Abstract
The peroxisome proliferator-activated receptor γ (PPARγ) is a nuclear transcription factor that controls the genes involved in metabolism and carcinogenesis. In the present study, we examined the alteration of gene expression in HCT-116 human colorectal cancer cells by PPARγ agonists: MCC-555 (5 µM), rosiglitazone (5 µM), and 15-deoxy-Δ12,14-prostaglandin J2 (1 µM). The long-oligo microarray data revealed a list of target genes commonly induced (307 genes) and repressed (32 genes) by tested PPARγ agonists. These genes were analyzed by Onto-Express software and KEGG pathway analysis and revealed that PPARγ agonists are involved in cell proliferation, focal adhesion, and several signaling pathways. Eight genes were selected to confirm the microarray data by RT-PCR and real-time PCR, from which CSTA, DAP13, TAF12, RIS1, CDKN3 and MAGOH were up-regulated, and KLHL11 and NCOA2 were down-regulated. This study elucidates the commonly induced genes modulated by tested PPARγ ligands involved in the different signaling pathways and metabolisms, probably mediated in a PPARγ-dependent manner in colorectal cancer cells and helps to better understand the pleiotropic actions of PPARγ ligands.
Keywords: MCC-555, rosiglitazone, colon cancer, PPAR, micro-array
Introduction
The peroxisome proliferator-activated receptors (PPARs) are members of the nuclear hormone receptor family and are activated by agonists that directly bind transcription factors to regulate the target genes (1). At present, three types of PPARs have been identified (PPARα, PPARβ/δ and PPARγ), each with different functions, tissue distributions and activations. Among them, PPARγ is highly expressed in adipose and colon tissue, and marginally expressed in the heart, skeletal muscle and liver tissues (2). The PPARγ activation by agonists regulates adipocyte differentiation and promotes lipid storage in mature adipocytes. The PPARγ agonists can also affect cell proliferation, differentiation, and apoptosis in a PPARγ-dependent and/or independent manner and thereby represent a potentially important therapeutic approach to cancer treatments (1). Thus, many studies describe beneficial effects of the PPARγ agonists for treatment of different types of cancer (3–7), including colorectal carcinoma (8,9).
Agonists of PPARγ include prostaglandins of the J series (PGJ2); the synthetic antidiabetic thiazolidinediones, such as troglitazone (TGZ), rosiglitazone (RGZ), and MCC-555; and oxidative metabolites of polyunsaturated fatty acids. Both TGZ and PGJ2 affect several pathways in a PPARγ-independent manner: TGZ induces the p53 pathway (10), inhibits cholesterol biosynthesis (11), inhibits translation initiation (12), and promotes antioxidant function (13), whereas PGJ2 induces apoptosis (14) and affects signaling pathways including Erk1/2 and NF-κB (15). We have also recently demonstrated that TGZ induces the early growth response gene (EGR-1) independently of PPARγ transcription factor activation (8) and that MCC-555 induces apoptosis in HCT-116 cells, independently of PPARγ (16). Thus, the anti-cancer activity of the PPARγ agonists should be explored both dependently and independently at the levels of transcription in several cancer types.
The objective of this study was to investigate the pattern of genes commonly modulated by three PPARγ agonists (MCC-555, RGZ and PGJ2) in human HCT-116 colorectal cancer cells. The long-oligo microarray was employed and the data revealed a list of target genes commonly induced by tested PPARγ agonists. The pattern of altered genes was evaluated by cluster analysis, and the commonly regulated genes were analyzed by ontology and signaling pathway analysis, to better understand the biological profiles involved in PPARγ dependency. In addition, common genes involved in up-regulation (307 genes) and down-regulation (32 genes) by PPARγ agonists were analyzed and were shown to play important roles in cell proliferation, apoptosis, cell adhesion, energy homeostasis, insulin metabolism and other signaling pathways. Eight genes were selected for further analysis, of which 6 displayed higher mRNA expression in PPARγ agonist-treated HCT-116 cells than in untreated control: (DAP13) 13 kDa differentiation-associated protein cystatin A (CSTA), NADH dehydrogenase 1 α subcomplex 12, 20 kDa TATA box binding protein (TBP)-associated factor (TAF12), Ras-induced senescence 1 (RIS1), cyclin-dependent kinase inhibitor 3 (CDKN3) and mago-nashi homolog (Drosophila) proliferation-associated (MAGOH). We also determined that two genes displayed down-regulation of mRNA expression: kelch-like 11 (KLHL-11) and nuclear receptor coactivator 2 (NCOA2). This study identified several human genes that were commonly expressed by PPARγ agonists in human colorectal cancer cells, and this report describes their possible cellular and physiological roles in PPARγ-dependent tumor growth.
Materials and methods
Cell lines
Human colorectal adenocarcinoma cell line HCT-116 was obtained from American Type Culture Collection (ATCC, Manassas, VA). The cells were maintained in McCoy's 5A modified medium supplemented with 10% fetal bovine serum, 100 IU penicillin, and 100 µg/ml streptomycin and grown in an atmosphere of 5% CO2 at 37°C.
Materials
5-[[6-[(2-fluorophenyl)-methoxy]-2-napthalenyl] methyl]-2,4-thiazolidinedione (MCC-555), rosiglitazone (RGZ) and 15-deoxy-Δ12,14-prostaglandin J2 (PGJ2) were purchased from Cayman Chemical Co. (An Arbor, MI). All other chemicals and reagents were purchased from Fischer Scientific, unless otherwise specified.
Plasmids, transient transfections and luciferase assay
The tk-PPREx3-Luc reporter plasmid, a PPAR-dependent luciferase construct, was generously provided by Dr Ron M. Evans (Salk Institute, La Jolla, CA). All transfection experiments were performed using Lipofectamine reagent (Invitrogen Corp., Carlsbad, CA) according to the manufacturer's instructions. Briefly, HCT-116 cells were seeded 2 ×105 cells per well in 12-well tissue culture plates in complete media overnight. Then the cells were co-transfected with 0.5 µg tk-PPREx3-Luc reporter plasmid and 0.05 µg pRL-null plasmid. After 5-h transfection, the media were replaced with complete media overnight, and cells were treated with different PPARγ agonists (5 µM MCC-555, 5 µM RGZ, or 1 µM PGJ2) for an additional 24 h. The luciferase assay was performed using the Dual-Glo luciferase assay kit (Promega Corp., Madison, WI) according to the manufacturer's protocol. Produced luminescence was measured by a luminometer (TD-20/20, Turner Designs, Sunnyvale, CA) and data were analyzed by the Student's t-test. The results at *p<0.05, **p<0.01 and ***p<0.001 were considered statistically significant.
RNA isolation for microarray analysis
HCT-116 cells were grown in 100-mm dishes (2×106) in complete media overnight, and then washed twice with PBS and treated with 5 µM MCC-555, 5 µM RGZ, or 1 µM PGJ2 in serum-free media for an additional 24 h. Total RNA was isolated from treated cell samples using an RNeasy mini kit (Qiagen, Valencia, CA).
Microarray labeling and hybridization
HEEBO long-oligo microarray slides were purchased from Microarray Inc. (Nashville, TN) based on a probe set designed by Illumina (San Diego, http://www.illumina.com) and Stanford University. The HEEBO long-oligo set contains about 39,000 probes for exonic regions and alternative spliced transcripts. Total RNA (1 µg) was labeled for each sample with a SuperScript III Labeling Kit (Invitrogen) according to the manufacturer's instructions. The purified probes from both control and treated samples were mixed and hybridized with the long-oligo microarray using a microarray hybridization kit (Corning, Inc., Corning, NY) on a MAUI hybridization station (Bio Micro Systems Inc., Salt Lake City, UT) according to the manufacturer's instruction. Dye swap experiments were included to eliminate dye-specific bias. For each sample set of treated vs. control, the treated mRNA was first labeled with Cy5 and the control with Cy3. In the reverse experiment, the labeling dyes were swapped. The two labeling reactions and microarray hybridizations were performed in parallel. Considering the reverse labeling experiments, a total of two biological replicates and two technical replicates of the experiment were performed.
Microarray data processing
After hybridization, the microarray slides were washed and scanned in a GenePix 4000 scanner (Axon Instrument, Union City, CA), and the images were processed by GenePix Pro 4.0 software (Axon Instrument). The resultant file was analyzed with Bioconductor (http://www.bioconductor.org), where local background subtraction and lowess normalization were carried out for each microarray slide. The biological replicates and technical replicates were considered differently in the analysis. Linear models from the limma package of Bioconductor were applied to derive a p-value and average of logarithm 2-based ratio. Changes in gene expression pattern were considered statistically significant at *p<0.05. A ratio cut-off of 1.5 and degree of freedom higher or equal to two (represents the changes of gene expression on at least three slide replicates of same experiment) were included as quality controls. The ratio cut-off of 1.5 in gene expression was selected because the action of PPARγ agonists are diverse in gene profiles; so to obtain the list of genes that are commonly down-regulated, we had to decrease the cut-off to a 1.5-fold instead of a 2-fold change as is commonly adopted for microarray data analysis.
In order to evaluate the microarray quality, the M vs. A plot was generated as a scatter plot of log intensity ratios M = log2 (R/G) vs. average log intensities A = log2 (RxG)/2, where R and G represent the fluorescence intensities in the red (R) and green (G) channels, respectively. Scatter plots were generated to compare the signal strength between slides to evaluate the reproducibility of the experiments. Hierarchical clustering was carried out to cluster among the genes and among the samples with Mev4.0 software from TIGR (The Institute of Genome Research). For the gene ontology analysis and identification of signaling pathways, we used Onto-Express software (Onto-Express and Pathway analysis, Intelligent Systems and Bioinformatics Laboratory, Detroit, MI).
Reverse transcription PCR
HCT-116 cells were grown in 60-mm dishes (1×106) in complete media overnight, and then washed twice with PBS and treated with MCC-555 (5, 10 and 50 µM), RGZ (5, 10 and 50 µM), or PGJ2 (1, 2 and 10 µM) in serum-free media for an additional 24 h. Total RNA was isolated from treated cell samples using an RNeasy mini kit (Qiagen), and then cDNA was synthesized from 1 µg of total RNA using an iScript cDNA Synthesis kit (Bio-Rad, Hercules, CA) according to the manufacturer's protocols. Synthesized cDNA (1 µg) was then added to a 20 µl PCR reaction mixture (GoTaq Green Master Mix PCR Reaction Mixture, Promega, Madison, WI) with each set of gene-specific primers. The primer sequences are described in Table I in detail. The final PCR products were loaded to agarose gels and photographed under UV light. The experiments were repeated in three independent replicates. The densitometries of the captured images were evaluated by Scion Image Software (Frederick, MD), and were shown on the final chart with mean ± SE values for each treatment. The Student's unpaired t-test was used to analyze the statistical significance.
Table I.
Primer sequences used for RT-PCR.
| Gene | Forward (F) and reverse (R) primers |
PCR product size (bp) |
|---|---|---|
| CSTA | F: 5′-ggcttatctgaggccaaacc-3′ | 217 |
| R: 5′-tttgtccgggaagacttttg-3′ | ||
| DAP13 | F: 5′-gcgaggcaagatggagttag-3′ | 248 |
| R: 5′-ttccatccacatcccagaat-3′ | ||
| TAF12 | F: 5′-gcaagtctagcaccctggag-3′ | 201 |
| R: 5′-attgctgtccattccctgac-3′ | ||
| RIS1 | F: 5′-cttccagttccgaaaagcag-3′ | 165 |
| R: 5′-ttggtcattttctgccatga-3′ | ||
| CDKN3 | F: 5′-catagccagctgctgtgaaa-3′ | 190 |
| R: 5′-cccggatcctcttaggtctc-3′ | ||
| SEC61B | F: 5′-tgttccagtattggttatgagtcttctg-3′ | 75 |
| R: 5′-cgagtgtacttgccccaaatg-3′ | ||
| MAGOH | F: 5′-ccaaagaggatgatgcattgtg-3′ | 77 |
| R: 5′-ttcatctccaatgacgatttcaag-3′ | ||
| KLHL-11 | F: 5′-tcggaaagaagcctaccgatatt-3′ | 80 |
| R: 5′-ctctacaacgaggttgtggcatag-3′ | ||
| NCOA2 | F: 5′-aatgcatcagcaacagcaag-3′ | 225 |
| R: 5′-ataagtgggctctggggagt-3′ | ||
| TBP | F: 5′-gcccgaaacgccgaatat-3′ | 73 |
| R: 5′-ccgtggttcgtggctctct-3′ | ||
| GAPDH | F: 5′-gaccacagtccatgccatcact-3′ | 560 |
| R: 5′-tccaccaccctgttgctgtag-3′ |
Quantitative real-time-PCR
Complementary DNA samples were diluted into 10 ng/µl, 0.1 ng/µl and 0.01 ng/µl concentration series. Three replicates of real-time PCR experiments were performed for each concentration using an ABI 7000 Sequence Detection System from AB Applied Biosystems (Foster City, CA) with the ABsolute QPCR SYBR-Green mix (ABgene House, Epsom, UK). The primers for target genes were designed by Primer Express software (AB Applied Biosystems) and the primers sequences are listed in Table I. The gene for TATA binding protein (TBP) was used as an internal control. After the real-time PCR experiments, Ct numbers were extracted for both reference genes and target genes with auto baseline and manual threshold. Amplification efficiency for the reactions was estimated as described previously (17), and multiple regression models were used to derive point estimation of ΔΔCt, p-value, standard error and 95% confidence intervals with the SAS 9.1 programs provided (SAS Institute).
Statistical analysis
We used SAS 9.1 (SAS Institute) or Student's unpaired t-test to analyze the data. Results were considered statistically significant at *p<0.05, **p<0.01 and ***p<0.001.
Results
Transcriptional activation of the PPARγ receptor by PPARγ agonists in HCT-116 cells
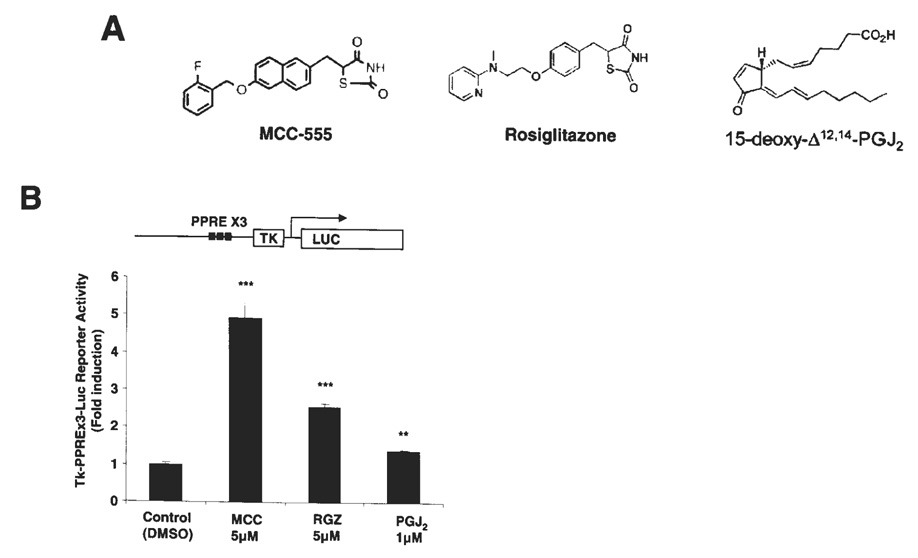
Agonists of the PPARγ receptor are known to possess anti-diabetic, anti-inflammatory and anti-cancer effects. In our study, we used three PPARγ agonists: two from the synthetic family of thiazolidinediones (MCC-555 and RGZ), and one naturally occurring metabolite of arachidonic acid (PGJ2) (Fig. 1A). To confirm that all three tested compounds are able to activate PPARγ and transcribe genes containing PPARγ binding sites in HCT-116 cells, a reporter construct, tk-PPREx3-Luc, was transfected and treated with the indicated PPARγ agonists, and luciferase activity was measured. It has been reported that HCT-116 cells highly express PPARγ (18). As shown in Fig 1B, treatment with 5 µM MCC-555 demonstrates the highest transcriptional activation of the PPARγ receptor by 4.93-fold induction (p<0.001) as compared to the other PPARγ agonists, RGZ and PGJ2 with 2.54- (p<0.001) and 1.37-fold (p<0.01) inductions, respectively. This result suggests that HCT-116 cells express functionally active PPARγ proteins that are responsible for PPARγ agonists.
Figure 1.
Transcriptional activation of the PPARγ receptor in HCT-116 cells by PPARγ agonists. (A) Molecular structures of three PPARγ agonists used in this study, MCC-555 (MCC), rosiglitazone (RGZ) and 15-deoxy-Δ12,14- prostaglandin J2 (PGJ2). (B) Transcriptional activation of the PPARγ receptor in HCT-116 cells transfected with tk-PPREx3-Luc reporter plasmid, and then treated with different PPARγ agonists, MCC-555 (5 µM), RGZ (5 µM) and PGJ2 (1 µM) for 24 h.
Transcriptional responses of PPARγ agonists in HCT-116 cells
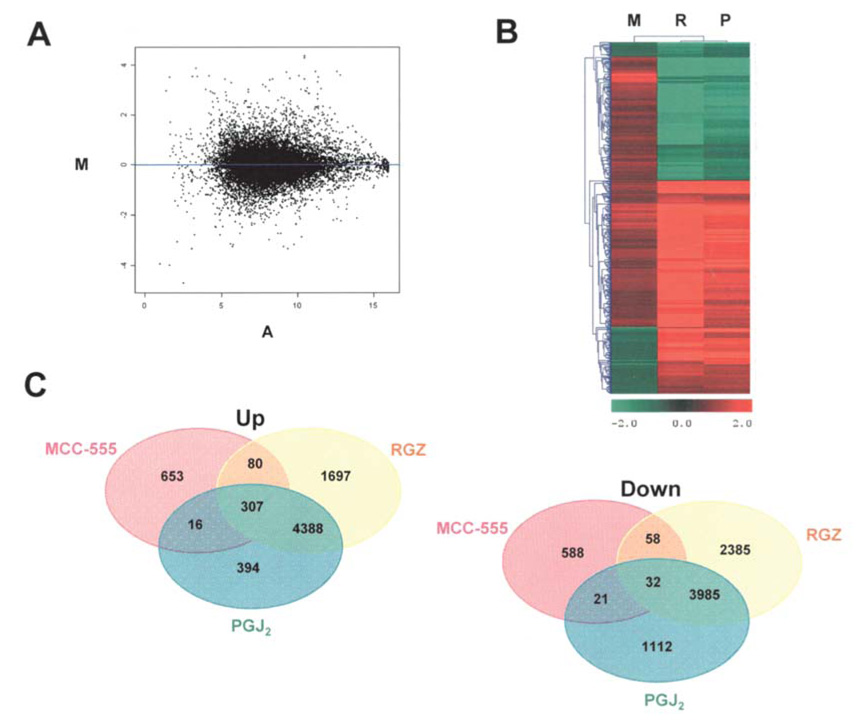
To examine the transcriptional responses of PPARγ agonists, gene expression patterns were compared in MCC- 555-, RGZ- and PGJ2-treated HCT-116 cells using longoligo microarrays containing the HEEBO oligo set. In our experiments, the cells were treated with 5 µM MCC-555, 5 µM RGZ and 1 µM PGJ2 for 24 h, and then total RNAs were isolated and processed for the microarray experiment as described in Materials and methods. Experiments for each treatment group were carried out in four replicates. Genes modulated by PPARγ agonists with statistical significance between the experimental groups (vehicle vs. treatments) were identified by SAS statistical software. Changes in gene expression patterns were considered statistically significant at p<0.05 if present in at least three replicates (degree of freedom 2). To evaluate the quality of microarray data, the M vs. A plot was generated to plot logarithm 2-based two color ratios against the logarithm 2-based two color signal strength multiples (Fig. 2A), indicating even distribution of down-regulated and up-regulated genes across different signal intensities. Scatter plots were also generated to compare the signal strengths between slides to evaluate the reproducibility of the experiments and microarray data quality (data not shown). Fig. 2B shows the hierarchical cluster analysis of the distance between genes regulated by PPARγ agonists. Interestingly, MCC-555 shows a unique pattern in gene expression, compared to that of RGZ and PGJ2. This result may support a previous report that MCC-555 is a dual agonist for PPARγ and PPARα, whereas RGZ and PGJ2 are PPARγ-specific agonists (16). As shown in Fig. 2C, microarray data showed that MCC-555 (5 µM) treatment up-regulated 1056 genes and down-regulated 699 genes, RGZ (5 µM) treatment up-regulated 6472 genes and down-regulated 6460 genes, and PGJ2 (1 µM) treatment up-regulated 5105 genes and down-regulated 5150 genes. Our results demonstrated that many genes were affected by PPARγ agonists, suggesting that PPARγ agonists play a role in the regulation of numerous signaling pathways in human colorectal cancer both in a PPARγ-dependent and independent manner. For further analysis, we focused on a group of genes commonly regulated by the tested PPARγ agonists: 307 genes were commonly up-regulated and 32 genes commonly down-regulated. The list of top 20 genes commonly altered by PPARγ agonists are shown in Table II and Table III with fold changes (ratios) and significant p-values. These genes were significantly modulated by all tested PPARγ agonists, suggesting that alteration of these genes may act in a PPARγ-dependent manner.
Figure 2.
Microarray data quality and analysis. (A) In order to evaluate the hybridization quality of microarrays, the M vs. A plot was generated as a scatter plot of log intensity ratios M = log2 (R/G) vs. average log intensities A = log2 (R×G)/2, where R and G represent the fluorescence intensities in the red and green channels, respectively. The M vs. A plot for the normalized average of MCC-555 treatments (4 slides) shows an even distribution of down-regulated and up-regulated genes across different signal intensities. (B) Hierarchical cluster map of gene expression. The expression matrix displays genes that were differentially expressed in HCT-116 cells treated by PPARγ agonists. For cluster analysis of the logarithm 2-based gene expression ratio for treatment by MCC-555, RGZ and PGJ2 (green color down-regulated genes, red color up-regulated genes), we used the following selection criteria: i) at least 1.5-fold difference between untreated and PPARγ agonist-treated; ii) p-value for difference in expression <0.05 (SAS test); and iii) appearance of the signal in at least three slide microarray replicates from each treatment. (C) Bend diagrams with the up- and down-regulated genes by MCC-555, RGZ and PGJ2 treatments.
Table II.
List of commonly 20 up-regulated genes by tested PPARγ ligands, MCC-555, RGZ and PGJ2 (p<0.05, fold change or ratio >1.5) in HCT-116 cells.
| Gene |
MCC-555 |
RGZ |
PGJ2 |
||||
|---|---|---|---|---|---|---|---|
| Symbol | Description | p-value | Ratio | p-value | Ratio | p-value | Ratio |
| CSTA | Cystatin A (stefin A) | 5.73E-05 | 4.238 | 5.80E-05 | 4.18 | 0.0307297 | 2.521 |
| UQCRB | Ubiquinol-cytochrome c reductase binding protein | 0.000269 | 3.079 | 0.0058807 | 5.967 | 0.0019013 | 4.975 |
| COX17 | COX17 homolog, cytochrome c oxidase assembly protein (yeast) | 8.76E-05 | 3.031 | 0.0049923 | 5.282 | 0.0153466 | 4.09 |
| COX7A2 | Cytochrome c oxidase subunit VIIa polypeptide 2 (liver) | 4.51E-05 | 3.011 | 0.0008634 | 6.178 | 0.0015228 | 3.651 |
| COMMD6 | COMM domain containing 6 | 0.000369 | 2.941 | 0.0013956 | 8.212 | 0.0009533 | 4.718 |
| LOC257039 | PREDICTED: similar to ribosomal protein S17 | 0.000358 | 2.935 | 0.0020001 | 6.81 | 0.0045161 | 5.811 |
| MT1J | Metallothionein 1J | 4.04E-05 | 2.917 | 0.0001288 | 5.131 | 0.0204116 | 2.519 |
| LOC392487 | PREDICTED: similar to ribosomal protein L31 | 0.000151 | 2.882 | 0.0055197 | 5.187 | 0.0029512 | 3.77 |
| LOC130865 | PREDICTED: similar to 60S ribosomal protein L26-like 1 | 0.000822 | 2.837 | 0.0254389 | 4.847 | 0.0091955 | 4.598 |
| LOC391581 | PREDICTED: similar to ribosomal protein L31 | 0.000445 | 2.835 | 0.0006942 | 8.253 | 0.0072957 | 3.713 |
| LOC440539 | PREDICTED: similar to ribosomal protein L37 | 0.000305 | 2.82 | 0.0003481 | 7.013 | 0.0020661 | 3.216 |
| RPL37A | Ribosomal protein L37a | 0.002622 | 2.768 | 9.05E-05 | 7.718 | 0.0003969 | 3.842 |
| LOC343153 | PREDICTED: similar to ribosomal protein L26 | 0.00061 | 2.761 | 0.0026409 | 5.46 | 0.0063362 | 2.706 |
| LOC402318 | PREDICTED: similar to 60S ribosomal protein L32 | 0.00186 | 2.749 | 0.0070023 | 7.32 | 0.012813 | 4.682 |
| LOC391035 | PREDICTED: similar to ribosomal protein S15a | 0.003748 | 2.724 | 0.0033576 | 8.008 | 0.0149866 | 6.855 |
| LOC402716 | PREDICTED: similar to 60S ribosomal protein L32 | 0.000397 | 2.709 | 0.0020859 | 6.651 | 0.0036407 | 3.235 |
| RPS21 | Ribosomal protein S21 | 0.00065 | 2.677 | 0.0001934 | 6.545 | 0.0039326 | 2.584 |
| LOC346950 | PREDICTED: similar to ribosomal protein L37 | 0.001874 | 2.648 | 3.20E-05 | 8.496 | 0.0003481 | 7.725 |
| COX17 | COX17 homolog, cytochrome c oxidase assembly protein (yeast) | 0.000491 | 2.645 | 0.0030994 | 5.341 | 0.0061663 | 2.955 |
| NDUFC1 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6 kDa | 0.000198 | 2.635 | 0.0015227 | 6.358 | 0.0032387 | 3.841 |
Table III.
List of commonly 20 down-regulated genes by tested PPARγ ligands, MCC-555, RGZ and PGJ2 (p<0.05, fold change or ratio <0.667) in HCT-116 cells.
| Gene |
MCC-555 |
RGZ |
PGJ2 |
||||
|---|---|---|---|---|---|---|---|
| Symbol | Description | p-value | Ratio | p-value | Ratio | p-value | Ratio |
| FAM13A1 | Family with sequence similarity 13,member A1 | 0.045278 | 0.399 | 0.0001102 | 0.307 | 0.0062549 | 0.485 |
| ARID5B | AT rich interactive domain 5B (MRF1-like) | 0.000568 | 0.428 | 0.0010759 | 0.41 | 0.0407399 | 0.587 |
| MDM2 | Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) | 0.041641 | 0.504 | 0.0001257 | 0.289 | 0.0091824 | 0.44 |
| TEF | Thyrotrophic embryonic factor | 0.001256 | 0.527 | 0.0030892 | 0.373 | 0.0211248 | 0.491 |
| NCOA2 | Nuclear receptor coactivator 2 | 0.011429 | 0.55 | 0.0011083 | 0.228 | 0.0034108 | 0.271 |
| FLJ36754 | P18SRP protein | 0.003659 | 0.568 | 0.0029843 | 0.374 | 0.0368733 | 0.599 |
| ATXN1 | Ataxin 1 | 0.009137 | 0.575 | 0.0012633 | 0.367 | 0.0187268 | 0.453 |
| KLHL11 | Kelch-like 11 (Drosophila) | 0.005889 | 0.581 | 7.68E-05 | 0.151 | 5.34E-05 | 0.2 |
| D2S448 | PREDICTED: melanoma associated gene | 0.002008 | 0.589 | 0.0264569 | 0.46 | 0.0218527 | 0.516 |
| ATP9A | ATPase, class II, type 9A (ATP9A), mRNA | 0.005898 | 0.589 | 0.0050552 | 0.288 | 0.0091849 | 0.384 |
| LOC91664 | Similar to zinc finger protein 268,clone IMAGE:3352268 | 0.001072 | 0.591 | 0.0009028 | 0.365 | 0.0435433 | 0.632 |
| PDE5A | Phosphodiesterase 5A, cGMP-specific | 0.008455 | 0.591 | 0.0002081 | 0.26 | 0.000969 | 0.241 |
| NLN | Neurolysin (metallopeptidase M3 family) | 0.014697 | 0.591 | 0.0013405 | 0.47 | 0.0288632 | 0.606 |
| FLJ10707 | PREDICTED: hypothetical protein FLJ10707 | 0.010421 | 0.592 | 0.0140784 | 0.257 | 0.0015614 | 0.267 |
| APPL | Adaptor protein containing pH domain, PTB domain and leucine zipper motif 1 | 0.003269 | 0.603 | 0.004938 | 0.307 | 0.0048149 | 0.41 |
| ABAT | 4-aminobutyrate aminotransferase | 0.00488 | 0.612 | 0.0073504 | 0.365 | 0.0336587 | 0.5 |
| SCN5A | Sodium channel, voltage-gated, type V,α (long QT syndrome 3) | 0.002446 | 0.613 | 0.0020712 | 0.442 | 0.0275364 | 0.579 |
| RECQL | RecQ protein-like (DNA helicase Q1-like) | 0.004036 | 0.618 | 0.0004371 | 0.406 | 0.0219375 | 0.536 |
| INPP5D | Inositol polyphosphate-5-phosphatase,145 kDa | 0.023678 | 0.618 | 0.0039209 | 0.481 | 0.0345667 | 0.607 |
| SUV420H1 | Suppressor of variegation 4–20 homolog 1 (Drosophila) | 0.039171 | 0.624 | 0.0003402 | 0.359 | 0.0221746 | 0.517 |
RT-PCR analysis of selected genes modulated by PPARγ agonists
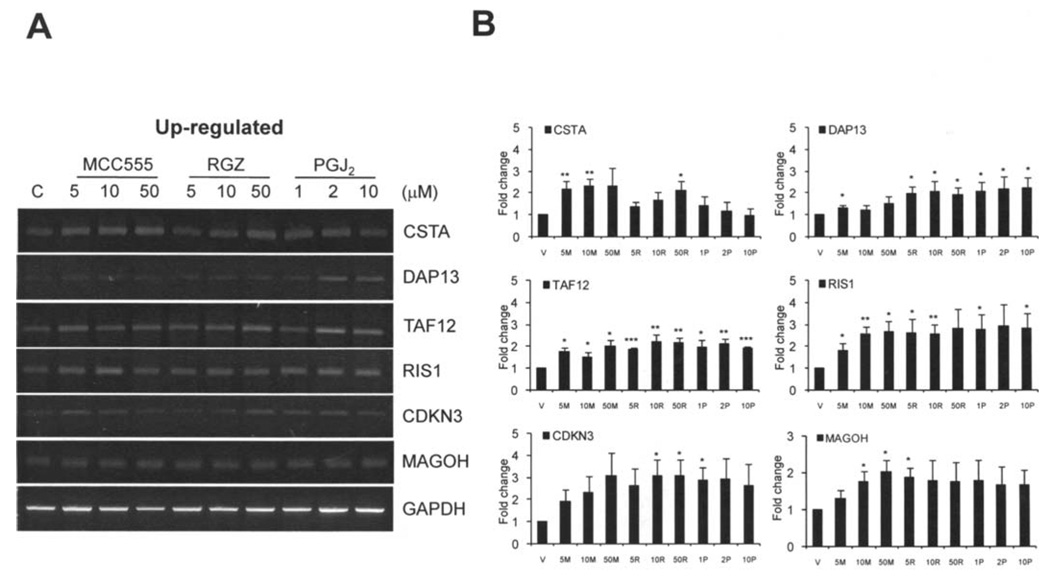
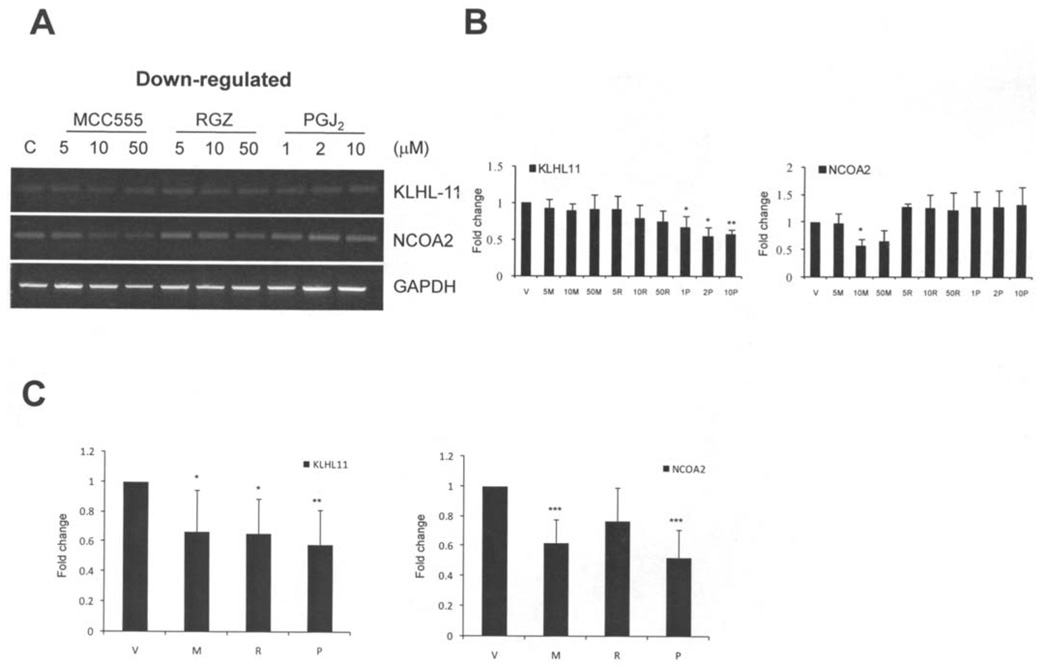
We have examined the expression of selected genes by semi-quantitative reverse transcriptase (RT-PCR) in HCT-116 cells treated by MCC-555, RGZ and PGJ2 for 24 h as described in Materials and methods. We checked six commonly induced and two commonly repressed genes from our microarray data, based on the highest induction, p-value, and potential involvement in tumorigenesis. The six up-regulated genes are CSTA, DAP13, TAF12, RIS1, CDKN3 and MAGOH. As shown in Fig. 3, results of RT-PCR showed that the expression levels of these six genes increased in HCT-116 cells treated with the indicated PPARγ agonists and consistent with the result of microarray data. In addition, we selected two down-regulated genes from microarray data and confirmed their expression by RT-PCR. As shown in Fig. 4, KLH11 and NCOA2 expression levels were suppressed by the indicated PPARγ agonists. However, we did not observe a significant reduction of these two genes in the presence of RGZ. Since RT-PCR is not sensitive enough to detect minute differences, we performed real-time PCR to confirm our microarray data for the down-regulation of gene expression. The real-time PCR results confirmed down-regulation of the KLHL-11 and NCOA2 genes (Fig. 4C). The PCR results are in general consistent with our microarray data, although RT-PCR results showed smaller differences than microarray data. Overall, these results confirmed that CSTA, DAP13, TAF12, RIS1, CDKN3 and MAGOH are induced by PPARγ agonists, whereas KLHL11 and NCOA2 are down-regulated by PPARγ agonists.
Figure 3.
RT-PCR analysis of the selected PPARγ agonist-induced genes in HCT-116 cells. (A) HCT-116 cells were treated by different PPARγ agonists for 24 h as described in Materials and methods in detail. Six up-regulated genes (CSTA, DAP13, TAF12, RIS1, CDKN3 and MAGOH) characterized from our microarray data were analyzed by RT-PCR. All genes showed increased expression levels by all treatments with MCC-555, RGZ and PGJ2. A housekeeping gene, GAPDH, was used for internal control and normalization. (B) Charts of the densitometry analysis of represented genes. Each chart represents the mean value ± SE of three independent experiments.
Figure 4.
RT-PCR analysis of the selected PPARγ agonist-repressed genes in HCT-116 cells. (A) HCT-116 cells were treated by different PPARγ agonists for 24 h as described in Materials and methods in detail. Two down-regulated genes (KLHL11 and NCOA2) characterized from our microarray data were analyzed by reverse transcriptase-PCR. Genes showed repressed expression levels by treatments with MCC-555, RGZ and PGJ2. (B) Charts of the densitometry analysis of represented genes. Each chart represents the mean values ± SE of three independent experiments. (C) Real-time PCR analysis of the down-regulated genes in HCT-116 cells treated with MCC-555 (5 µM), RGZ (5 µM) and PGJ2 (1 µM) for 24 h. Charts represent the mean values ± SE of three independent experiments with three dilutions.
Biological profiles of PPARγ agonist-induced genes
The final pattern of altered genes was evaluated by cluster analysis (Fig. 2B), and the commonly regulated genes were analyzed by ontology and signaling pathway analysis to better understand the biological phenomena involved in PPARγ dependency. Functional analysis of genes with a ≥1.5-fold change in expression (p<0.05 with degree of freedom 2) was performed using the Onto-Express software to identify biological process and molecular function categories affected by PPARγ agonists. The data analysis showed distribution of biological process (143 genes), molecular function (157 genes), and cellular component classes (144 genes) with involved genes regulated by PPAR agonists. In addition, molecular function classes were associated with antioxidant activity, binding, catalytic activity, enzyme regulator activity.
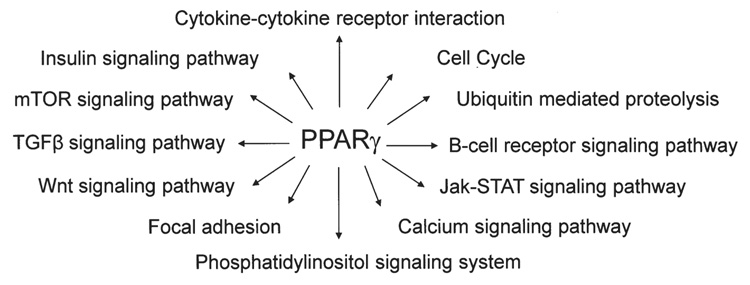
Furthermore, the functional importance of PPARγ-induced target genes was analyzed by Pathway Express Software and reflected in major KEGG pathways containing previously well-identified and established genes. As shown in Fig. 5 the pathways affected by PPARγ agonists were cell cycle (CHEK2, MDM2 and RBX1), cytokine-cytokine receptor interaction (IL18 and VEGFB), ubiquitin-mediated proteolysis (UBE2E1 and RBX1), phosphatidylinositol signaling system (INPP5D), insulin signaling pathway (INPP5D), Wnt signaling pathway (RBX1), TGF-β signaling pathway (RBX1), focal adhesion (VEGFB), calcium signaling pathway (SLC25A4) and colorectal cancer pathway (APPL).
Figure 5.
Scheme of pleiotropic action of PPARγ on various signaling pathways in human HCT-116 colorectal cancer cells.
Discussion
Colorectal cancer is the third most common cancer and the third leading cause of cancer-related mortality in the United States according to the American Cancer Society. However, over the past decade, colorectal cancer incidence and mortality rates have decreased compared to other cancers, and it is believed that chemoprevention research on human colorectal cancer has significantly contributed to this reduced risk. It is well known that the use of non-steroidal anti-inflammatory drugs (NSAIDs) decreases the risk of colorectal cancer and is very effective for chemoprevention. Data from our and other laboratories indicate that PPARγ agonists also induce apoptosis and have anti-tumorigenic effects in colorectal cancer (8,9). PPARγ agonists play a pivotal role in anti-tumorigenesis in a PPARγ-dependent and -independent manner (1), and that is why both pathways should be considered to investigate their roles. In this regard, our data may provide an up-to-date list of genes on which PPARγ agonists act in a PPARγ-dependent and -independent manner in human colorectal cancer cells. In our experiments, we used the HEEBO long-oligo set which contains about 39,000 probes for exonic regions, and the alternative spliced transcripts microarray screening technique to identify the genes and signaling pathways commonly regulated by three different PPARγ agonists in human HCT-116 colorectal adenocarcinoma cells. As shown in this study, the expression of several genes was modulated either positively (307) or negatively (32) by three PPARγ agonists in HCT-116 cells, suggesting that both activation and repression can play an important role of PPARγ agonists in tumorigenesis, cell growth and differentiation. MCC-555 treatment showed a unique pattern of gene expression with the least number of modified genes compared to the RGZ or PGJ2 treatments (Fig. 2B and C). One possible explanation is that MCC-555 also binds to PPARα and possesses distinct properties compared to other PPARγ agonists. Indeed, we have shown that MCC-555 increases tumor suppressor protein NAG-1 at the post-transctiptional level, whereas TGZ affects it at the transcriptional level (16).
We have identified some of the genes that are growth-related, or involved in tumor invasion, metastasis and apoptosis. Cystatin A or stefin A (CSTA) has been known to belong to family I of cysteine proteinase inhibitors, which are commonly down-regulated in many cancers. A large body of literature has accumulated to suggest that stefins correlate with malignancy of various murine and human tumors (19), such as lung, gastric, brain, colorectal and malignant melanoma (20). Cystatins act as tumor suppressor genes and positively relate with survival probability (21). Our data showed that the CSTA gene was up-regulated by all tested PPARγ agonists, which can have a beneficial effect on the treatment of colorectal cancer. Another tumor-growth related gene, CDKN3, which belongs to the dual specificity protein phosphatase family, was identified as a cyclin-dependent kinase (Cdk) inhibitor. CDKN3 has been shown to interact with the dephosphorylated Cdk-2 kinase, and thus prevent its activation (22). Our data indicate that all tested PPARγ agonists induced the expression of the CDKN3 gene, which may be a potential candidate for a colorectal cancer target gene to inhibit cell cycle progression.
Unlike the above-mentioned genes CSTA and CDKN3, MAGOH, TAF12, RIS1 and DAP13 genes are potentially new candidate genes associated with cancer. MAGOH is one of the eight PPARγ-regulated genes reported in this study, and its protein product is the human homolog of the Drosophila mago-nashi protein (23). Magoh protein plays important roles in mRNA splicing functions in the nucleus and cytoplasm. Indeed, both null mutation in flies and disruption of mRNA by RNAi in C.elegans show embryonic lethality, indicating that magoh is essential for viability (24). In our study, the expression of the MAGOH gene was up-regulated in human HCT-116 colorectal cancer cells; however, its exact biological function and regulation by PPARγ agonists in colorectal cancer remains to be elucidated. In contrast, little is known about DAP13 [also known as NADH dehydrogenase (ubiquinone) 1 α subcomplex 12, NDUFA12], except that it is described as a theoretical product of a gene identified in a lung adenocarcinoma cell line that had been induced to differentiate by treatment with all trans-retinoic acid (25).
Colorectal tumors develop mostly through several pathways, which are characterized by mutations in several suppressor genes (APC, SMAD and TP53) and oncogenes (KRAS2, MYC) and in chromosome instability. Ras-induced senescence 1 (RIS1) is a novel gene identified by Barradas et al (26), and is up-regulated in association with Ras senescence. The RIS1 gene is located at chromosomal position 3p21.3, previously defined for its tumor-suppressor activity (27); this region frequently exhibits the loss of heterozygosity in tumors, which is what confers this gene a putative role as a tumor suppressor. Moreover, mutations in the RIS1 gene are associated with a worse prognosis and development of metastasis in colorectal tumors (28). Another gene regulated by PPARγ agonists is TAF12 RNA polymerase II, a TATA box binding protein (TBP)-associated factor, which is an important part of the protein complex controlling the transcription by RNA polymerase II, and controlling the cell cycle and apoptosis (29). It has been shown that TAF12 interacts with ATF7, a transcription factor, and that this interaction can be modulated by another TAF protein, TAF4 (30).
Other genes characterized in our study are KLHL11 and NCOA2 that are down-regulated by PPARγ agonists. KLHL11 is the human ortholog of the identified zebrafish gene, kelchlike (klhl), an important gene involved in embryo-genesis. The human KLHL gene is specifically expressed in the muscles and heart, which suggests a role in muscle cytoarchitecture (31). Our microarray data also showed the down-regulation of nuclear receptor co-activator 2 gene, NCOA2, which is a family member of p160 kDa proteins involved in regulation of nuclear receptors, such as estrogen receptors and androgen receptors (32). It has been reported that NCOA2 and other cofactors including SRC-1, p300/ CBP, and Tip60 are up-regulated in advanced prostate cancer (32), suggesting that suppression of its expression is beneficial to prostate and other cancers.
Co-activators and co-repressors modulate the activity of PPARγ with influence on tumor formation. The pattern of commonly PPARγ agonist-altered genes were analyzed using the Onto-Express software, and signaling pathway analysis by Pathway Express Software was done to reflect the major KEGG pathways to better understand the biological phenomena involved in PPARγ dependency (Fig. 5). However, KEGG pathway analyses were not able to recognize the new genes involved in well established signaling pathways. Microarray data and ontology report analysis showed that PPARγ plays an important role in cellular processes, development, growth, physiological processes, regulation of biological process, response to stimulus, antioxidant activity, binding, catalytic activity, enzyme regulator activity, signal transducer activity, structural molecular activity, transcriptional regulator activity and transporter activity.
A better understanding of the PPARγ receptor and its molecular signaling pathway opens up new therapeutic prospects in treatment of colorectal cancer. Our microarray data of PPARγ-induced genes showed that PPARγ agonists induced pleiotropic effects on the transcriptional program of human colorectal adenocarcinoma cells, involving metabolic (e.g., amino acid and lipid metabolism) and hormonal (e.g., sterol biosynthesis), cell organization, and regulatory pathways (e.g., cytoskeletal proteins). This raises the possibility that these additional mechanisms might contribute to the anti-cancer activity of nuclear receptor agonists, perhaps in a tumor-specific and tissue-specific manner. From all of the above, it is clear that the influence of PPARγ on cell cycle proliferation, differentiation and apoptosis is complex. These effects depend on the concentrations of agonists, the cell type, and/or the mutational events that predispose cancer development. The full understanding of the mechanisms requires additional laboratory studies to address the role of PPARγ in tumorigenesis.
Acknowledgments
The authors thank Misty R. Bailey (The University of Tennessee, Knoxville, TN) for her technical assistance. We also thank Ron M. Evans (The Salk Institute, La Jolla, CA) for providing the tk-PPREx3-Luc reporter plasmid for our experiments. This study was supported by a grant from the National Institutes of Health (NIH-R01-CA108975) to S.J.B.
Abbreviations
- PPAR
peroxisome proliferator-activated receptor
- PGJ2
15-deoxy-Δ12,14-prostaglandin J2
- RGZ
rosiglitazone
- PPRE
peroxisome proliferator response element
- TGZ
troglitazone
References
- 1.Wang T, Xu J, Yu X, Yang R, Han ZC. Peroxisome proliferator-activated receptor gamma in malignant diseases. Crit Rev Oncol Hematol. 2006;58:1–14. doi: 10.1016/j.critrevonc.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 2.Vidal-Puig AJ, Considine RV, Jimenez-Linan M, et al. Peroxisome proliferator-activated receptor gene expression in human tissues. Effects of obesity, weight loss and regulation by insulin and glucocorticoids. J Clin Invest. 1997;99:2416–2422. doi: 10.1172/JCI119424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bren-Mattison Y, van Putten V, Chan D, Winn R, Geraci MW, Nemenoff RA. Peroxisome proliferator-activated receptor-gamma [PPAR(gamma)] inhibits tumorigenesis by reversing the undifferentiated phenotype of metastatic non-small-cell lung cancer cells (NSCLC) Oncogene. 2005;24:1412–1422. doi: 10.1038/sj.onc.1208333. [DOI] [PubMed] [Google Scholar]
- 4.Vignati S, Albertini V, Rinaldi A, et al. Cellular and molecular consequences of peroxisome proliferator-activated receptor-gamma activation in ovarian cancer cells. Neoplasia. 2006;8:851–861. doi: 10.1593/neo.06433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elstner E, Muller C, Koshizuka K, et al. Ligands for peroxisome proliferator-activated receptorgamma and retinoic acid receptor inhibit growth and induce apoptosis of human breast cancer cells in vitro and in BNX mice. Proc Natl Acad Sci USA. 1998;95:8806–8811. doi: 10.1073/pnas.95.15.8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarraf P, Mueller E, Jones D, et al. Differentiation and reversal of malignant changes in colon cancer through PPARgamma. Nat Med. 1998;4:1046–1052. doi: 10.1038/2030. [DOI] [PubMed] [Google Scholar]
- 7.Konopleva M, Elstner E, McQueen TJ, et al. Peroxisome proliferator-activated receptor gamma and retinoid X receptor ligands are potent inducers of differentiation and apoptosis in leukemias. Mol Cancer Ther. 2004;3:1249–1262. [PubMed] [Google Scholar]
- 8.Baek SJ, Wilson LC, Hsi LC, Eling TE. Troglitazone, a peroxisome proliferator-activated receptor gamma (PPAR gamma) ligand, selectively induces the early growth response-1 gene independently of PPAR gamma. A novel mechanism for its anti-tumorigenic activity. J Biol Chem. 2003;278:5845–5853. doi: 10.1074/jbc.M208394200. [DOI] [PubMed] [Google Scholar]
- 9.Bull AW. The role of peroxisome proliferator-activated receptor gamma in colon cancer and inflammatory bowel disease. Arch Pathol Lab Med. 2003;127:1121–1123. doi: 10.5858/2003-127-1121-TROPPR. [DOI] [PubMed] [Google Scholar]
- 10.Okura T, Nakamura M, Takata Y, Watanabe S, Kitami Y, Hiwada K. Troglitazone induces apoptosis via the p53 and Gadd45 pathway in vascular smooth muscle cells. Eur J Pharmacol. 2000;407:227–235. doi: 10.1016/s0014-2999(00)00758-5. [DOI] [PubMed] [Google Scholar]
- 11.Wang M, Wise SC, Leff T, Su TZ. Troglitazone, an antidiabetic agent, inhibits cholesterol biosynthesis through a mechanism independent of peroxisome proliferator-activated receptor-gamma. Diabetes. 1999;48:254–260. doi: 10.2337/diabetes.48.2.254. [DOI] [PubMed] [Google Scholar]
- 12.Palakurthi SS, Aktas H, Grubissich LM, Mortensen RM, Halperin JA. Anticancer effects of thiazolidinediones are independent of peroxisome proliferator-activated receptor gamma and mediated by inhibition of translation initiation. Cancer Res. 2001;61:6213–6218. [PubMed] [Google Scholar]
- 13.Davies GF, Khandelwal RL, Wu L, Juurlink BH, Roesler WJ. Inhibition of phosphoenolpyruvate carboxykinase (PEPCK) gene expression by troglitazone: a peroxisome proliferator-activated receptor-gamma (PPARgamma)-independent, antioxidant-related mechanism. Biochem Pharmacol. 2001;62:1071–1079. doi: 10.1016/s0006-2952(01)00764-x. [DOI] [PubMed] [Google Scholar]
- 14.Nikitakis NG, Siavash H, Hebert C, Reynolds MA, Hamburger AW, Sauk JJ. 15-PGJ2, but not thiazolidinediones, inhibits cell growth, induces apoptosis, and causes downregulation of Stat3 in human oral SCCa cells. Br J Cancer. 2002;87:1396–1403. doi: 10.1038/sj.bjc.6600618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eligini S, Banfi C, Brambilla M, et al. 15-deoxy-delta 12,14-Prostaglandin J2 inhibits tissue factor expression in human macrophages and endothelial cells: evidence for ERK1/2 signaling pathway blockade. Thromb Haemost. 2002;88:524–532. [PubMed] [Google Scholar]
- 16.Yamaguchi K, Lee SH, Eling TE, Baek SJ. A novel peroxisome proliferator-activated receptor gamma ligand, MCC-555, induces apoptosis via posttranscriptional regulation of NAG-1 in colorectal cancer cells. Mol Cancer Ther. 2006;5:1352–1361. doi: 10.1158/1535-7163.MCT-05-0528. [DOI] [PubMed] [Google Scholar]
- 17.Yuan JS, Reed A, Chen F, Stewart CN., Jr Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nixon JB, Kamitani H, Baek SJ, Eling TE. Evaluation of eicosanoids and NSAIDs as PPARgamma ligands in colorectal carcinoma cells. Prostaglandins Leukot Essent Fatty Acids. 2003;8:323–330. doi: 10.1016/s0952-3278(03)00023-1. [DOI] [PubMed] [Google Scholar]
- 19.Keppler D. Towards novel anti-cancer strategies based on cystatin function. Cancer Lett. 2006;235:159–176. doi: 10.1016/j.canlet.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 20.Kos J, Lah TT. Cysteine proteinases and their endogenous inhibitors: target proteins for prognosis, diagnosis and therapy in cancer (review) Oncol Rep. 1998;5:1349–1361. doi: 10.3892/or.5.6.1349. [DOI] [PubMed] [Google Scholar]
- 21.Strojan P, Budihna M, Smid L, et al. Prognostic significance of cysteine proteinases cathepsins B and L and their endogenous inhibitors stefins A and B in patients with squamous cell carcinoma of the head and neck. Clin Cancer Res. 2000;6:1052–1062. [PubMed] [Google Scholar]
- 22.Yeh CT, Lu SC, Chao CH, Chao ML. Abolishment of the interaction between cyclin-dependent kinase 2 and Cdk-associated protein phosphatase by a truncated KAP mutant. Biochem Biophys Res Commun. 2003;305:311–314. doi: 10.1016/s0006-291x(03)00757-5. [DOI] [PubMed] [Google Scholar]
- 23.Zhao XF, Colaizzo-Anas T, Nowak NJ, Shows TB, Elliott RW, Aplan PD. The mammalian homologue of mago-nashi encodes a serum-inducible protein. Genomics. 1998;47:319–322. doi: 10.1006/geno.1997.5126. [DOI] [PubMed] [Google Scholar]
- 24.Boswell RE, Prout ME, Steichen JC. Mutations in a newly identified Drosophila melanogaster gene, mago-nashi, disrupt germ cell formation and result in the formation of mirror-image symmetrical double abdomen embryos. Development. 1991;113:373–384. doi: 10.1242/dev.113.1.373. [DOI] [PubMed] [Google Scholar]
- 25.Skehel JM, Fearnley IM, Walker JE. NADH:ubiquinone oxidoreductase from bovine heart mitochondria: sequence of a novel 17.2-kDa subunit. FEBS Lett. 1998;438:301–305. doi: 10.1016/s0014-5793(98)01317-9. [DOI] [PubMed] [Google Scholar]
- 26.Barradas M, Gonos ES, Zebedee Z, et al. Identification of a candidate tumor-suppressor gene specifically activated during Ras-induced senescence. Exp Cell Res. 2002;273:127–137. doi: 10.1006/excr.2001.5434. [DOI] [PubMed] [Google Scholar]
- 27.Kok K, Naylor SL, Buys CH. Deletions of the short arm of chromosome 3 in solid tumors and the search for suppressor genes. Adv Cancer Res. 1997;71:27–92. doi: 10.1016/s0065-230x(08)60096-2. [DOI] [PubMed] [Google Scholar]
- 28.Iglesias D, Fernandez-Peralta AM, Nejda N, et al. RIS1, a gene with trinucleotide repeats, is a target in the mutator pathway of colorectal carcinogenesis. Cancer Genet Cytogenet. 2006;167:138–144. doi: 10.1016/j.cancergencyto.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Martinez E. Multi-protein complexes in eukaryotic gene transcription. Plant Mol Biol. 2002;50:925–947. doi: 10.1023/a:1021258713850. [DOI] [PubMed] [Google Scholar]
- 30.Hamard PJ, Dalbies-Tran R, Hauss C, Davidson I, Kedinger C, Chatton B. A functional interaction between ATF7 and TAF12 that is modulated by TAF4. Oncogene. 2005;24:3472–3483. doi: 10.1038/sj.onc.1208565. [DOI] [PubMed] [Google Scholar]
- 31.Wu YL, Gong Z. A novel zebrafish kelchlike gene klhl and its human ortholog KLHL display conserved expression patterns in skeletal and cardiac muscles. Gene. 2004;338:75–83. doi: 10.1016/j.gene.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 32.Powell SM, Christiaens V, Voulgaraki D, Waxman J, Claessens F, Bevan CL. Mechanisms of androgen receptor signalling via steroid receptor coactivator-1 in prostate. Endocr Relat Cancer. 2004;11:117–130. doi: 10.1677/erc.0.0110117. [DOI] [PubMed] [Google Scholar]