To identify a rare (e.g., diseased or foreign) cell in a complex mixture, or to understand the proteomic complexity[1,2] of cells, one needs to be able to measure simultaneously and quantitatively a large number of proteins or other biomarkers that may be present in a complex sample. This is a difficult task and is beyond the reach of current capabilities. To address a problem of this complexity, we have begun to develop a high-sensitivity assay[3–6] based upon elemental tags that will enable the simultaneous measurement of many proteins in a single sample. The advantage of this approach lies in the large number of available elements and isotopes (potentially greater than 79) found in low abundance in biological systems, which will allow multiple tags to be used simultaneously. Inductively coupled plasma mass spectrometry (ICP-MS) is an ideal technique for detecting and quantifying these tags, as ICP-MS provides excellent resolution between the tag masses and an exceptional dynamic range (nine orders of magnitude).[7] This method allows one to overcome some of the limitations of currently available fluorescent tagging approaches.[8] These limitations arise from the spectral overlap of different dyes and the difficulty in measuring simultaneously targets that differ in abundance by an order of magnitude or more. Other benefits of ICP-MS detection include the high sensitivity, which is comparable to that of radioimmunoassays or chemiluminescent assays,[3] insensitivity of elemental tags to photobleaching and storage time, as well as the stability of the tagged sample so that it can be stored or shipped for analysis. We discuss herein the development of a new class of elemental tags for ICP-MS detection and their use for tagging of antibodies chosen to allow specific recognition of distinguishing cell surface markers. By using this technique it should be possible to achieve detection limits on the order of parts per billion, which will allow the detection of cell surface markers with copy numbers as low as 100.
Our experimental design is presented in Figure 1. The assay is based upon the concept of a water-soluble polymer bearing multiple metal-chelating ligands. The polymer contains a terminal maleimide group for coupling to cysteine -SH groups on the Fc portion of an antibody. It is now well established that attaching tags to antibodies through -SH groups (generated by selective reduction of disulfide bonds) is much more likely to preserve antibody activity than, for example, the random covalent attachment of tags to the amino group of lysines. The chelating ligand is chosen to form high-affinity complexes with lanthanide (Ln3+) ions. These elements satisfy our requirement for low natural abundance and a wide selection of elements and isotopes. The use of a metal-chelating polymeric tag allows us to incorporate multiple numbers of a given ion, which leads to an increase in the sensitivity of the method, since the ICP-MS signal increases linearly with the number of atoms of a given element. Another important feature of our design is that the same polymer can be attached to a variety of different antibodies. Prior to the assay, each type of antibody can be treated with a different lanthanide ion. In this way we can create a family of element-labeled antibodies that can, in principle, be analyzed simultaneously in a single assay. Herein we describe our first success at implementing this approach.
Figure 1.
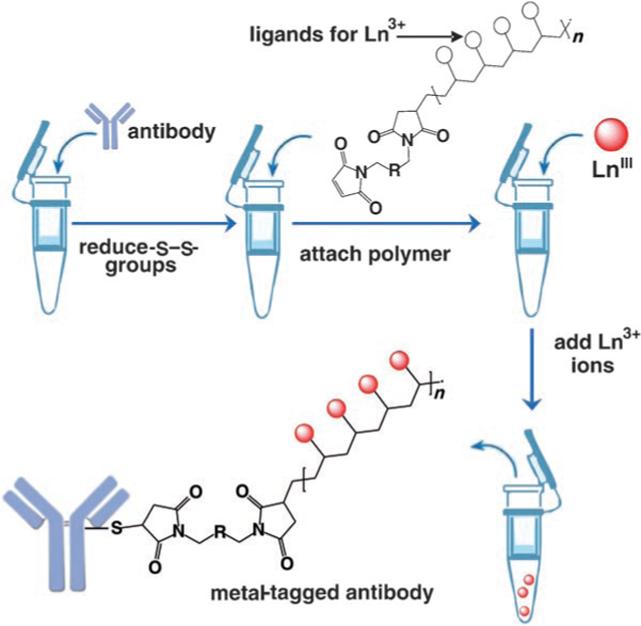
Experimental design for tagging antibodies with metal-chelating polymers. The antibody of interest is subjected to selective reduction of -S—S-groups to produce reactive -SH groups, which are reacted with the terminal maleimide groups of a polymer bearing metal-chelating ligands along its backbone. The polymer-bearing antibodies are purified, treated with a given lanthanide ion, and then purified again. Each type of antibody is labeled with a different element.
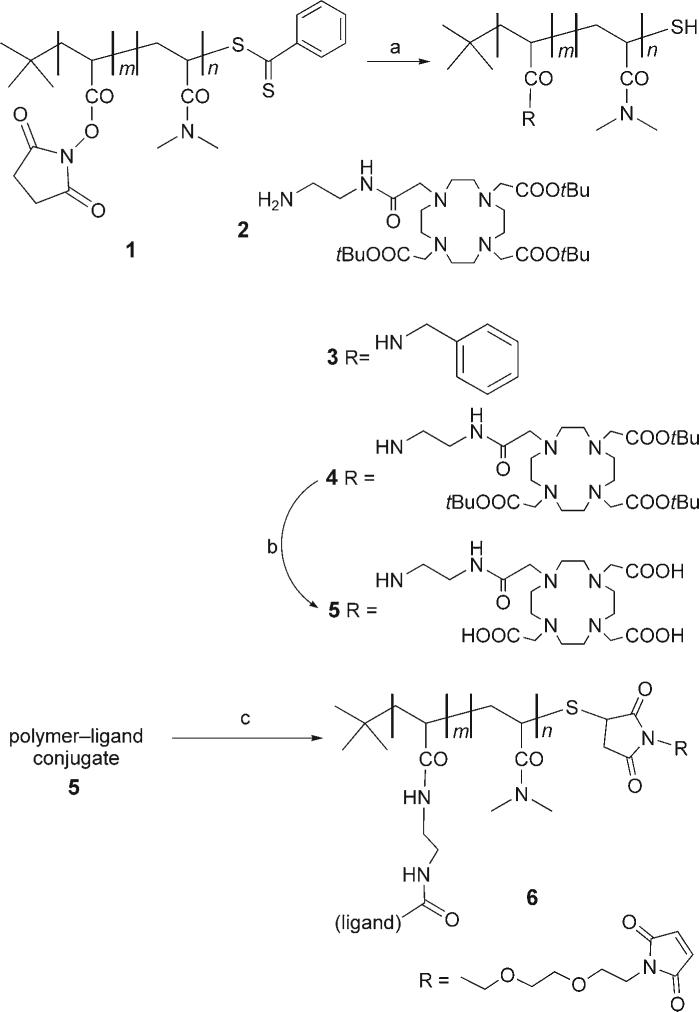
The ligand-functionalized polymer was synthesized as shown in Scheme 1 (for experimental details, see the Supporting Information). We began with the synthesis of an N,N-dimethylacrylamide (DMA) and N-acryloxysuccinimide (NAS) random copolymer (1) by reversible addition-fragmentation chain transfer (RAFT) polymerization following a published procedure.[9] Our reaction employed 2,2′-azobis(2-methylbutyronitrile) as the initiator, and tert-butyldithiobenzoate as the RAFT agent. In this way we obtained polymers with a tert-butyl group at one end and a dithiobenzoate group at the other terminus. Both groups were useful for characterizing the polymer by 1H NMR measurements.
Scheme 1.
a) Et3N, DMF, benzylamine or 2, 14 h; b) TFA 95 %, 14 h; c) 1) DTT (20 mm), Na2HPO4 (50 mm, pH 8.5), 50 °C, 1 h; 2) 2,2′-(ethylenedioxy)bis(ethylmaleimide), DMF/H2O, 1 h, 22 °C.
Random poly(DMA-co-NAS) with a target content of 60 mol % NAS was synthesized in N,N-dimethylformamide (DMF) at 60 °C. The molecular weight (Mn) and the polydispersity index (Mw/Mn) were measured by gel-permeation chromatography (0.2 wt % LiCl in N-methylpyrrolidone at 80 °C) with polystyrene standards for the calibration curve. The apparent number-average molecular weight (Mn) for copolymer 1 was 8000 g mol−1 with a narrow polydispersity (Mw/Mn = 1.15). To obtain absolute Mn values, the polymer was characterized by 1H NMR spectroscopy in CDCl3. The peakat δ = 0.9 ppm can be assigned to the protons on the tert-butyl end group. The broad peakat δ = 1.2−2.2 ppm can be assigned to the protons of the methylene groups on copolymer main chain. Integration of these two peaks allows us to calculate the degree of polymerization (DP) of the copolymers. The DP for 1 was 52, which corresponds to Mn = 7500. The composition of NAS units was obtained by treating 1 with excess benzylamine to give 3, which was characterized by 1H NMR spectroscopy in CD2Cl2. The purified polymer 3 showed a similar peak to that of starting polymer 1 corresponding the tert-butyl end group at δ = 0.9 ppm. The broad peaks at δ = 6.9−7.6 and δ = 3.8−4.8 ppm are due to protons from the phenyl and CH2 units, respectively, of the aminobenzyl groups. The composition of the polymer can be determined by comparing the integration of these two peaks with that of the tert-butyl end group. Assuming that each NAS unit was converted into a benzylamide group, we find 63 mol % NAS units, which corresponds to 33 units per chain.
As the metal-chelating ligand, we chose 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA). This ligand has a high affinity for the lanthanide metals and a low exchange rate.[10] We synthesized a DOTA derivative (2) functionalized with a pendant primary amine by following literature procedures.[11] The DOTA ligand was condensed with the activated NHS ester groups along the polymer backbone. Under the reaction conditions, the terminal dithioester derivative was cleaved to the primary thiol, as shown in Scheme 1. Conjugate 4 was treated with trifluoroacetic acid (TFA) to remove the protecting groups to give the polymer 5. The polymer was treated with a reducing buffer containing DL-dithiothreitol (DTT) to reduce any disulfide bonds formed during these manipulations. The reaction mixture was adjusted to pH 8.5 and treated with bismaleimide to afford maleimide-functionalized polymer–ligand conjugate 6.
The sharp singlet at 6.95 ppm in the 1H NMR spectrum of 6 (Figure 2) was assigned to the vinylogous protons of the maleimide group. This assignment was confirmed by treating the solution in the NMR tube with a small excess of 2-aminoethanethiol, which led to the disappearance of this signal. Furthermore, the ratio of integration of this peak relative to that of the tert-butyl protons at δ = 0.90 ppm derived from the initiator is 2:9, which suggests essentially quantitative recovery of thiol end groups on the polymer, which were in turn completely converted into the maleimide functionality. This is an important result, not only for the development of our assay; other research groups have been interested in quantifying the thiol end group content of polymers synthesized by RAFT. Many research groups have reported less than full recovery of thiol functionality following hydrolysis of the RAFT end group. Part of the problem may have been the assay used to detect the -SH groups.[12–14]
Figure 2.
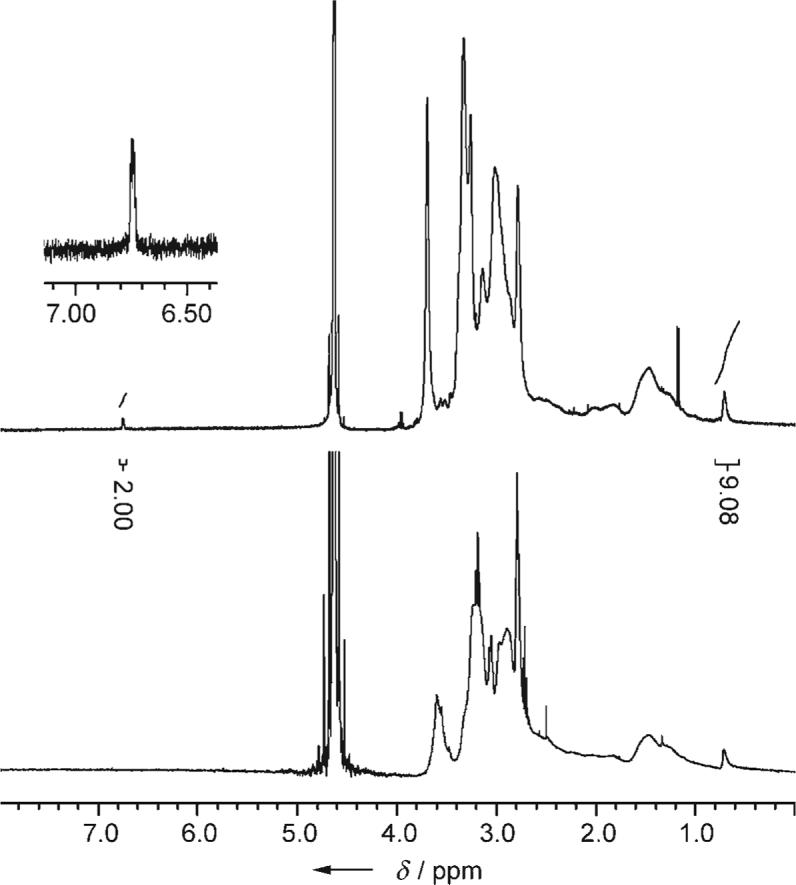
1H NMR spectrum of maleimide conjugate 6 in D2O prior to (top) and after (bottom) reaction with 2-aminoethanethiol.
Antibodies were labeled with the DOTA-containing polymer tag 6 through free cysteine residues generated by partial reduction of the antibody, as depicted in Figure 1. The antibodies were reduced, washed in a centrifugal concentrator, and then a 10-fold excess of polymer tag was added, and the mixture was incubated at 37 °C for 1 hour. The antibody–tag conjugate was subsequently washed and combined with a solution (μm concentration) of the desired lanthanide chloride.
The potential of our antibody polymer conjugates was first evaluated with a europium-labeled mouse antibody against the CD45 antigen. A mouse IgG labeled with an element tag in the same way was generated to be used as a negative control (IgG-Eu). The specificity measurements and titrations of elemental-tagged antibody CD45-Eu (0.7 mg mL−1) were performed on KG-1a cells. CD45 is one of the more abundant antigens expressed on these mononuclear cells. CD45-Eu was washed, serially diluted twofold (starting at 1:25), and then added to the live cell suspension. The cells were incubated and then washed several times by low-speed centrifugation, and the cellular pellets were dissolved in ultrapure concentrated HCl. An equal volume of Ir (1 ppb) was added to each tube as an internal standard, and the solution was analyzed by ICP-MS. The results are presented in Figure 3 as the normalized response, whereby the measured isotope intensities are divided by the corresponding intensity of the Ir reference. The binding of CD45-Eu to KG-1a cells follows a saturation curve, whereas the nonspecific binding of IgG-Eu displays a linear dependence. There is at least two orders of magnitude difference between the specific antibody binding and the nonspecific IgG binding. By using an element-tagged antibody, we obtained a 100−200-fold increase of signal over the nonspecific IgG control at non-saturating antibody concentrations.
Figure 3.
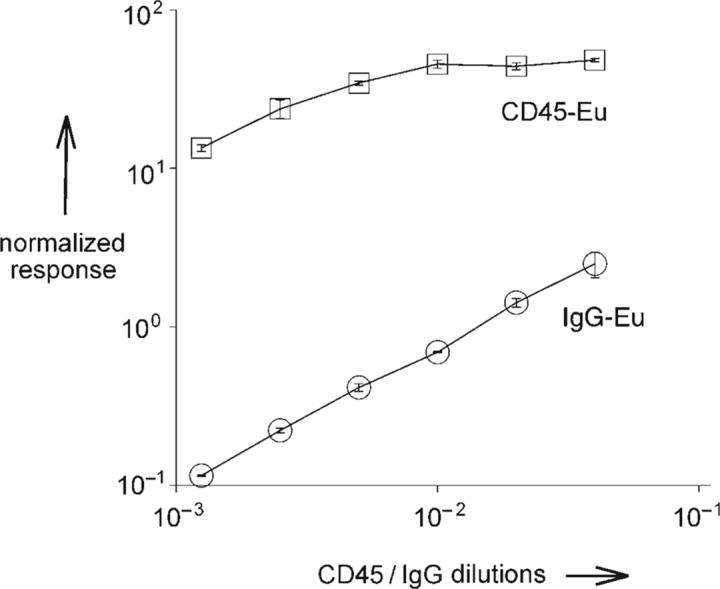
Titration of element-tagged antibody against cell surface antigen. KG-1a cells (1 × 106 cells per sample, run in triplicate) were incubated with increasing concentrations of CD45-Eu antibody. Separately, the same number of KG-1a cells were treated with mouse IgG-Eu.
Next, the potential of this tagging method in a multiplexed assay was evaluated. Monoclonal antibodies to leukemia cell surface markers were labeled with five different lanthanide elements according to the protocol described above: CD33-Pr, CD34-Tb, CD38-Ho, CD45-Eu, and CD54-Tm. Two cell lines representing myeloid (KG-1a) and monocytic (THP-1) acute leukemia were compared. Each cell line has its own characteristic level of marker expression.[12,13] An equal number of cells were distributed into triplicate tubes for each antibody separately, and one set of tubes was prepared for a mix of all of the antibodies. The same number of cell samples was set up for nonspecific binding of element-tagged mouse IgG prepared similarly to the specific antibodies: IgG-Pr, IgG-Tb, IgG-Ho, IgG-Eu, and IgG-Tm. Cells were treated with tagged antibodies, and the washed cells were fixed in a 3.7 % solution of formaldehyde in phosphate-buffered saline and stained with a Rh3+-containing DNA metallointercalator[15] for cell enumeration and signal normalization. The ICP-MS results are shown in Figure 4. One important result is that in both cases, the signals obtained using a single antibody were very similar to those obtained with all five antibodies mixed together. This result demonstrates that there is no signal interference between detection channels in the mixed samples. More specifically, this experiment establishes that the metal ions do not dissociate and reassociate with DOTA ligands on other polymer chains during the timescale of the analysis. The results also indicate that the two cell types differ dramatically in the expression of CD33 (THP-1 is 500-fold higher than KG-1a) and CD34 (KG-1a is 100-fold higher than THP-1). This difference in level of expression is characteristic of these cell lines. The signal-to-noise ratio for the CD33 antigen is above 2, which indicates an accurate measurement of the low CD33 signal. We were able in a single assay to obtain quantitative information about two different protein markers that differ by a factor of 500 in degree of expression. Both cell lines express CD45 and CD54 antigens at comparable levels, whereas THP-1, as a more differentiated cell type, shows 10-fold higher CD38 expression than KG-1a, a primitive hematopoietic progenitor cell.
Figure 4.
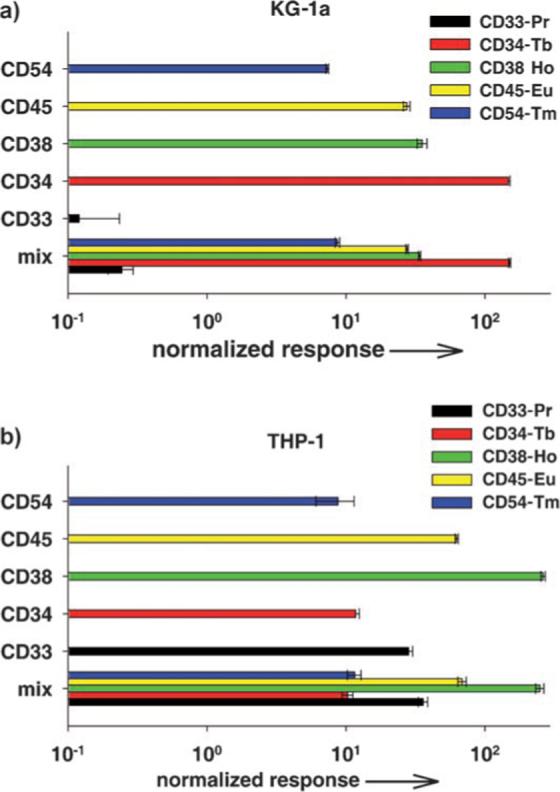
Multiplex analysis of antigen expression by two acute leukemia cell lines. a) KG-1a cells were probed with five element-tagged antibodies to cell surface antigens: CD33-Pr, CD34-Tb, CD38-Ho, CD45-Eu, and CD54-Tm. Background controls included element-tagged mouse IgG-Pr, IgG-Tb, IgG-Ho, IgG-Eu, and IgG-Tm. Triplicate samples with 1 × 106 cells per tube were set up for reaction with each antibody separately and with a mix of all five antibodies together as well as controls. The cells were stained, fixed, and then treated with a RhIII-containing metallointercalator for cell enumeration and signal normalization. Washed cell pellets were dissolved in concentrated HCl, combined with an equal volume of Ir standard solution (1 ppb), and analyzed by ICP-MS. Results are presented as normalized response with respect to Ir, Rh, and background signals from nonspecific IgG binding. b) THP-1 cell line treated as described for KG-1a.
In conclusion, we have developed a novel elemental-tagging procedure that, in conjunction with ICP-MS analysis, allows the multiplexed detection of proteins on cells. The use of polymer-based elemental tags offers the important advantage that each tag carries many copies of a given element, which greatly increases the assay sensitivity. Because of this sensitivity, we were able to detect in a single assay two different cell surface markers (CD33 and CD34 in KG-1a) that differed by a factor of around 500 in their abundance. Our simultaneous assay of five cell surface markers opens the door for larger multiplex analyses that will allow “fingerprint” detection of individual types of cell lines. This approach appears to have many advantages over conventional fluorescence-based detection.
Supplementary Material
Footnotes
This project was funded in part by Genome Canada through the Ontario Genomics Institute and Ontario Cancer Research Network and NIH grant #GM076127−01A1
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Robert Kinach, Institute of Biomaterials and Biomedical Engineering University of Toronto 164 College Street, Room 407, Toronto, ON M5S 3G9 (Canada) Fax: (+ 1) 416−978−4317.
Olga Ornatsky, Institute of Biomaterials and Biomedical Engineering University of Toronto 164 College Street, Room 407, Toronto, ON M5S 3G9 (Canada) Fax: (+ 1) 416−978−4317.
Vladimir Baranov, Institute of Biomaterials and Biomedical Engineering University of Toronto 164 College Street, Room 407, Toronto, ON M5S 3G9 (Canada) Fax: (+ 1) 416−978−4317 E-mail: vladimir.baranov@utoronto.ca.
Mark Nitz, Department of Chemistry University of Toronto 80 St. George St., Toronto ON M5S 3H6 (Canada) Fax: (+ 1) 416−978−0541 E-mail: mnitz@chem.utoronto.ca
Mitchell A. Winnik, Department of Chemistry University of Toronto 80 St. George St., Toronto ON M5S 3H6 (Canada) Fax: (+ 1) 416−978−0541 E-mail: mwinnik@chem.utoronto.ca
References
- 1.Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, Radich J, Anderson G, Hartwell L. Nat. Rev. Cancer. 2003;3:243. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 2.Melton L. Nature. 2004;429:101. doi: 10.1038/429101a. [DOI] [PubMed] [Google Scholar]
- 3.Baranov VI, Quinn Z, Bandura DR, Tanner SD. Anal. Chem. 2002;74:1629. doi: 10.1021/ac0110350. [DOI] [PubMed] [Google Scholar]
- 4.Baranov VI, Quinn ZA, Bandura DR, Tanner SD. J. Anal. At. Spectrom. 2002;17:1148. [Google Scholar]
- 5.Ornatsky O, Baranov VI, Bandura DR, Tanner SD, Dick J. J. Immunol. Methods. 2006;308:68. doi: 10.1016/j.jim.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 6.Quinn ZA, Baranov VI, Tanner SD, Wrana JL. J. Anal. At. Spectrom. 2002;17:892. [Google Scholar]
- 7.Tanner SD, Baranov VI, Bandura DR. Spectrochim. Acta Part B. 2002;57:1361. [Google Scholar]
- 8.Perfetto SP, Chattopadhyay PK, Roederer M. Nat. Rev. Immunol. 2004;4:648. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- 9.Relogio P, Charreyre M-T, Farinha JPS, Martinho JMG, Pichot C. Polymer. 2004;45:8639. [Google Scholar]
- 10.Parker D, Dickins RS, Puschmann H, Crossland C, Howard JAK. Chem. Rev. 2002;102:1977. doi: 10.1021/cr010452+. [DOI] [PubMed] [Google Scholar]
- 11.André JP, Geraldes CFGC, Martins JA, Merbach AE, Prata MIM, Santos AC, Lima J. J. P. d., Tóth É. Chem. Eur. J. 2004;10:5804. doi: 10.1002/chem.200400187. [DOI] [PubMed] [Google Scholar]
- 12.Nakayama M, Okano T. Biomacromolecules. 2005;6:2320. doi: 10.1021/bm050232w. [DOI] [PubMed] [Google Scholar]
- 13.Scales CW, Convertine AJ, McCormick CL. Biomacromolecules. 2006;7:1389. doi: 10.1021/bm060192b. [DOI] [PubMed] [Google Scholar]
- 14.Schilli CM, Müller AHE, Rizzardo E, Thang SH, Chong YK. In: Controlled/LivingRadical Polymerization. Matyjaszewski K, editor. American Chemical Society; Washington, D.C.: 2003. p. 603. [Google Scholar]
- 15.Barton JK. Pure Appl. Chem. 1998;70:873. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.