Abstract
Type I myotonic dystrophy (DM1) is caused by a triplet repeat expansion in the 3′-untranslated region (UTR) of the dystrophia myotonia protein kinase (DMPK) gene. Pathogenesis is closely linked with production of a toxic RNA from the mutant allele, which interferes with function of several RNA-binding proteins, including CUGBP1. Here we show that expression of a mutant DMPK 3′-UTR containing 960 CUG repeats is sufficient to increase expression and stability of an mRNA encoding the potent proinflammatory cytokine, tumor necrosis factor (TNF). CUGBP1 specifically recognizes sequences within the TNF 3′-UTR that are dissimilar from its canonical UG-rich binding site. Depletion of CUGBP1 from mouse myoblasts results in increased abundance of TNF mRNA through stabilization of the transcript. Moreover, activation of the protein kinase C pathway by treatment with phorbol ester, which has been shown previously to result in CUGBP1 phosphorylation, also causes TNF mRNA stabilization. Our results suggest that the elevated serum TNF seen in DM1 patients may be derived from muscle where it is induced by expression of toxic DMPK RNA. Importantly, overexpression of this potent cytokine could contribute to the muscle wasting and insulin resistance that are characteristic of this debilitating disease.
Myotonic dystrophy type I (DM1)3 is a debilitating autosomal dominant disorder caused by a triplet CTG repeat expansion in the 3′-untranslated region (UTR) of the dystrophin myotonia protein kinase (DMPK) gene (1–3). The disease is characterized by myotonia, progressive muscle wasting, insulin resistance, and cardiac conduction defects (4). Although some of the symptoms of DM1 may be attributed to reduced levels of the DMPK protein, much of the disease pathogenesis is recapitulated by expression of the mutant DMPK 3′-UTR (5, 6), or even by elevated expression of the wild type DMPK 3′-UTR (7), in otherwise normal mice. This is, at least in part, because transcripts containing the expanded CUG repeat accumulate in nuclear foci where they sequester cellular proteins (8).
The function of two RNA-binding proteins, CUGBP1 (CUG-binding protein 1) and MBNL (Muscleblind), is profoundly affected in DM1 patient cells. MBNL associates with the repeat-containing mRNA and is confined to nuclear foci (8), whereas CUGBP1 is overexpressed and hyperphosphorylated in the nucleus (9, 10). There is also evidence that CUGBP1 may be depleted from the cytoplasm (10). Interestingly, CUGBP1 and MBNL are both splicing regulators that play antagonistic roles in splice site selection. Consistent with this, several clinically relevant mRNAs, including the muscle-specific chloride channel (11), the insulin receptor (12), and cardiac troponin T (13), exhibit aberrant splice patterns in DM1 patient cells.
Several studies have indicated that aberrant function of CUGBP1 is a key factor in development of DM1 (5, 6, 14), and it is therefore important to consider other cellular functions of this protein that may be affected. It has been known for some time that CUGBP1 regulates cytoplasmic events in addition to modulating splicing in the nucleus. Specifically, CUGBP1 binds to the 5′-UTR of C/EBPβ and p21 mRNAs and affects translation efficiency (15, 16). Reduced p21 translation in DM1 because of aberrant CUGBP1 function has been linked with impaired differentiation of muscle cells (15). CUGBP1 has also recently been implicated as an mRNA destabilizing factor associated with short lived mRNAs (17), and we previously showed that CUGBP1 is involved in the decay of TNF mRNA in vitro (18).
Although aspects of DM1 pathogenesis may be linked to aberrant splicing or translation of clinically relevant mRNAs, several important symptoms have yet to receive an adequate molecular explanation. For example, muscle wasting, a major symptom of DM1, cannot be readily explained by any of the splicing changes identified to date. Previous studies have suggested that levels of the TNF cytokine are elevated in DM1 patients (19, 20), and excess TNF has been strongly linked with muscle wasting, insulin resistance, and cardiac dysfunction, all of which are seen in DM1. However, it remains unclear whether excess TNF production is a primary symptom caused directly by the repeat expansion or is an inflammatory response to the disease state.
Here we show that expression of a mutant DMPK 3′-UTR containing 960 CUG repeats in mouse myoblasts is sufficient to increase the expression of TNF mRNA. Significantly, this is achieved through stabilization of the TNF transcript. As mentioned above, CUGBP1 is a protein whose function is affected in DM1. We show that CUGBP1 recognizes multiple sequence elements within the TNF 3′-UTR and binds with high affinity. Moreover, depletion of CUGBP1 from mouse myoblasts results in stabilization of TNF mRNA. Finally, activation of protein kinase C (PKC), which has been shown to phosphorylate CUGBP1 (9), also stabilizes the TNF transcript. We conclude that the expression of expanded CUG repeat RNA in DM1 interferes with the cytoplasmic function of CUGBP1, resulting in a significant increase in TNF mRNA abundance and stability in muscle cells. Changes in TNF expression in muscle could therefore contribute to aspects of DM1 pathogenesis.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and Selection of Cell Lines—Murine C2C12 myoblasts (ATCC CRL1772) were maintained at 37 °C, 5% CO2 at or below 60% confluency in Dulbecco's modified Eagle's media containing 10% fetal bovine serum, penicillin (10 units/ml), and streptomycin (10 μg/ml). Plasmid DNA was treated with Mirus endotoxin removal agent prior to being transfected into C2C12 cells. Transfection was accomplished with Lipofectamine™ 2000 (Invitrogen) using a reverse transfection approach that gives ∼70–80% efficiency (21).
For selection of the LKO-1 and CUGBP1 KD C2C12 cell lines, either empty LKO-1 plasmid or LKO-1 plasmid containing an shRNA sequence against the 3′-UTR of murine CUGBP1 (Sigma MISSION clone ID NM_198683) was transfected into C2C12 cells. Two days post-transfection, cells were switched to media containing puromycin (1.5 μg/ml). Single colonies were selected, and expression of CUGBP1 was assessed by qRT-PCR (using primers CUGBP-forward 5′-GATCAGTGCAGCGTCTGTGT-3′ and CUGBP-reverse 5′-GTGTTGAGGTTCCCAGAGGA-3′) and corroborated by Western blot.
For transient siRNA-mediated knockdown of CUGBP1 expression, a CUGBP1 siRNA (5′-UUUGGCUGCACUAGCUGCU-3′(22)) or control siRNA (Silencer® Negative Control 1; Ambion) was transfected into C2C12 cells using Mirus TKO reagent according to the manufacturer's instructions. TNF mRNA half-life and CUGBP1 expression levels were assessed after 24 h.
For experiments involving TPA treatment, 100 ng/ml of TPA (Sigma) or an equivalent volume of DMSO was added, and cells were incubated for 3 h prior to either cell fractionation or half-life measurement.
Half-life Analysis—Transcription inhibition was achieved by addition of actinomycin-D (8 μg/ml; Sigma) for a period of 20 min. Cells were collected in TRIzol™ (Invitrogen) at the indicated time points post-transcription inhibition. Total RNA was isolated, and 1–3 μg were reverse-transcribed using Improm II reverse transcriptase (Promega) and reverse primers specific for either GAPDH or TNF. GAPDH and TNF mRNA levels were determined by real time PCR using a Bio-Rad MyIQ thermocycler and Bio-Rad SYBR-Green Supermix. Primers were GAPDH-forward 5′-TCACCACCATGGAGAAGGC-3′, GAPDH-reverse 5′-GCTAAGCAGTTGGTGGTGCA-3′, TNF-forward 5′-CATCTTCTCAAAATTCGAGTGACAA-3′, and TNF-reverse 5′-TGGGAGTAGACAAGGTACAACCC-3′. Data were analyzed using the ΔΔCt method (23). Data shown represents the mean values from at least three independent experiments; error bars represent mean ± S.E.
Gel Shift Analysis—RNAs were in vitro transcribed and labeled with [α-32P]UTP using SP6 RNA polymerase from pGem TNF (18), pGem ARE (24), pGem TNFΔ (18), and pGem4 plasmids. pGem TNF contains 250 nts of 3′-UTR sequence flanking the ARE of TNF (Fig. 1A) (18). pGem ARE contains just the 34-nt ARE from TNF mRNA. pGem TNFΔ is derived from pGem TNF by deletion of 53 nts containing the ARE, and pGem4 is a cloning vector from Promega and was used to generate the control Gem RNA. All plasmids were linearized with HindIII prior to transcription.
FIGURE 1.
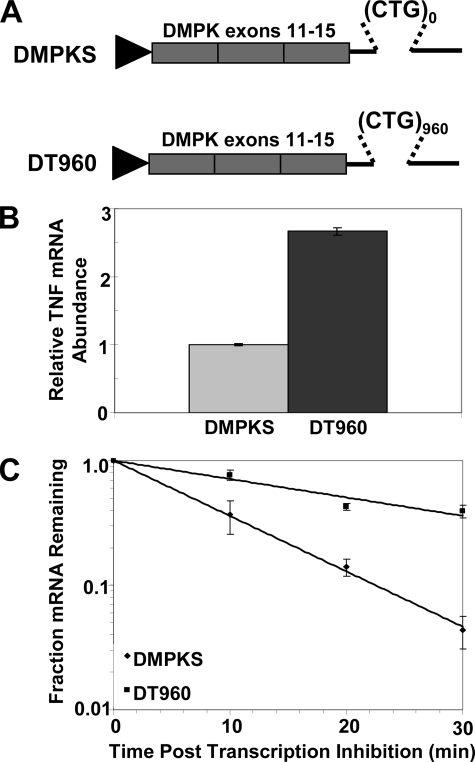
Expression of DMPK 3′-UTR containing 960 CUG repeats stabilizes TNF mRNA. A, schematic depicting DMPKS and DT960 constructs used to express normal and expanded repeat DMPK 3′-UTR. B, relative abundance of TNF mRNA was determined in C2C12 cells transiently transfected with either DMPKS or DT960 plasmids. C, half-life of TNF mRNA was determined by qRT-PCR in C2C12 cells following transfection with DMPKS or DT960 plasmids. The graphs represent cumulative data from multiple independent experiments.
Increasing amounts of recombinant CUGBP1 purified as described (18) were incubated with 3 fmol of the indicated RNA in the presence of 20 units of RNase inhibitor, 0.15 mm spermidine, 20 mm HEPES (pH 7.9), 8% glycerol, 100 mm KCl, and 2 mm MgCl2 for 5 min at 30 °C in a total volume of 10 μl. Low molecular weight heparin (Sigma) was added to a final concentration of 4 μg/μl. Samples were chilled on ice for an additional 5 min, and 2 μl of loading buffer (0.5% bromphenol blue, 0.5% xylene cyanol, 30% glycerol) was added, followed by electrophoresis at room temperature on 5% native polyacrylamide gels in 1× TBE buffer at ∼10 V/cm. Gels were dried and then exposed to phosphor screen and visualized by Phosphor-Imaging using a Typhoon Trio Imager (GE Healthcare) or FX Personal Imager (Bio-Rad) and the accompanying software.
The fraction of RNA bound was calculated by quantifying the amount of RNA associated with protein and dividing it by the total amount of RNA in each lane. Graphpad Prism version 5.0 (Graphpad Software, San Diego) was used to plot the graphs shown, and dissociation constants (Kd) were defined as the protein concentration required to achieve half-maximal binding at equilibrium.
Fractionation of Cell Membranes—Cell fractionation was performed as described (25). Briefly, LKO-1 cells and CUGBP1 KD cells were plated on 100-mm dishes 24 h before the experiment. Following 3 h of incubation with TPA or DMSO, cells were washed with phosphate-buffered saline and harvested by cell scraping in phosphate-buffered saline. The cells were pelleted by centrifugation at 1000 × g for 5 min and then resuspended in 300 μl of buffer A containing 10 mm KCl, 10 mm HEPES (pH 7.9), 0.5 mm dithiothreitol, and 1.5 mm MgCl2. After 30 min of incubation on ice, the cell suspension was lysed by passing through a 25-gauge needle 10 times without damaging nuclei. Lysates were spun in a microcentrifuge at 12,000 rpm at 4 °C for 5 min to pellet nuclei and membranes. The pellet was incubated with 100 μl of buffer A containing 1% Triton X-100 for 30 min to solubilize the membrane proteins. The samples were then centrifuged for 5 min at 12,000 × g, and the resulting supernatants containing solubilized membranes were used in Western blots to detect phosphorylated PKC.
Western Blot Analysis—For CUGBP1 Westerns, 40 μg of whole protein extract prepared by lysis of cells in RIPA buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1.0% deoxycholate, 1% Triton X-100, 1 mm EDTA, and 0.1% SDS) was separated on a 10% SDS-polyacrylamide gel and blotted to polyvinylidene difluoride membrane. CUGBP1 was detected using monoclonal antibody 3B1 (1:20,000 dilution; Santa Cruz Biotechnology, catalog number 20003). GAPDH was used as a loading control (1:20,000 dilution; Chemicon mAB374).
For detection of phosphorylated PKC by Western blotting, 10 μg of membrane fraction proteins prepared as described above were separated on a 10% SDS-polyacrylamide gel and blotted to polyvinylidene difluoride membrane. Phosphorylated PKC was detected using rabbit anti-pPKC antibodies that recognize phosphorylated PKCα and PKCβII (1:2000 dilution; Santa Cruz Biotechnology, catalog number sc12356-R). Abundance was normalized to that of the transferrin receptor (1:2000 dilution; Zymed Laboratories Inc., catalog number 13-6800). In all cases, anti-mouse or anti-rabbit horseradish peroxidase-conjugated secondary antibodies were employed as appropriate, and detection was by the Supersignal Pico West kit (Pierce) or by ECL plus (GE Healthcare) using either x-ray film or a Bio-Rad VersaDoc imager.
RESULTS
Expression of Mutant DMPK 3′-UTR Stabilizes TNF mRNA in Myoblasts—We wished to test whether the altered cellular conditions induced by an expanded CUG repeat in the DMPK gene can result in changes in TNF expression. We chose to examine TNF mRNA abundance in C2C12 mouse myoblasts because of the following: (i) muscle cells express DMPK, and muscle is the main tissue affected in DM1 patients; (ii) expression of a mutant DMPK 3′-UTR containing expanded CUG repeats has been shown to result in impaired differentiation and accumulation of nuclear foci in this cell line (26, 27); and (iii) C2C12 cells constitutively express TNF at low levels (28). We investigated the effects of expressing DMPK 3′-UTR containing 960 CUG repeats (DT960) or completely lacking CUG repeats (DMPKS) on TNF mRNA abundance. We used two constructs derived from the human DMPK gene that have been described previously (13, 29) because they maintain the CUG repeats in their natural context. This is a critical factor in mouse and cell culture models of DM1 (5, 27). The 960 repeats in the DT960 construct were selected to approximate the number of CUG repeats known to cause adult onset DM1 in patients (80–1000 repeats (1–3)). In contrast, the DMPKS construct has zero repeats rather than the five repeats seen in the normal gene because constructs expressing the normal DMPK 3′-UTR can cause disease when overexpressed in mice (7).
Abundance of TNF mRNA was determined by qRT-PCR following transient transfection of DT960 and DMPKS into C2C12 cells. As shown in Fig. 1B, the abundance of TNF mRNA was reproducibly increased greater than 2-fold in the DT960-transfected cells when compared with those transfected with DMPKS. Significantly, this increase in TNF mRNA abundance is observed only with the DT960 construct despite the fact that it is expressed at lower levels than DMPKS (supplemental Fig. 1). This indicates that the presence of the expanded CUG repeat is essential for increased TNF expression. Although both DT960 and DMPKS constructs are expressed at higher levels than would be seen in patient cells (30), previous studies suggest that the repeat context is more critical than the number of repeats or their expression level with regards to disease pathogenesis (5, 27, 31).
We hypothesized that decay of the TNF transcript may be affected in the DT960-transfected cells, and we therefore measured the stability of the transcript following inhibition of transcription with actinomycin D. Total RNA samples were taken at the indicated time points after transcription inhibition, and abundance of TNF mRNA for each was determined by qRT-PCR and normalized to GAPDH mRNA levels. GAPDH mRNA is stable over the time course utilized here. In the DMPKS-transfected cells, TNF mRNA degraded rapidly with a half-life of 6.8 ± 0.1 min. This rapid decay is typical of a transcript containing an ARE and is comparable with the decay rate of TNF mRNA in untransfected cells (see below). In contrast, in the DT960-transfected cells TNF mRNA was dramatically stabilized, degrading with a half-life of 20.4 ± 1.8 min (Fig. 1C). These data demonstrate that expression of the mutant DMPK 3′-UTR is sufficient to induce stabilization of TNF mRNA in C2C12 myoblasts.
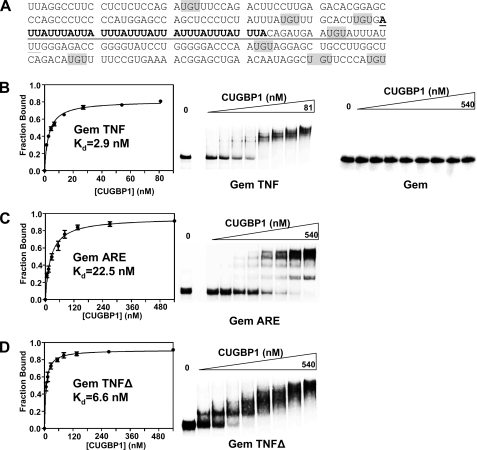
CUGBP1 Binds to the TNF 3′-UTR with High Affinity—We showed previously, by UV cross-linking, that CUGBP1 binds to a 250-nt fragment of the TNF 3′-UTR in HeLa extracts (18). Binding of CUGBP1 results in rapid deadenylation in vitro through recruitment of the PARN deadenylase (18). These observations, together with the fact that CUGBP1 function is known to be aberrant in DM1, strongly suggested that CUGBP1 was likely linked with the TNF mRNA stabilization seen in DT960-transfected cells. However, close examination of the 250 nts of the region previously shown to bind CUGBP1 (Fig. 2A) does not reveal any elements that closely resemble characterized high affinity CUGBP1-binding sites such as the extensive UG-rich sequences identified by systematic evolution of ligands by exponential enrichment (32). A related protein, CUGBP2/ETR3, has been reported to bind to the AU-rich element (ARE) in the COX-2 mRNA 3′-UTR (33), so it seemed possible that CUGBP1 might recognize the well characterized AU-rich element in the TNF 3′-UTR. To determine whether CUGBP1 binds the TNF ARE, we employed a gel shift assay using recombinant CUGBP1 and radiolabeled in vitro a fragment of the TNF 3′-UTR corresponding to the 250-nt region flanking the AU-rich element (Gem TNF). Gem TNF RNA or a control RNA derived from the polylinker of pGem4 (Gem) was incubated with increasing amounts of recombinant CUGBP1 protein, and the complexes were separated on a native polyacrylamide gel (Fig. 2B). CUGBP1 did not bind detectably to the Gem RNA but recognized the Gem TNF RNA with high affinity (Kd = 2.9 ± 0.2 nm). Next, we assessed binding of CUGBP1 to the 34-nt AU-rich element itself (Gem ARE). As shown in Fig. 2C, CUGBP1 does bind to Gem ARE but with ∼8-fold reduced affinity (Kd = 22.5 ± 2.4 nm) when compared with the 250-nt fragment. Consistent with this observation, when we examined binding of CUGBP1 to a variant in which the ARE and 19 nt of flanking sequence had been deleted (Gem TNFΔ; Fig. 2D), the binding affinity was only slightly reduced (Kd = 6.6 ± 0.7 nm). Taken together these data indicate that high affinity binding of CUGBP1 to Gem TNF may occur through simultaneous recognition of multiple sequence elements, including the ARE and perhaps UGU trimers distributed throughout the flanking regions.
FIGURE 2.
CUGBP1 recognizes the AU-rich element and flanking regions within the TNF 3′-UTR. A, sequence of the 250-nt region flanking the ARE in the TNF 3′-UTR. The 34-nt AU-rich sequence contained within Gem ARE is in boldface. The region deleted in Gem TNFΔ is underlined. UGU trimers that may be recognized by CUGBP1 are highlighted in gray. B, gel shift analysis of CUGBP1 binding to Gem TNF and Gem RNAs. Increasing nanomolar amounts of CUGBP1 protein were incubated with either Gem TNF or Gem RNAs, and complexes were separated on a native polyacrylamide gel. Binding was plotted as fraction of RNA bound against nm of CUGBP1 to determine the dissociation constant (Kd). C, gel shift analysis CUGBP1 binding to Gem ARE RNA. D, gel shift analysis of CUGBP1 binding to Gem TNFΔ RNA.
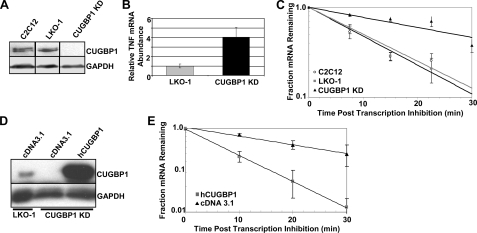
CUGBP1 Is Required for Rapid Degradation of TNF mRNA in C2C12 Myoblasts—We next wanted to examine whether decay of TNF mRNA was affected by depletion of CUGBP1. C2C12 cells were transfected either with empty vector (LKO-1) or with a plasmid expressing an shRNA targeting the 3′-UTR of all CUGBP1 mRNA isoforms. Stable cell lines were selected and screened for reduced expression of CUGBP1 by Western blot. A stable cell line exhibiting >90% knockdown of CUGBP1 (CUGBP1 KD) was utilized in all subsequent experiments (Fig. 3A). We measured abundance of TNF mRNA by qRT-PCR in the knockdown and control (LKO-1) cell lines and normalized it to GAPDH mRNA levels. As shown in Fig. 3B, TNF mRNA abundance is increased ∼4-fold in the knockdown cell line. To measure the half-life of the TNF mRNA in normal C2C12, control (LKO-1), and CUGBP1 KD cells, we first treated the cells with actinomycin D to inhibit transcription. Samples were taken at various times after transcription inhibition, and total RNA was prepared. Abundance of TNF mRNA at each time point was determined by qRT-PCR and normalized to GAPDH mRNA levels, as before. The decay of TNF mRNA in each of the three cell lines (C2C12, LKO-1, and CUGBP1 KD) is depicted in Fig. 3C. In the normal C2C12 cell line TNF is very unstable as expected and decays with a half-life of 10.1 ± 0.9 min. A similar decay rate is seen in the control LKO-1 cell line (t½ = 9.4 ± 0.4 min). However, the CUGBP1 KD cells show dramatic stabilization of the TNF transcript (t½ = 27.0 ± 3.5 min), indicating that CUGBP1 is indeed required for rapid decay.
FIGURE 3.
Depletion of CUGBP1 results in stabilization of TNF mRNA in mouse myoblasts. A, Western blot depicting levels of CUGBP1 protein in untreated C2C12 cells and in C2C12 cells stably transfected with either empty vector (LKO-1) or CUGBP1 shRNA (CUGBP1 KD). Note that all samples were run on the same gel and exposed for the same time; lanes have been juxtaposed for esthetic purposes. B, relative abundance of TNF mRNA was determined by qRT-PCR in the LKO-1 and CUGBP1 KD cells and normalized to GAPDH mRNA abundance. C, rate of decay of TNF mRNA was assessed in C2C12, LKO-1, and CUGBP1 KD cells, following inhibition of transcription with actinomycin D. mRNA abundance at each time point was measured by qRT-PCR and normalized to GAPDH mRNA levels. D, LKO-1 and CUGBP1 KD cells were transiently transfected with either empty vector (cDNA3.1) or a vector expressing human CUGBP1 (hCUGBP1). Expression of CUGBP1 was determined by Western blot and normalized to GAPDH levels. E, half-life of TNF mRNA was determined in CUGBP1 KD cells transiently transfected with either hCUGBP1 or empty vector (cDNA3.1).
We utilized two approaches to confirm that the effect on TNF mRNA decay was directly due CUGBP1 knockdown. First, we transfected either empty vector (pcDNA3.1) or a plasmid encoding the human CUGBP1 protein into the CUGBP1 KD cells. As the human CUGBP1 transcript is resistant to the shRNA expressed in these cells, hCUGBP1 was expressed at high levels (Fig. 3D). In the CUGBP1 KD cells that received empty vector, TNF mRNA was stable decaying with a half-life of 30.4 ± 1.26 min; however, transfection of the hCUGBP1 plasmid restored TNF mRNA decay to the normal rate (t½ = 9.5 ± 0.1 min; Fig. 3E).
We also used an siRNA targeting a different region of the mouse CUGBP1 transcript to transiently knockdown CUGBP1 expression and found that TNF was stabilized under these conditions as well (supplemental Fig. 2). These results demonstrate that stabilization of TNF mRNA in the CUGBP1 knockdown cell line is directly because of loss of CUGBP1 function.
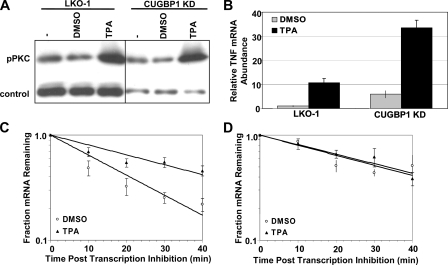
Activation of Protein Kinase C Stabilizes TNF mRNA through Modulation of CUGBP1 Function—A recent study by Cooper and co-workers (9) demonstrated that CUGBP1 is hyperphosphorylated and overexpressed in DM1 patient cells. Furthermore, phosphorylation of CUGBP1 appears to be mediated by the PKC pathway. We therefore investigated the effect of activating the PKC pathway on TNF mRNA abundance and stability by treating the control (LKO-1) and CUGBP1 KD cells with the phorbol ester TPA. As shown in Fig. 4A, 3 h of TPA treatment did indeed activate PKC as measured by increased association of the phosphorylated form of the PKC protein (pPKC) with cellular membranes. TPA treatment resulted in increased abundance of TNF mRNA in both the control and the CUGBP1 KD cell lines (Fig. 4B). Moreover, TPA-treated control cells exhibited stabilization of TNF mRNA to a similar level as seen in the CUGBP1 knockdown cells (DMSO t½ = 14.9 ± 1.2 min, TPA t½ = 32.7 ± 2.8 min; Fig. 4C). It is likely that both transcription rate and stability of the TNF mRNA are elevated under these drug-treated conditions as the ∼2-fold increase in half-life is not sufficient to explain the 10-fold increase in TNF mRNA abundance seen in the LKO-1 control cells.
FIGURE 4.
Phorbol ester stabilizes TNF mRNA through abrogation of CUGBP1 function. A, Western blot showing increased association of pPKCα with membranes in LKO-1 and CUGBP1 KD cells following 3 h of treatment with TPA. pPKCα levels were normalized to transferrin receptor abundance. B, relative abundance of TNF mRNA was determined by qRT-PCR in LKO-1 and CUGBP1 KD cells following 3 h of treatment with TPA or DMSO. C, half-life TNF mRNA was determined by qRT-PCR in LKO-1 cells following DMSO or TPA treatment. D, half-life TNF mRNA was determined by qRT-PCR in CUGBP1 KD cells following DMSO or TPA treatment.
We next investigated the effect of TPA on TNF half-life in the CUGBP1 knockdown C2C12 cell line. As expected, TNF mRNA was stable in the CUGBP1 KD cells when treated with DMSO (t½ = 31.4 ± 4.1 min; Fig. 4D), but the transcript was not further stabilized by TPA treatment (t½ = 33.3 ± 3.0 min). Thus, in the absence of CUGBP1, TPA treatment no longer impacts stability of the TNF transcript suggesting that disruption of CUGBP1 function is required for stabilization of TNF mRNA in response to TPA.
We note that the increased abundance of TNF mRNA in the CUGBP1 KD cells cannot be completely explained by stabilization of the transcript in the TPA-treated cells (Fig. 4). The transcription of TNF must also be higher in CUGBP1 KD cells than in the LKO-1 cells. This may be linked to indirect effects of constitutive overexpression of TNF or because of the role of CUGBP1 in regulating expression of other genes at the level of splicing, mRNA stability, or translation.
In summary, our results show that CUGBP1 binds to the TNF 3′-UTR with high affinity. Abrogation of CUGBP1 function either by knockdown, by activation of the PKC pathway, or by expression of the mutant DMPK 3′-UTR, results in stabilization and increased abundance of TNF mRNA. These results have important implications for our understanding of both TNF expression in muscle and DM1 pathogenesis as discussed below.
DISCUSSION
Our findings provide interesting insights with regards to regulation of TNF expression. CUGBP1 joins several other RNA-binding proteins, including tristetraprolin, AUF1, TIA-1, TIAR, and HuR, as a regulator of TNF mRNA stability. Significantly, CUGBP1 is important for mediating efficient TNF mRNA degradation primarily in unstimulated C2C12 cells but not under conditions (such as phorbol ester treatment; Fig. 4) where TNF expression is induced. Indeed our results indicate that stabilization of TNF mRNA in response to TPA treatment involves disruption of CUGBP1 function. In this respect, CUGBP1 may be important in restricting expression of this potent cytokine to the low levels required for its beneficial effects on muscle metabolism. It will be intriguing to see whether the CUGBP1-dependent regulation we observe in C2C12 cells also occurs in macrophages and monocytes, which are major producers of TNF.
Our previous results indicated that CUGBP1 interacts directly with the PARN deadenylase to facilitate removal of the poly(A) tail and thereby destabilize the TNF transcript (18). TPA treatment and expression of mutant DMPK 3′-UTR are both reported to result in phosphorylation of CUGBP1 (9). In principle, such a phosphorylation event could affect CUGBP1 function either by preventing its association with the TNF mRNA or by interfering with the CUGBP1-PARN interaction. CUGBP1 has been reported to be restricted to the nucleus in DM1 patient cells (10), which would prevent it from binding TNF mRNA and mediating mRNA decay in the cytoplasm. However, by Western blotting of fractionated cell extracts, we have been unable to detect any change in CUGBP1 abundance or nuclear versus cytoplasmic localization either in TPA-treated cells or in cells expressing the DT960 mRNA (data not shown). We therefore currently favor the hypothesis that either interaction of CUGBP1 with other proteins is altered or its RNA-binding capacity is affected thus preventing it from mediating TNF mRNA decay.
CUGBP1 is somewhat unique among the factors previously implicated in regulating TNF mRNA stability as it principally recognizes sequences that flank the AU-rich element rather than the ARE itself (Fig. 2). Indeed, the binding of CUGBP1 to TNF mRNA warrants further investigation as there are no predicted high affinity CUGBP1-binding sites (sequences of 30 nt or less containing at least four nonoverlapping UGU motifs (17, 32)) within the TNF 3′-UTR. We propose that the RNA secondary structure may facilitate CUGBP1 binding by bringing UGU sequences dispersed throughout the region into close proximity. Notably, our gel shift assays are consistent with multiple CUGBP1 monomers associating with all three RNA substrates up to a maximum of approximately four CUGBP1 molecules. This is clearest in Fig. 2D (Gem TNFΔ) where the complex increases in size in a stepwise fashion as additional CUGBP1 molecules bind, but multiple complexes can also be seen associating with the other two RNAs. Interestingly, a recent study found that EDEN-BP, the Xenopus homolog of CUGBP1, oligomerizes, and that self-interaction was necessary for high affinity binding (34).
In the light of a recent report suggesting that CUGBP1 may be an important regulator of decay of a large number of transcripts containing UG-rich elements in their 3′-UTRs (17), it seems likely that the cytoplasmic function of CUGBP1 in mRNA turnover has a more extensive impact on DM1 pathogenesis than previously thought. However, identification of CUGBP1-binding sites is clearly not as simple as pinpointing extensive UG-rich regions. Thus further investigation will be necessary to enable accurate prediction of CUGBP1 targets. Experiments are underway to identify other transcripts that are stabilized in CUGBP1 knockdown cells and in cells expressing the mutant DMPK 3′-UTR.
Finally, our findings may have important implications for patients with DM1. We suggest that rather than being produced by immune cells as a response to the disease state, the elevated TNF observed in sera of DM1 patients (19, 20) may actually be derived from the skeletal muscle as a direct result of expression of mutant DMPK 3′-UTR in this tissue. Although skeletal muscle is not generally thought of as a major cytokine producer, like most tissues it does in fact produce TNF at low levels. When one considers that skeletal muscle is the most abundant tissue in the body, it is quite possible that elevated production of TNF by muscle could impact serum levels of this cytokine. Moreover, muscle is exquisitely sensitive to TNF; low levels of TNF stimulate myogenesis, and excess TNF can inhibit the process (35). TNF also induces insulin resistance and protein turnover in muscle cells (36). We suggest that aspects of DM1 pathogenesis, specifically muscle wasting and insulin resistance, may be caused or exacerbated by autocrine effects of chronic excess TNF production by the muscle itself.
Supplementary Material
Acknowledgments
We thank Dr. Tom Cooper for the hCUGBP1 expression plasmid and the DT960 and DMPKS constructs. We are also grateful to Dr. Michael Tamkun for the anti-transferrin receptor antibody. In addition, we thank Jim Zumbrunnen of the Colorado State University Statistics Department for assistance with data analysis and Lesley Jones for selecting stable cell lines. Finally, we thank members of the Wilusz laboratory and Dr. Shobha Vasudevan for critical review of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM 072481 (to J. W.). This work was also supported by Muscular Dystrophy Association Award 4326 (to C. J. W.), and by Colorado State University College of Veterinary Medicine and Biomedical Sciences (to C. J. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: DM1, myotonic dystrophy type I; ARE, AU-rich element; TNF, tumor necrosis factor; UTR, untranslated region; PKC, protein kinase C; TPA, 12-O-tetradecanoylphorbol-13-acetate; GAPDH, glycer-aldehyde-3-phosphate dehydrogenase; PARN, poly(A) ribonuclease; DMPK, dystrophia myotonica protein kinase; qRT, quantitative reverse transcription; siRNA, short interfering RNA; shRNA, short hairpin RNA; nt, nucleotide.
References
- 1.Mahadevan, M., Tsilfidis, C., Sabourin, L., Shutler, G., Amemiya, C., Jansen, G., Neville, C., Narang, M., Barcelo, J., O'Hoy, K., et al. (1992) Science 255 1253–1255 [DOI] [PubMed] [Google Scholar]
- 2.Brook, J. D., McCurrach, M. E., Harley, H. G., Buckler, A. J., Church, D., Aburatani, H., Hunter, K., Stanton, V. P., Thirion, J. P., Hudson, T., Sohn, R., Zemelman, B., Snell, R. S., Rundle, S. A., Crow, S., Davies, J., et al. (1992) Cell 68 799–808 [DOI] [PubMed] [Google Scholar]
- 3.Fu, Y. H., Pizzuti, A., Fenwick, R. G., Jr., King, J., Rajnarayan, S., Dunne, P. W., Dubel, J., Nasser, G. A., Ashizawa, T., de Jong, P., et al. (1992) Science 255 1256–1258 [DOI] [PubMed] [Google Scholar]
- 4.Machuca-Tzili, L., Brook, D., and Hilton-Jones, D. (2005) Muscle Nerve 32 1–18 [DOI] [PubMed] [Google Scholar]
- 5.Orengo, J. P., Chambon, P., Metzger, D., Mosier, D. R., Snipes, G. J., and Cooper, T. A. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 2646–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang, G. S., Kearney, D. L., De, B. M., Taffet, G., and Cooper, T. A. (2007) J. Clin. Investig. 117 2802–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahadevan, M. S., Yadava, R. S., Yu, Q., Balijepalli, S., Frenzel-McCardell, C. D., Bourne, T. D., and Phillips, L. H. (2006) Nat. Genet. 38 1066–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mankodi, A., Urbinati, C. R., Yuan, Q. P., Moxley, R. T., Sansone, V., Krym, M., Henderson, D., Schalling, M., Swanson, M. S., and Thornton, C. A. (2001) Hum. Mol. Genet. 10 2165–2170 [DOI] [PubMed] [Google Scholar]
- 9.Kuyumcu-Martinez, N. M., Wang, G. S., and Cooper, T. A. (2007) Mol. Cell 28 68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts, R., Timchenko, N. A., Miller, J. W., Reddy, S., Caskey, C. T., Swanson, M. S., and Timchenko, L. T. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 13221–13226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charlet, B., Savkur, R. S., Singh, G., Philips, A. V., Grice, E. A., and Cooper, T. A. (2002) Mol. Cell 10 45–53 [DOI] [PubMed] [Google Scholar]
- 12.Savkur, R. S., Philips, A. V., and Cooper, T. A. (2001) Nat. Genet. 29 40–47 [DOI] [PubMed] [Google Scholar]
- 13.Philips, A. V., Timchenko, L. T., and Cooper, T. A. (1998) Science 280 737–741 [DOI] [PubMed] [Google Scholar]
- 14.Ho, T. H., Bundman, D., Armstrong, D. L., and Cooper, T. A. (2005) Hum. Mol. Genet. 14 1539–1547 [DOI] [PubMed] [Google Scholar]
- 15.Timchenko, N. A., Iakova, P., Cai, Z. J., Smith, J. R., and Timchenko, L. T. (2001) Mol. Cell. Biol. 21 6927–6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Timchenko, N. A., Cai, Z. J., Welm, A. L., Reddy, S., Ashizawa, T., and Timchenko, L. T. (2001) J. Biol. Chem. 276 7820–7826 [DOI] [PubMed] [Google Scholar]
- 17.Vlasova, I. A., Tahoe, N. M., Fan, D., Larsson, O., Rattenbacher, B., Sternjohn, J. R., Vasdewani, J., Karypis, G., Reilly, C. S., Bitterman, P. B., and Bohjanen, P. R. (2008) Mol. Cell 29 263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moraes, K. C., Wilusz, C. J., and Wilusz, J. (2006) RNA (N. Y.) 12 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansson, A., Carlstrom, K., Ahren, B., Cederquist, K., Krylborg, E., Forsberg, H., and Olsson, T. (2000) J. Clin. Endocrinol. Metab. 85 3169–3176 [DOI] [PubMed] [Google Scholar]
- 20.Mammarella, A., Ferroni, P., Paradiso, M., Martini, F., Paoletti, V., Morino, S., Antonini, G., Gazzaniga, P. P., Musca, A., and Basili, S. (2002) J. Neurol. Sci. 201 59–64 [DOI] [PubMed] [Google Scholar]
- 21.Mercer, S. E., Ewton, D. Z., Deng, X., Lim, S., Mazur, T. R., and Friedman, E. (2005) J. Biol. Chem. 280 25788–25801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Timchenko, N. A., Patel, R., Iakova, P., Cai, Z. J., Quan, L., and Timchenko, L. T. (2004) J. Biol. Chem. 279 13129–13139 [DOI] [PubMed] [Google Scholar]
- 23.Vandesompele, J., De, P. K., Pattyn, F., Poppe, B., Van, R. N., De, P. A., and Speleman, F. (2002) Genome Biol. 3 [DOI] [PMC free article] [PubMed]
- 24.Gao, M., Wilusz, C. J., Peltz, S. W., and Wilusz, J. (2001) EMBO J. 20 1134–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, W., Feifel, E., Holcomb, T., Liu, X., Spitaler, N., Gstraunthaler, G., and Curthoys, N. P. (1998) Am. J. Physiol. 275 F361–F369 [DOI] [PubMed] [Google Scholar]
- 26.Amack, J. D., Paguio, A. P., and Mahadevan, M. S. (1999) Hum. Mol. Genet. 8 1975–1984 [DOI] [PubMed] [Google Scholar]
- 27.Amack, J. D., and Mahadevan, M. S. (2001) Hum. Mol. Genet. 10 1879–1887 [DOI] [PubMed] [Google Scholar]
- 28.Jove, M., Planavila, A., Sanchez, R. M., Merlos, M., Laguna, J. C., and Vazquez-Carrera, M. (2006) Endocrinology 147 552–561 [DOI] [PubMed] [Google Scholar]
- 29.Ho, T. H., Savkur, R. S., Poulos, M. G., Mancini, M. A., Swanson, M. S., and Cooper, T. A. (2005) J. Cell Sci. 118 2923–2933 [DOI] [PubMed] [Google Scholar]
- 30.Krahe, R., Ashizawa, T., Abbruzzese, C., Roeder, E., Carango, P., Giacanelli, M., Funanage, V. L., and Siciliano, M. J. (1995) Genomics 28 1–14 [DOI] [PubMed] [Google Scholar]
- 31.Le, M. G., Ezzeddine, N., Capri, M., and it-Ahmed, O. (2008) Plos ONE 3 e1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marquis, J., Paillard, L., Audic, Y., Cosson, B., Danos, O., Le, B. C., and Osborne, H. B. (2006) Biochem. J. 400 291–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukhopadhyay, D., Houchen, C., Kennedy, S., Dieckgraefe, B. K., and Anant, S. (2003) Mol. Cell 11 113–126 [DOI] [PubMed] [Google Scholar]
- 34.Cosson, B., Gautier-Courteille, C., Maniey, D., Ait-Ahmed, O., Lesimple, M., Osborne, H. B., and Paillard, L. (2006) Biol. Cell 98 653–665 [DOI] [PubMed] [Google Scholar]
- 35.Chen, S. E., Jin, B., and Li, Y. P. (2007) Am. J. Physiol. 292 C1660–C1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, Y. P., and Reid, M. B. (2001) Curr. Opin. Rheumatol. 13 483–487 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.