/rna.638707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rheinberger H-J, Nierhaus KH. Adjacent codon–anticodon interactions of both tRNAs present at the ribosomal A and P or P and E sites. FEBS Lett. 1986;204:97–99. doi: 10.1016/0014-5793(86)81393-x. [DOI] [PubMed] [Google Scholar]
- 17.Gnirke A, Geigenmüller U, Rheinberger H-J, Nierhaus KH. The allosteric three-site model for the ribosomal elongation cycle. J Biol Chem. 1989;264:7291–7301. [PubMed] [Google Scholar]
- 18.Jenner L, Rees B, Yusupov M, Yusupova G. Messenger RNA conformations in the ribosomal E site revealed by x-ray crystallography. EMBO Rep. 2007;8:846–850. doi: 10.1038/sj.embor.7401044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schilling-Bartetzko S, Bartetzko A, Nierhaus KH. Kinetic and thermodynamic parameters for transfer RNA binding to the ribosome and for the translocation reaction. J Biol Chem. 1992;267:4703–4712. [PubMed] [Google Scholar]
- 20.Warner JR, Rich A. The number of soluble RNA molecules on reticulocyte polyribosomes. Proc Natl Acad Sci USA. 1964;51:1134–1141. doi: 10.1073/pnas.51.6.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Remme J, Margus T, Villems R, Nierhaus KH. The third ribosomal tRNA-binding site, the E site, is occupied in native polysomes. Eur J Biochem. 1989;183:281–284. doi: 10.1111/j.1432-1033.1989.tb14925.x. [DOI] [PubMed] [Google Scholar]
- 22.Nierhaus KH. Solution of the ribosomal riddle: How the ribosome selects the correct aminoacyl-tRNA out of 41 similar contestants. Mol Microbiol. 1993;9:661–669. doi: 10.1111/j.1365-2958.1993.tb01726.x. [DOI] [PubMed] [Google Scholar]
- 23.Valle M, et al. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J. 2002;21:3557–3567. doi: 10.1093/emboj/cdf326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stark H, et al. Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat Struct Biol. 2002;15:15–20. doi: 10.1038/nsb859. [DOI] [PubMed] [Google Scholar]
- 25.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 26.Tats A, Remm M, Tenson T. Highly expressed proteins have an increased frequency of alanine in the second amino acid position. BMC Genomics. 2006;7:28. doi: 10.1186/1471-2164-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stenstrom CM, et al. Codon bias at the 3′-side of the initiation codon is correlated with translation initiation efficiency in Escherichia coli. Gene. 2001;263:273–284. doi: 10.1016/s0378-1119(00)00550-3. [DOI] [PubMed] [Google Scholar]
- 28.Varshavsky A. The N-end rule: Functions, mysteries, use. Proc Natl Acad Sci USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solbiati J, et al. Processing of the N termini of nascent polypeptide chains requires deformylation prior to methionine removal. J Mol Biol. 1999;290:607–614. doi: 10.1006/jmbi.1999.2913. [DOI] [PubMed] [Google Scholar]
- 30.Bartetzko A, Nierhaus KH. A simple Mg2+/NH4+/polyamine system for poly(U) dependent poly(Phe) synthesis with near in vivo characteristics. Methods Enzymol. 1988;164:650–658. doi: 10.1016/s0076-6879(88)64075-4. [DOI] [PubMed] [Google Scholar]
- 31.Robert F, Brakier-Gingras L. A functional interaction between ribosomal proteins S7 and S11 within the bacterial ribosome. J Biol Chem. 2003;278:44913–44920. doi: 10.1074/jbc.M306534200. [DOI] [PubMed] [Google Scholar]
- 32.Trimble MJ, Minnicus A, Williams KP. tRNA slippage at the tmRNA resume codon. RNA. 2004;10:805–812. doi: 10.1261/rna.7010904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson DN, Nierhaus KH. The E-site Story: The importance of maintaining two tRNAs on the ribosome during protein synthesis. Cell Mol Life Sci. 2006;63:2725–2737. doi: 10.1007/s00018-006-6125-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersen BF, et al. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature. 2006;433:663–668. doi: 10.1038/nature05126. [DOI] [PubMed] [Google Scholar]
- 35.Korostelev A, et al. Interactions and dynamics of the Shine Dalgarno helix in the 70S ribosome. Proc Natl Acad Sci USA. 2007;104:16840–16843. doi: 10.1073/pnas.0707850104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaminishi T, et al. A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine–Dalgarno interaction. Structure (London) 2007;15:289–297. doi: 10.1016/j.str.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 37.Yusupova G, et al. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 38.de Smit MH, van Duin J. Translational standby sites: How ribosomes may deal with the rapid folding kinetics of mRNA. J Mol Biol. 2003;331:737–743. doi: 10.1016/s0022-2836(03)00809-x. [DOI] [PubMed] [Google Scholar]
- 39.Gualerzi CO, et al. Initiation factors in the early events of mRNA translation in bacteria. Cold Spring Harb Symp Quant Biol. 2001;66:363–376. doi: 10.1101/sqb.2001.66.363. [DOI] [PubMed] [Google Scholar]
- 40.Chang B, Halgamuge S, Tang SL. Analysis of SD sequences in completed microbial genomes: Non-SD-led genes are as common as SD-led genes. Gene. 2006;373:90–99. doi: 10.1016/j.gene.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 41.Ma J, Campbell A, Karlin S. Correlations between Shine–Dalgarno sequences and gene features such as predicted expression levels and operon structures. J Bacteriol. 2002;184:5733–5745. doi: 10.1128/JB.184.20.5733-5745.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vimberg V, Tats A, Remm M, Tenson T. Translation initiation region sequence preferences in Escherichia coli. BMC Mol Biol. 2007;8:100. doi: 10.1186/1471-2199-8-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pestova TV, et al. Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Londei P. Evolution of translational initiation: New insights from the archaea. FEMS Microbiol Rev. 2005;29:185–200. doi: 10.1016/j.femsre.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 46.Moll I, Grill S, Gualerzi CO, Blasi U. Leaderless mRNAs in bacteria: Surprises in ribosomal recruitment and translational control. Mol Microbiol. 2002;43:239–246. doi: 10.1046/j.1365-2958.2002.02739.x. [DOI] [PubMed] [Google Scholar]
- 47.Triana-Alonso FJ, Dabrowski M, Wadzack J, Nierhaus KH. Self-coded 3′-extension of run-off transcripts produces aberrant products during in vitro transcription with T7 RNA polymerase. J Biol Chem. 1995;270:6298–6307. doi: 10.1074/jbc.270.11.6298. [DOI] [PubMed] [Google Scholar]
- 48.Schäfer MA, et al. Codon–anticodon interaction at the P Site is a prerequisite for tRNA interaction with the small ribosomal subunit. J Biol Chem. 2002;277:19095–19105. doi: 10.1074/jbc.M108902200. [DOI] [PubMed] [Google Scholar]
- 49.Pawlik R, Littlechild J, Pon C, Gualerzi C. Purification and properties of Escherichia coli translational initiation factors. Biochem Intl. 1981;2:421–428. [Google Scholar]
- 50.Dinos G, et al. Dissecting the ribosomal inhibition mechanisms of edeine and pactamycin: The universally conserved residues G693 and C795 regulate P-site tRNA binding. Mol Cell. 2004;13:113–124. doi: 10.1016/s1097-2765(04)00002-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.