Abstract
Phosphodiesterase 5 (PDE5) controls intracellular levels of cGMP through its regulation of cGMP hydrolysis. Hydrolytic activity of the C-terminal catalytic domain is increased by cGMP binding to the N-terminal GAF A domain. We present the NMR solution structure of the cGMP-bound PDE5A GAF A domain. The cGMP orientation in the buried binding pocket was defined through 37 intermolecular nuclear Overhauser effects. Comparison with GAF domains from PDE2A and adenylyl cyclase cyaB2 reveals a conserved overall domain fold of a six-stranded β-sheet and four α-helices that form a well defined cGMP binding pocket. However, the nucleotide coordination is distinct with a series of altered binding contacts. The structure suggests that nucleotide binding specificity is provided by Asp-196, which is positioned to form two hydrogen bonds to the guanine ring of cGMP. An alanine mutation of Asp-196 disrupts cGMP binding and increases cAMP affinity in constructs containing only GAF A causing an altered cAMP-bound structural conformation. NMR studies on the tandem GAF domains reveal a flexible GAF A domain in the absence of cGMP, and indicate a large conformational change upon ligand binding. Furthermore, we identify a region of ∼20 residues directly N-terminal of GAF A as critical for tight dimerization of the tandem GAF domains. The features of the PDE5 regulatory domain revealed here provide an initial structural basis for future investigations of the regulatory mechanism of PDE5 and the design of GAF-specific regulators of PDE5 function.
Intracellular concentrations of the second messengers cAMP and cGMP are tightly regulated by the rate of synthesis through cyclases and hydrolysis through cyclic nucleotide phosphodiesterases (PDEs)3 (1). The cGMP-specific, cGMP-binding phosphodiesterase, PDE5, is one of eleven identified PDE families. It has been characterized as the major cGMP-hydrolyzing PDE in numerous tissues such as lung, platelets, pulmonary artery smooth muscle cells, and the penile corpus cavernosum (2, 3). The abundance of PDE5 in smooth muscles and its role in regulating their contractile tone has made PDE5 an important drug target for the treatment of erectile dysfunction and pulmonary hypertension (2), leading to the development of potent PDE5 inhibitors, such as tadalafil (Cialis™) (4), vardenafil (Levitra™) (5), and sildenafil (Viagra™ and Revatio™) (6).
All mammalian PDE families are dimeric and contain a homologous, but distinct C-terminal catalytic domain, whereas they differ more in their family-specific N-terminal regulatory domains. The regulatory regions of PDE2, -5, -6, -10, and -11 contain tandem GAF domains (acronym derived from the first identified GAF proteins, namely mammalian cGMP-regulated PDEs, Anabaena adenylyl cyclases and the Escherichia coli transcription factor FhlA) (7). GAF domains represent a large family of signaling and sensory domains found in numerous proteins that display a wide variety of functions, including light detection in plants (8), sodium sensing (9) and, as recently reported, enzymatic activity (10). Structural similarities between the ∼150 residue GAF domains and the PAS domain family indicate a close evolutionary relationship between these two domain families, possibly forming a superfamily of photo-transducing and signaling domains with common evolutionary origin (11, 12). The regulatory domain of PDE5 contains two GAF domains (according to the nomenclature, the first domain is labeled GAF A, the second one GAF B) and a preceding phosphorylation site at Ser-92 (numbering from murine PDE5A1) (Fig. 1A). Regulation of PDE5 is controlled via protein kinase G-dependent phosphorylation (13, 14) and allosteric cGMP binding to GAF A. Although the details of the mechanisms by which PDE5 activity is regulated remain to be defined, both events are thought to increase the catalytic activity of PDE5 through conformational changes (15, 16).
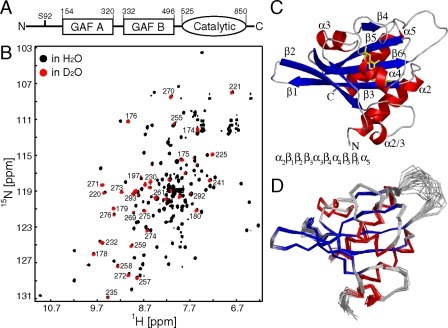
FIGURE 1.
A, domain organization of mouse PDE5A1. Construct borders for GAF A and GAF B are shown. S92 indicates the phosphorylation site. B,[1H,15N]HSQC spectrum of cGMP-bound PDE5 GAFA154-320 recorded in 90% H2O/10% D2O(black) and ∼5 days after dissolution of lyophilized GAFA154-320 in 99.9% D2O(red). Approximately 40 unexchanged backbone amide proton resonances were detected. Assignments of selected unexchanged backbone amides are shown. Residue numbers correspond to the full-length mouse PDE5A1. The majority of the non-exchanging amides are located in the anti-parallel β-sheet, with some in helices α3, α4, and α5 (supplemental Fig. S1). C, ribbon diagram of PDE5A GAF A in the presence of cGMP. α-Helices are shown in red, β-strands in blue, and loops in gray. cGMP is shown in yellow sticks. Note: The N-terminal α-helix observed in the structure is named “α2” with analogy to other GAF domain structures. D, backbone traces of the 20 lowest energy structures of PDE5 GAF A.
Though homologous in sequence and topologically similar, not all PDE GAF domains bind cyclic nucleotides and may instead be involved in dimerization or play other unknown regulatory roles. There is no experimental evidence that PDE5 GAF B binds cGMP (or in fact any small molecule). Instead, it has been suggested to be critical for dimerization of the holoenzyme PDE5 (17). Recently, it has been shown that the presence of GAF B affects the affinity of the catalytic domain for PDE5 inhibitors (18), indicating that GAF B potentially alters catalytic site activity through inter-domain interactions.
The question of how events that involve the regulatory N-terminal region of PDE5 regulate the catalytic domain and its activity remains to be answered. Detailed structural information of the involved elements at high resolution is necessary to understand the complexity of the allosteric regulation and the corresponding conformational changes. Whereas high resolution structures of the catalytic domain of PDE5 have been determined in the absence and presence of the three above mentioned inhibitors (19-22), a three-dimensional structure of full-length PDE5, or of any full-length PDE, remains elusive. To date, structural information on the PDE5 holoenzyme is limited to electron microscopy images at 28 Å resolution (23). These suggest an extended dimeric structure with distinct domains, in which the subunits are aligned in a head-to-head fashion. Neither GAF domain appears to be in contact with the putative catalytic domain. A crystal structure of the tandem GAF domains of mouse PDE2A is currently the only published high resolution structure of PDE GAF domains (24). In contrast to PDE5, this structure revealed that GAF A is involved in dimerization, whereas GAF B has a cGMP molecule bound. Furthermore, no structure of an unliganded cyclic nucleotide binding GAF domain has been reported, and little is known about the cyclic nucleotide-dependent conformational change and mechanism of binding.
Here, we present an NMR solution structure of the cGMP-bound GAF A domain from PDE5A. Although similar in overall domain fold, the nucleotide binding pocket differs from previously solved structures of cNMP-bound GAF domains. NMR studies of the tandem GAF domains of PDE5 reveal a dynamic GAF A domain in the absence of cGMP indicating that cGMP binding causes large conformational changes as cGMP locks GAF A into a less flexible and more structurally defined state. We also identify the N-terminal region directly proximal to GAF A as a critical dimerization interface of the regulatory domain. The structure presented here is the first reported NMR solution structure of any GAF domain and gives insight into the nucleotide binding mechanism. The structural details will be useful for the design of a GAF-specific small molecule inhibitor or activator and assist further investigations of the structural basis of PDE5 regulation and the conformational consequences of cGMP binding.
MATERIALS AND METHODS
Construct Design—Several constructs were designed as truncations of full-length mouse PDE5A1 through a modified two-step site-directed mutagenesis protocol to efficiently loop out large DNA segments of the expression vector (25) and verified by sequencing. The expression vector is a derivative of the pMW172 vector with a C-terminal LE(H)6 tag and an N-terminal Met. Our initial construct design for GAF A was based on sequence homology with PDE2A GAF B and included residues 125-320 (GAFA125-320). However, this construct was not optimal for NMR, because we were able to only obtain incomplete backbone NMR assignments (26). To optimize the construct for the determination of the solution structure we performed limited trypsin proteolysis and identified a very stable domain that included residues 154-320. Homology modeling with SWISS-MODEL (27) based on the crystal structure of mouse PDE2A GAF B (24) revealed the presence of a putative hydrophobic patch on the surface of the PDE5A GAF A domain, which may be involved in an unidentified intermolecular interaction. Two residues on this surface (Ala-295 and Ile-302) were mutated to Glu. Addition of these surface charges improved the NMR behavior and increased expression levels, yielding a construct suitable for solution structure determination via NMR. This construct was termed GAFA154-320. Two constructs containing the tandem GAF domains (GAF A plus GAF B) were designed: GAFAB134-496 (residues 134-496) and GAFAB154-496 (residues 154-496). A construct containing only the GAF B domain (GAFB332-496, residues 332-496) was also used in this study. The D196A mutation was introduced through site-directed mutagenesis into GAFA154-320, GAFA125-320, and GAFAB154-496.
NMR Experiments and Resonance Assignments—All NMR experiments were performed at 37 °C on an in-house Bruker 500 MHz DMX or on Varian INOVA (600, 800, or 900 MHz) spectrometers at Pacific Northwest National Laboratories, Richland, WA. Data were processed and analyzed using NMRDraw (28) and NMRView (29). 1 mm [15N,13C,2H]GAFA154-320 in NMR buffer with 10% D2O was used for triple-resonance experiments. 1.2 mm [15N,13C]GAFA154-320 in NMR buffer in 99.9% D2O was used for side-chain resonance assignments and 13C-edited NOESY data collection. A 15N-edited NOESY was collected on 1.2 mm [15N,13C]GAFA154-320 in NMR buffer with 10% D2O. Backbone resonances were manually assigned using the following TROSY-based triple-resonance experiments (30, 31): heteronuclear [1H,15N]HSQC, HNCA, HN(CO)CA, HNCACB, and HN(CO)CACB, HNCO. Side-chain resonances were assigned from HCCH-COSY, HCCH-TOCSY, HC(CO)NH-TOCSY, 13C-edited NOESY (mixing time = 80 ms), and 15N-edited NOESY (mixing time = 80 ms). Assignments are deposited at BioMagResBank (accession no. 15734). Abovementioned NOESY experiments were used to derive NOE-based distance restraints. Intermolecular NOEs between cGMP (natural abundance) and the protein were detected in the NOESY experiments and assigned manually.
Structure Calculation—CYANA 2.1 (32) was used for assignments of NOEs with the NOEassign module (which is based on CANDID (33)) and for the initial structure calculation of the protein without consideration of cGMP. Dihedral backbone angles restraints were predicted from backbone assignments and calculated with TALOS (34). Only dihedral angle restraints with good fits were included in the calculations. Initially, twelve inter-strand hydrogen bond restraints for β-sheet formation were included based on slowly exchanging amide-protons as determined through H-D exchange experiments and 15N-edited NOESY (perdeuterated protein, mixing time = 200 ms). Several rounds of manual NOE assignments and iterations of CYANA yielded a set of structures with an average backbone r.m.s.d. of 0.72 Å (residues 155-302) but a collapsed cGMP binding pocket. To dock cGMP based on intermolecular NOEs into the center of the protein structure, calculations with CYANA-derived input structures, CNS 1.1 (35) and the all-hydrogen PARALLHDG5.3 force field (36) were carried out. The coordinates for cGMP were obtained from the crystal structure of the GAF domains of PDE2A (PDB code: 1mc0) (24). Upon addition of hydrogen atoms with PRODRG2 (37), topology and parameter files for cGMP were calculated with XPLO2D (38). The 20 lowest energy structures out of 100 accepted structures were analyzed with PROCHECK-NMR (39) and deposited at the Protein Data Bank (PDB code 2k31). Structures were visualized by PyMOL.4
Quantification of Co-purified Cyclic Nucleotides—Purified protein (200 μm, expressed and purified without addition of cAMP or cGMP) was denatured in 6 m GdnHCl (added in dry form) in 25 mm sodium phosphate, pH 6.5, and 1 mm β-mercaptoethanol at 95 °C. 200 μl was injected onto a Superose-12 column and eluted in 6 m GdnHCl in 25 mm sodium phosphate, pH 6.5. Absorbance at 260 nm was recorded and used to quantify the area of the peak determined with the Galaxie 1.7 software (Varian). Extinction coefficients for the protein constructs at 280 nm were calculated with the ProtParam tool (41) and extrapolated to 260 nm by measuring the 260 nm/280 nm absorption ratio. Extinction coefficients at 260 nm for cAMP and cGMP used in the calculations were 15,000 and 12,320 m-1 cm-1, respectively. To determine the stoichiometry of cGMP binding to GAFAB154-496 WT, 400 μm protein was incubated with 1 mm cGMP for 30 min at 22 °C. Free nucleotide was removed by a HiTrap desalt column (Amersham Biosciences), and bound cGMP was quantified through GdnHCl denaturation and by using a size exclusion column as described above. Additional materials and methods are provided in the supplemental data.
RESULTS AND DISCUSSION
cGMP-bound PDE5A GAF A Has a Well Defined GAF Domain Fold—The cGMP-bound GAFA154-320 domain of PDE5A is highly soluble and, as determined by the rotational correlation time, behaves mainly as a monomer of 20 kDa at concentrations up to 1 mm at 37 °C (see below). The two-dimensional [1H,15N]HSQC spectrum of GAFA154-320 shows good chemical shift dispersion, and the number of signals is consistent with protein size and amino acid content (Fig. 1B). Nearly complete backbone assignments were obtained for residues 154-302, whereas 18 C-terminal residues were either not observed and/or remained unassigned. Resonance assignments within the structured core of GAFA154-320 are essentially identical to those observed for a longer GAFA125-320 construct (26), thus confirming that the structure is not significantly affected by the truncation and two surface mutations made to optimize the construct (see “Materials and Methods”). However, the completeness of resonance assignments extends further at the N and C termini compared with the longer construct. Importantly, ∼95% of the side-chain chemical shifts of residues 154-302 were assigned by analysis of TOCSY, COSY, and NOESY data.
The solution structure of the cGMP-bound GAFA154-320 was determined through a two-step procedure combining the structure calculation software packages CYANA (32) and CNS (35) (see “Materials and Methods”). Slowly solvent-exchanging amide protons (Fig. 1B and supplemental Fig. S1) within the β-sheet were used to assign inter-strand hydrogen bond restraints and were confirmed through long range 1HN-1HN NOEs observed in a 15N-edited NOESY acquired on perdeuterated protein. Twelve such inter-strand hydrogen bonds were included in the initial structure calculation process performed by CYANA, and the total number of hydrogen bond restraints within secondary structure elements was successively increased to 50 for the final structure calculation in CNS (additional restraints and structural statistics are provided in supplemental Table S1). Intermolecular NOEs between GAF A and natural abundance cGMP observed in a 13C-edited NOESY (37 NOEs total) were input to define the ligand orientation.
The PDE5A GAF A solution structure consists of a central six-stranded anti-parallel β-sheet and four α-helices with an order of the secondary structure elements of αβββαβαββα (Fig. 1C). The topology of the six β-strands forming the anti-parallel β-sheet is 3-2-1-6-5-4. The core structure is well defined by NMR observables; the 20 lowest energy structures align with a mean global backbone r.m.s.d. of 0.53 ± 0.10 Å for residues 157-277 and 285-302 (Fig. 1D). Residues 278-284 comprise a glycine-rich loop between β6 and α5 that is poorly defined by NMR observables.
PDE5 GAF A Displays Evolutionary Conserved GAF Domain Fold—Comparison of the domain topology with the crystal structures of cGMP-bound GAF B domain from PDE2A (24) and cAMP-bound GAF B domain5 from the Anabaena adenylyl cyclase cyaB2 (42) reveals that all adopt a conserved fold with a six-stranded amphipathic β-sheet, two α-helices that cap the binding pocket (α3 and α4), and two α-helices on the backside of the β-sheet (α2 and α5) (Fig. 2A). PDE2A GAF B and cyaB2 GAF B superimpose structurally with Cα r.m.s.d. values of 2.26 Å and 2.00 Å with PDE5A GAF A, respectively. The structural conservation is remarkable considering the more than two-billion-year evolutionary distance (12) and the low sequence conservation among the three cNMP-binding GAF domains (20.7% identity) (Fig. 2B).
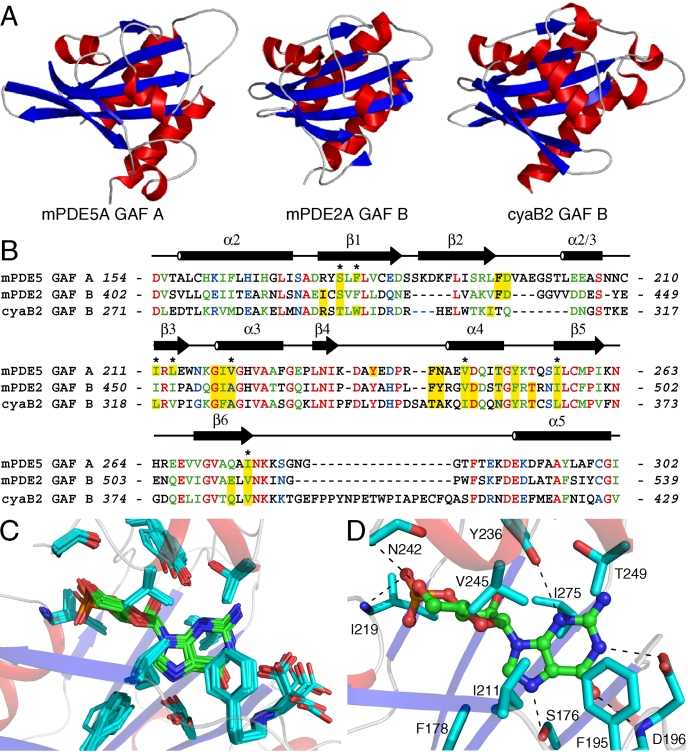
FIGURE 2.
A, comparison of cNMP-binding GAF domains from PDE5A, PDE2A, and cyaB2. The structures of PDE5A GAF A, PDE2A GAF B (1mc0), and cyaB2 GAF B (1ykd) were aligned using PyMOL.4 The r.m.s.d. of the α-carbons of the aligned region is 2.26 Å (PDE5-PDE2) and 2.00 Å (PDE5-cyaB2). α-Helices are shown in red, β-strands in blue, and loops in gray. B, sequence alignment of mouse PDE5A GAF A, mouse PDE2A GAF B, and cyaB2 GAF B. Identical residues are highlighted in red, strongly similar residues in green, and weakly similar residues in blue. Residues within the cNMP binding pocket are highlighted in yellow. Alignment was performed with ClustalW (40). Asterisks highlight PDE5 residues that were involved in measured intermolecular NOEs. Secondary structure elements from PDE5A are indicated. C, overlay of cGMP and binding residues from the 20 lowest energy structures of PDE5A GAF A after alignment of residues 157-277 and 285-302. D, PDE5A GAF A binding pocket. Binding residues are displayed as sticks with carbon atoms in cyan. Hydrogen bonds are shown as black dashed lines.
Several notable differences are apparent when comparing the GAF domain structures. First, the β1- and β2-strands of PDE5A are significantly longer than in the other structures resulting in longer dimensions for the GAF domain (Fig. 2, A and B). Second, the β2-β3 connecting loop is significantly longer in PDE5A (17 residues) than in PDE2A GAF B (10 residues) and cyaB2 GAF B (7 residues) (Fig. 2B). Additionally, the loop of PDE5A contains an α-helical turn (Leu-203 through Ser-207, labeled “α2/3” in Fig. 1C) that is not present in either of the two other structures (see below). Third, the loop that connects β6 and α5 varies in length among the three domains. PDE2A has a short and well defined loop, whereas the loop in PDE5A is glycine-rich and appears disordered in the solution structure. In cyaB2, an insert of ∼15-20 residues extends the loop and contains short stretches of helical secondary structure.
Although variable in length and definition, the β5-α6 loop connects a conserved Lys and Phe, which are both part of the “NKFDE” motif that is conserved among cNMP-binding GAF domains. Although initially thought to be involved in cGMP binding (43, 44), the motif is not within the binding pocket of any of the determined GAF domain structures. It has been proposed to be essential for stabilizing the closed domain fold (24, 42) and/or for allosteric signaling by transmitting conformational changes to the catalytic domain (45). In the ensemble of the 20 lowest energy structures of PDE5A GAF A, the side chains of Asn-276, Phe-285, Asp-289, and Glu-290 (i.e. N, F, D, and E of the motif) are well defined and mainly buried, whereas the side chain of Lys-277 is solvent-exposed and appears flexible. The roles of the disordered loop, its extension in cyaB2, and the conserved “NKFDE” motif remain enigmatic.
Nucleotide Binding Site of PDE5A GAF A—The scarcity of carbon-bound hydrogens in cGMP posed a challenge for defining the ligand binding pocket from NMR observations. Only five ribose protons and one guanine proton are observable by NMR. We detected the proton resonances of cGMP bound to [15N,13C]GAFA154-320 in a 13C-filtered one-dimensional spectrum acquired in D2O and assigned them by comparison to the one-dimensional proton NMR spectrum of free cGMP in D2O. This enabled us to manually assign 37 peaks in the 13C-edited NOESY spectrum as protein-ligand NOEs that remained unassigned after multiple rounds of automated assignments by CYANA (using only protein resonance assignments). Intermolecular NOEs between protein and cGMP involved the side chains of Ser-176, Phe-178, Ile-211, Leu-213, Val-220, Val-245, Ile-256, and Ile-275 (labeled with asterisks in Fig. 2B). The mean r.m.s.d. of heavy cGMP atoms in a structurally aligned ensemble of the 20 lowest energy structures was 0.40 ± 0.11 Å, indicating that the cGMP binding pocket is well defined by the NMR observables (Fig. 2C).
In PDE5A GAF A, cGMP adopts an anti-conformation, similar to cGMP in PDE2A (24) and cAMP in cyaB2 (42), and is deeply buried in the binding pocket (supplemental Fig. S2A). Only the N2 amino group of guanine is solvent-accessible, similar to the solvent accessibility of cGMP in PDE2A GAF B. Notably, the negatively charged 3′,5′-cyclic phosphate group is completely buried and points toward the positive helix dipole at the N-terminal end of helix α3. In addition, the backbone amides of Asn-242 and Ile-219 are within hydrogen bond distance (3.1-3.4 Å in 18 of the 20 lowest energy conformers and 2.8-3.4 Å, 16/20, respectively) of the phosphate oxygens6 (Fig. 2D). The hydroxyl groups of Ser-176 and Tyr-236 are within hydrogen bond distance of the unprotonated N7 (2.7-3.2 Å, 18/20) and N3 (2.7-3.4 Å, 15/20) of the guanine ring, respectively. The guanine ring is packed against hydrophobic side chains: Phe-195 is stacked against the guanine ring, whereas Phe-178, Ile-211, Tyr-236, Val-245, Thr-249, and Ile-275 provide additional hydrophobic packing. Asp-196 is oriented to provide two important hydrogen bonds to the guanine ring. Its backbone amide group is within hydrogen bond distance of the O6 carbonyl of cGMP (2.8-3.4 Å, 18/20). This role is consistent with the slow solvent exchangeability of the amide proton of Asp-196, observed in H-D exchange experiments. Additionally, the side-chain carboxyl group is within hydrogen bond distance of the protonated N1 of cGMP in 11 of the 20 lowest energy structures (2.8-3.4 Å, 11/20). This hydrogen bound orientation of Asp-196 is similar to the position of Asp-439 in PDE2A GAF B and therefore most likely the relevant orientation as it provides additional binding stability and nucleotide selectivity (see below).
Structural Determinants of Nucleotide Selectivity and Affinity—The regulatory GAF domains of PDE5A and PDE2A are selective for cGMP over cAMP, whereas those of cyaB2 selectively bind cAMP. The tandem GAF domains of PDE5A have a several thousand-fold preference for cGMP over cAMP (IC50 of 2.2-3.8 nm and 8-20 μm, respectively, Fig. 4D), whereas the tandem GAF domains of PDE2A are less selective with only a ∼20-fold preference for cGMP over cAMP (IC50 of 7 nm and 146 nm, respectively) (46). Although both have high affinity for cGMP, PDE5A has a higher selectivity for cGMP due to much lower cAMP binding affinity.
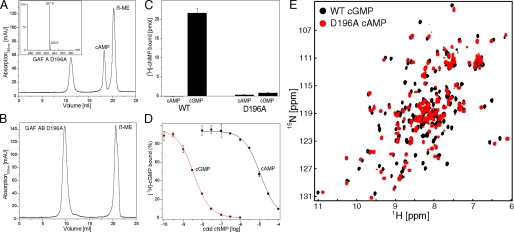
FIGURE 4.
Characterization of D196A mutant. A, SEC profile of PDE5A GAFA125-320 in 6 m GdnHCl. Inset shows the liquid chromatography-mass spectrometry spectrum from a small molecule peak. B, SEC profile of PDE5A GAFAB154-496 in 6 m GdnHCl. C, cyclic nucleotide binding to GAFAB154-496 WT and D196A. 0.5 μm protein and 1 μm [3H]cNMP were incubated in 50 μl for 16 h at 37 °C before being filtered and counted. Data shown are representative for three independent experiments. Values are mean ± S.D. of duplicate determinations. D, selectivity for cAMP and cGMP binding of tandem GAF domains. Various concentrations of unlabeled cAMP and GMP were incubated with GAFAB154-496 (1 nm) and [3H]cGMP (2 nm). Binding was determined using filter binding assays. Data shown are representative for three independent experiments. Values are mean ± S.D. of duplicate determinations. E,[1H,15N]HSQC spectra of cGMP-bound GAFA154-320 (black) and cAMP-bound GAFA154-320 D196A (red).
To investigate the basis of cyclic nucleotide selectivity and altered affinity, we structurally aligned and analyzed the cGMP binding pockets of PDE5A GAF A and PDE2A GAF B (supplemental Fig. S2C) (24) with the cAMP binding pocket of cyaB2 GAF B (supplemental Fig. S2D) (42). Residues within the cyclic nucleotide binding pocket are highlighted in the sequence alignment (Fig. 2B).
cGMP and cAMP have different substitutions at the pyrimidine ring positions C2, N1, and C6. Asp-196 in PDE5A and Asp-439 in PDE2A both provide similar hydrogen bonds from backbone amide and side-chain carboxyl group to this region of cGMP (24). In contrast, the hydroxyl group of cyaB2 Thr-293 makes hydrogen bond contacts with N6 (and N7) of cAMP (42). Arg-291 of cyaB2 makes contact to N1 of cAMP while the backbone of Thr-309, the equivalent residue to Asp-196 and Asp-439, is out of range to make hydrogen bond contact with cAMP. In PDE2A, the N2 amino group of cGMP is bound by a water-mediated hydrogen bond with Thr-488 and is the only solvent-accessible region of the ligand. In PDE5A, N2 is similarly solvent-accessible, but no hydrogen bond acceptor is nearby. Our NMR data cannot provide experimental evidence for water-mediated hydrogen bonds.
A striking difference between the three GAF domains is the orientation of Tyr-236 in PDE5, which is conserved as Tyr-475 in PDE2A GAF B and Tyr-344 in cyaB2 GAF B. In PDE5, the aromatic side chain points into the binding pocket, and its hydroxyl group is within hydrogen bond distance to N3 of the guanine ring while the aromatic ring is parallel to the sugar ring. In contrast, the side chains of Tyr-475 and Tyr-344 point away from the pocket and are partially solvent-exposed.
The loop between β2 and β3 may also contribute to nucleotide preference. PDE5A GAF A has a short helical turn (α2/3) within its β2-β3 loop that does not exist in the cAMP-binding cyaB2 or in the low selectivity PDE2A. Two residues play a crucial role in stabilizing the loop in PDE5A. The side chain of Leu-203 is buried and packed against α2, whereas Val-197 is buried and has a non-exchangeable backbone amide proton (Fig. 1B), which is oriented toward the β1-strand as a hydrogen bond donor. Together, these residues stabilize the position of Asp-196 to make contact with cGMP, indirectly affecting nucleotide selectivity. They may also stabilize the closed conformation of PDE5A GAF A and contribute to the very slow dissociation rate of cGMP.
The α2/3-helical turn and the associated longer loop could be a hallmark of higher cGMP selectivity. Supporting this hypothesis is the fact that GAF A from PDE6, which has been reported to bind cGMP preferentially over cAMP by >100,000-fold (47) possesses an extended loop of 18-19 residues (supplemental Fig. S3). Its crystal structure reveals a short α-helical turn in a similar position to the one in PDE5A GAF A.7 In contrast, the crystal structure of cAMP-bound GAF B domain of PDE10A (52) contains an extended loop (supplemental Fig. S3) but does not form helix α2/3. The loop points away from the binding pocket and does not provide stabilization similar to that observed in PDE5A.
In summary, comparison of the three binding pockets reveals that the majority of the protein-ligand hydrogen bond network is conserved. Based on subtle structural differences, we hypothesize that two main factors control nucleotide selectivity. First, a conserved Asp at position 196 provides critical hydrogen bonds to cGMP and increased selectivity for cGMP over cAMP. This Asp is not present in cyaB2. The role of Asp-196 for cGMP specificity in PDE5A was further investigated by mutational analysis (see below). Second, the length of the β2-β3 connecting loop and the presence of a short α2/3-helix may affect selectivity through stabilizing a closed binding pocket that locks the cGMP-contacting residues into position.
cGMP-dependent Conformational Change of PDE5 GAF A Domain—The cGMP-bound GAF structure presented here displays a compact “closed” domain conformation, in which the ligand is almost entirely buried. Because there is no clear way for the cGMP to dissociate, a conformational change that opens the binding site to solvent is expected. This may suggest that the apo-GAF domain is in an “open” conformation. To date, the only structures available of apo-GAF domains are for domains that are not known to bind a ligand. Such structures, for example PDE2A GAF A (24) and the yeast protein of unknown function, YKG9 (48), are in a closed conformation that could not accommodate a nucleotide ligand. To investigate the nature of the cGMP-induced conformational change in GAF A and characterize the structural effects of cGMP binding, we performed two-dimensional NMR experiments.
The apo-GAF A domain has limited solubility, and only low amounts of protein are found in the soluble fraction of a bacterial lysate. Nevertheless, we were able to produce sufficient purified unlabeled apo-GAFA125-320 to collect one-dimensional proton NMR spectra (∼0.1 mm apo-GAFA125-320). The spectrum has broad signals between 7 and 8 ppm, with no peaks downfield of 8.5 ppm indicative of amide groups in random-coil conformation (Fig. 3A) and, unlike cGMP-bound GAF A (Fig. 3B), lacks upfield-shifted methyl peaks characteristic of a well folded protein.
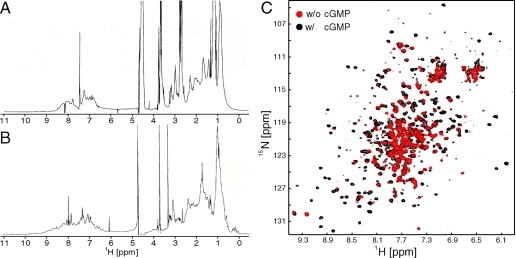
FIGURE 3.
cGMP-dependent conformational change of GAF domains. A, one-dimensional proton NMR spectrum of apo-GAFA125-320. B, one-dimensional proton NMR spectrum of cGMP-bound GAFA125-320. C,[1 H,15N]TROSY-HSQC spectra of cGMP-bound PDE5 GAFAB154-496 recorded in the absence (red) and presence (black) of cGMP.
In contrast to constructs containing only GAF A, apo-GAFAB154-496, a 40-kDa construct that contains both GAF domains, is soluble and stable over several hours at 37 °C. In the absence of cGMP, the [1H,15N]TROSY HSQC NMR spectrum contains significantly fewer resonances than expected based on the number of residues in GAFAB154-496, and the peaks observed display a range of intensities (Fig. 3C, red spectrum). Upon addition of cGMP, many new resonances appear in the spectrum (Fig. 3C, black spectrum). Far-UV CD spectra indicate that the apo- and cGMP-bound forms of GAFAB154-496 have similar secondary structure content (data not shown). Overlay of the NMR spectra of the single GAF domains, GAFA154-320 and GAFB332-496, onto the spectra of GAFAB154-496 clearly identifies the majority of resonances that appear upon cGMP binding as GAF A resonances and the majority of peaks observable in the spectrum of apo-GAFAB154-496 as GAF B resonances (supplemental Fig. S5). A subset of GAF B resonances is visible in the apo-GAFAB154-496 spectrum (labeled by rectangles in supplemental Fig. S5E), whereas other resonances are broadened beyond detection (labeled by circles in supplemental Fig. S5E). Thus the chemical environment of some GAF B amide groups is affected by apo-GAF A. This could be either a direct effect in which GAF A interacts with some part of GAF B or an indirect effect, in which the dynamic state of GAF A is translated further to GAF B and causes affected GAF B resonances to broaden.
Taken together, these observations reveal several features of the tandem GAF domains of PDE5A. First, the two domains behave largely independent of each other in the presence of cGMP, because the spectra of the individual domains overlay well with the spectrum of the cGMP-bound tandem GAF domains (supplemental Fig. S5C). Second, the GAF domains affect each other in the cGMP-free state. GAF B solubilizes apo-GAF A, and a subset of its resonances is broadened in the spectrum of the apo tandem domains, suggesting that GAF A and B interact with each other. This hypothesis is further supported by the significant modulation of nucleotide binding properties of GAF A by GAF B as shown in D196A mutant constructs (see below). This suggests that GAF B stabilizes the open state of GAF A and shifts the equilibrium toward the apo-GAF A domain, which, in turn, lowers the ligand binding affinity. Third, the extreme broadening of GAF A resonances indicates that the ligand-free domain samples multiple conformations on a timescale that is intermediate on the NMR timescale. That the CD spectra of the tandem GAF domains are essentially identical for the apo- and ligand-bound states indicates that the empty GAF A has its secondary structural elements formed, whereas the NMR broadening suggests that they do not adopt a defined tertiary structure in the absence of cGMP.
The results are consistent with a model, in which the empty GAF A is dynamic as opposed to existing in a single open conformation with a preformed binding pocket. GAF B stabilizes this dynamic state of GAF A by direct interaction, which is released through binding of cGMP to GAF A. cGMP stabilizes the closed conformation, which essentially buries the ligand and protects it from solvent. A possible opening mechanism could involve the β2-β3 connecting loop and helix α4. H-D exchange data indicate that the backbone amides of α3 are mainly solvent-protected while the amides of α4 are more exchangeable (as indicated by lower intensities in supplemental Fig. S1). This suggests that α4 undergoes more frequent excursions from the closed cGMP-bound state that allow solvent accessibility, whereas α3 remains in its buried position. If both the loop and α4 are removed from the structural coordinates, cGMP is solvent-accessible, and a path in and out of the binding pocket is formed. In such a scenario, the positively polarized pocket provided by α3 and the β-sheet is still intact and could attract the negatively charged phosphate group of the cyclic nucleotide.
Nucleotide Specificity Switch through D196A Mutation—The structure of the binding pocket indicates that Asp-196 may provide two hydrogen bonds to the guanine ring of cGMP at the edge of the base where it differs from cAMP. To investigate the importance of this interaction in mediating selectivity, we mutated Asp-196 to Ala in GAFA125-320. Unexpectedly, GAFA125-320 D196A could be purified from bacteria without addition of cyclic nucleotide and was stable at high concentrations. The one-dimensional proton NMR spectrum of D196A revealed upfield-shifted methyl groups and downfield-shifted amides indicative of a well folded protein (supplemental Fig. S4). Unfolding of GAFA125-320 D196A in 6 m GdnHCl under reducing conditions and subsequent size exclusion chromatography revealed the presence of a bound small molecule (Fig. 4A). The UV spectrum is characteristic of cAMP (data not shown), and the mass to charge ratio (m/z) determined by electronspray mass spectrometry was 327.9, identical to the cAMP standard (Fig. 4A, inset). Because we did not add cAMP during the protein purification, we hypothesize that GAFA125-320 D196A binds endogenous cAMP from E. coli with high affinity. The very slow off-rate hampered our ability to determine the cyclic nucleotide binding affinities of GAFA125-320 D196A, because we were unable to measure cAMP (or cGMP) binding to GAFA125-320 D196A in [3H]cNMP filter binding assays.
In marked contrast to GAFA125-320 D196A, no cyclic nucleotide co-purifies with the tandem GAFAB154-496 D196A (Fig. 4B), suggesting that the presence of GAF B lowers the cAMP affinity, similar to what has been observed for cGMP affinities in wild-type constructs (17). As indicated by our NMR data for GAFAB154-496 WT, GAF B presumably interacts with the apo-GAF A domain and influences its nucleotide binding properties. Because GAFAB154-496 D196A does not contain bound cAMP, we attempted to determine binding affinities for cAMP and cGMP. We did not detect binding of either cAMP or cGMP above baseline using a [3H]cNMP filter binding assay. Even at micromolar concentrations and 16 h of incubation at 37 °C no binding was detected (Fig. 4C). Thus the D196A mutation lowers cGMP binding affinity of the tandem GAF domains below the detectable range of the filter binding assay. Remarkably, in the context of the tandem GAF domains, the cAMP affinity was also lower than the detection limits.
Because GAFAB154-496 WT was stable and soluble in the absence of cGMP, we were able to measure the relative nucleotide affinity for cGMP and cAMP using [3H]cNMP filter binding competition assays as previously described (49). GAFAB154-496 WT (1 nm) was incubated with [3H]cGMP (2 nm) in the absence and presence of various concentrations of unlabeled cAMP or cGMP. Although cAMP competes with cGMP for binding with an IC50 value of 8-20 μm (n = 3), the IC50 for cGMP was 2.0-3.8 nm (n = 3), indicating a 2,000- to 10,000-fold preference for cGMP over cAMP (Fig. 4D).8 The preference is significantly larger than the previously reported ∼100-fold selectivity for the tandem GAF domains from human PDE5A (residues 1-539) (17). This may be due to the different construct borders and/or species origin or may indicate an affinity-altering role of the N-terminal ∼150 residues.
To assess the structural and functional effects of the D196A mutation, we compared the [1H,15N]HSQC spectra of GAFA154-320 D196A and GAFA154-320 WT (Fig. 4E). These spectra are significantly different from each other, with a majority of the peaks perturbed. Although the extent of the spectral differences is much larger than would be expected for a localized change in structure and may well represent a large difference in the structure of the two species, care should be taken in interpreting this observation. Small changes in the orientation of bound nucleotide and/or aromatic side chains could have dramatic effects on the chemical shifts of nearby resonances.
Taken together, the D196A point mutation displays intriguing features. As predicted by the structure it disrupted cGMP binding. In the isolated GAF A domain, it also increased the binding affinity for cAMP. In contrast, in the context of the tandem GAF domains, and presumably in the context of the full-length PDE5, the affinity for either nucleotide was lowered beyond detection of the [3H]cNMP filter binding assay. This implies that the presence of GAF B lowers the affinity and/or increases the off-rate for cAMP.
Dimerization Interface of the Tandem GAF Domains—All PDEs are reported to be dimeric, although the functional role of dimerization remains unknown. Dimerization is not required for catalytic activity, because the isolated catalytic domain of PDE5 is monomeric but displays near maximal activity (19, 50). The N-terminal regulatory region with its tandem GAF domains has previously been identified as the dimer interface (17, 51). To identify regions within the tandem GAF domains of PDE5A that are critical for dimerization, we determined the sedimentation coefficient and Stokes radii of a series of well behaved GAF domain constructs by analytical ultracentrifugation (AUC) and analytical size exclusion chromatography (SEC). Molecular weights were calculated from Stokes radii and the sedimentation coefficients and used to assign the oligomeric state of each construct under the conditions studied (Table 1).
TABLE 1.
Physical characteristics of the GAF domains from PDE5A at 25 °C
Stokes radii were determined by analytical SEC. Sedimentation coefficients were determined by AUC. Values are given as the mean ± S.D.
| mPDE5A1 construct | Predicted molecular mass | Stokes radius | Sedimentation coefficient | Calculated molecular mass | Assigned quaternary structure |
|---|---|---|---|---|---|
| kDa | Å | s | kDa | ||
| GAFA154-320 | 20.04 | 22.32 ± 0.39 (n = 3) | 3.35 ± 0.07 (n = 3) | 30.84 | Dimer |
| 2.25 ± 0.17 (n = 3) | 20.70 | Monomer | |||
| GAFA125-320 | 22.98 | 27.29 ± 0.46 (n = 3) | 3.70 ± 0.09 (n = 3) | 41.46 | Dimer |
| 2.32 ± 0.13 (n = 3) | 26.10 | Monomer | |||
| GAFB332-496 | 19.98 | 27.46 ± 0.28 (n = 3) | 3.35 ± 0.07 (n = 3) | 40.70 | Dimer |
| GAFAB154-496 | 39.86 | 39.51 ± 0.87 (n = 3) | 4.59 ± 0.17 (n = 3) | 74.90 | Dimer |
| 29.56 ± 0.93 (n = 3) | 3.32 ± 0.05 (n = 3) | 40.49 | Monomer | ||
| GAFAB134-496 | 42.10 | 45.45 ± 1.02 (n = 3) | 4.46 ± 0.09 (n = 4) | 83.72 | Dimer |
The AUC data are consistent with the existence of an equilibrium mixture of monomeric and dimeric species for both GAF A constructs (GAFA154-320 and GAFA125-320). Thus, under the experimental conditions (400 μm protein, 25 °C), the GAF A constructs are in a monomer-dimer equilibrium, indicative of a dissociation constant in the 10-4 m range. In contrast, SEC profiles showed only single peaks, most likely from dilution that occurs in the course of the experiment. Calculation of the molecular weight for GAFA154-320 and GAFA125-320 using the measured Stokes radius will therefore be somewhat skewed. The Stokes radii for the two GAF A constructs differ by ∼5Å. Because the additional 29 residues contained within GAFA125-320 would not be predicted to result in this large difference in Stokes radius, the result indicates that the dimer formed by GAFA154-320 is weaker and separates during SEC, whereas GAFA125-320 forms a dimer with stronger affinity. NMR behavior of the constructs at 37 °C and ∼400 μm is consistent with this conclusion. Although the HSQC spectrum of GAFA154-320 showed narrow line widths (Fig. 1B), consistent with its monomeric molecular weight, a comparable signal-to-noise ratio could only be detected in a TROSY-HSQC spectrum for GAFA125-320, consistent with its dimeric molecular weight. To confirm the oligomeric state of GAFA154-320 under NMR conditions of ∼1 mm and 37 °C, we determined its rotational correlation time from 15N-spin relaxation parameters. The obtained value, 13.3 ns, is slightly higher than expected for proteins in the 20-kDa range, indicating that a small fraction of GAFA154-320 is dimeric. Thus, the residues included in our GAF A constructs are not sufficient to form a tight dimer of GAF A at 37 °C, indicating that additional determinants are responsible for the observed dimer of the PDE5 holoenzyme.
In contrast to the GAF A domain, SEC and AUC show that GAFB332-496 exists as a dimeric species of ∼40 kDa, indicating that GAF B is capable of forming a strong homodimer. Analysis of our two tandem GAF AB constructs revealed an additional region that plays a role in PDE5 dimerization. Specifically, deletion of residues 134-153 alters the oligomeric state of the tandem GAF domains significantly. Although GAFAB134-496 behaves as a single dimeric species in AUC and SEC, GAFAB154-496 exists in both monomeric and dimeric forms of approximately equal populations at 400 μm (supplemental Fig. S7).
Based on our results, we conclude that residues 134-153 of mouse PDE5A are critical for high affinity dimerization of the tandem GAF domain and most likely of the PDE5 holoenzyme. Secondary structure predictions indicate that these residues form helical secondary structure. A three-dimensional helical model of the sequence reveals an amphipathic α-helix with large hydrophobic surface areas, consistent with involvement in dimerization. The putative α1-helix and the GAF B domain are the key determinants of tandem GAF dimerization and most likely form the native PDE5A parallel dimer interface. Our finding contrasts with the PDE2A dimerization revealed by the crystal structure of the PDE2A GAF domains, in which an inter-GAF domain α-helix and GAF A, but not GAF B, are involved in dimerization (12). Our results extend a previously published report, in which the authors investigated the dimerization interface of the regulatory region of PDE5A1 (17) at picomolar concentrations (rather than millimolar, as in our study). They characterized both single GAF domains as tight homodimers. Nevertheless, the domain constructs used contained extended elements of the connecting helix and the N-terminus, so that exact localization of the dimerization interface could not be identified. Also, this study did not present the oligomeric state of a construct that contains residues 156-331 of human PDE5A1 (17), a construct very similar to GAFA154-320, which in our study was in equilibrium between the monomeric and dimeric forms. Another study revealed that the N-terminal 46 residues of GAF B are critical for the dimerization of a construct containing GAF B and the catalytic domain (18). As truncation of these 46 residues is expected to disrupt the six-stranded β-sheet of GAF B and therefore the structural integrity of the domain, the interpretation of these results must be viewed with care.
Concluding Remarks—We present the NMR solution structure of the PDE5 GAF A domain and provide the first structural insight into the regulatory region of PDE5 at high resolution. The structure of the cGMP-bound GAF A domain reveals a conserved GAF domain fold and a distinct nucleotide binding environment. It also elucidates major determinants of the selectivity for cGMP of PDE5 GAF A, especially the prominent Asp-196 and the β2-β3 connecting loop. A dynamic apo-GAF domain that lacks tertiary structure and is stabilized by GAF B suggests that GAF A undergoes a major conformational change upon cGMP binding that presumably affects the overall conformation of the PDE5 holoenzyme. The identification of the dimerization interface within the tandem GAF domains will allow design of stable and soluble constructs with more accurate domain boundaries. Our structure also provides the basis for future investigations of the regulatory mechanism of PDE5 and a template for the design of GAF-specific small molecule inhibitors or activators.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. X. Tang for the expression vector of full-length mouse PDE5A1, Dr. C. Catalano for use of the AUC instrument, and D. Rhee for initial characterization of the D196A mutation. NMR experiments were performed, in part, in the Environmental Molecular Sciences Laboratories (a national scientific user facility sponsored by the Department of Energy (DOE) Biological and Environmental Research) located at the Pacific North-west National Laboratories and operated for DOE by Battelle.
The atomic coordinates and structure factors (code 2k31) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grant 1 P01 HL44948 (to R. E. K. and J. A. B.). This work was also supported by a Boehringer Ingelheim Fonds Ph.D. Scholarship (to C. C. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental text, references, Figs. S1-S7, and Table S1.
Footnotes
The abbreviations used are: PDE, 3′,5′-cyclic nucleotide phosphodiesterase; GAF, mammalian cGMP-dependent PDEs, Anabaena adenylyl cyclases, and E. coli FhlA; cNMP, cyclic nucleotides; NOE, nuclear Overhauser effect; GdnHCl, guanidine hydrochloride; r.m.s.d., root mean square deviation; AUC, analytical ultracentrifugation; SEC, size exclusion chromatography; WT, wild type; NOESY, nuclear Overhauser effect spectroscopy; TROSY, transverse relaxation-optimized spectroscopy.
W. L. DeLano (2002) PyMOL, DeLano Scientific, San Carlos, CA.
Of the two cAMP-bound GAF domains of cyaB2, GAF B aligns with a lower r.m.s.d. to PDE5A GAF A and was therefore used for comparisons.
Whether Ile-219 and Asn-242 have a lower rate of solvent exchange could not be determined by the H-D exchange experiments, because the amide peak of Ile-219 has low intensity in H2O, whereas the amide peak of Asn-242 is overlapped.
S. E. Martinez, personal communication.
We note that the stoichiometry of cGMP binding to GAFAB154-496 WT was consistently 0.95-1.0 mol of cGMP per mol of GAFAB154-496 (supplemental Fig. S6), indicating that virtually all protein molecules purified by our protocol were competent to bind ligand.
References
- 1.Beavo, J. A., and Brunton, L. L. (2002) Nat. Rev. Mol. Cell. Biol. 3 710-718 [DOI] [PubMed] [Google Scholar]
- 2.Bender, A. T., and Beavo, J. A. (2006) Pharmacol. Rev. 58 488-520 [DOI] [PubMed] [Google Scholar]
- 3.Francis, S. H., Zoraghi, R., Kotera, J., Ke, H., Bessay, E. P., Blount, M. A., and Corbin, J. D. (2007) in Cyclic Nucleotide Phosphodiesterases in Health and Disease (Beavo, J. A., Francis, S. H., and Housley, M. D., eds) pp. 131-164, CRC Press, Taylor and Francis Group, Boca Raton, FL
- 4.Padma-Nathan, H., McMurray, J. G., Pullman, W. E., Whitaker, J. S., Saoud, J. B., Ferguson, K. M., and Rosen, R. C. (2001) Int. J. Impot. Res. 13 2-9 [DOI] [PubMed] [Google Scholar]
- 5.Bischoff, E., Niewoehner, U., Haning, H., Es Sayed, M., Schenke, T., and Schlemmer, K. H. (2001) J. Urol. 165 1316-1318 [PubMed] [Google Scholar]
- 6.Boolell, M., Allen, M. J., Ballard, S. A., Gepi-Attee, S., Muirhead, G. J., Naylor, A. M., Osterloh, I. H., and Gingell, C. (1996) Int. J. Impot. Res. 8 47-52 [PubMed] [Google Scholar]
- 7.Aravind, L., and Ponting, C. P. (1997) Trends Biochem. Sci. 22 458-459 [DOI] [PubMed] [Google Scholar]
- 8.Wagner, J. R., Brunzelle, J. S., Forest, K. T., and Vierstra, R. D. (2005) Nature 438 325-331 [DOI] [PubMed] [Google Scholar]
- 9.Cann, M. (2007) Mol. Microbiol. 64 461-472 [DOI] [PubMed] [Google Scholar]
- 10.Lin, Z., Johnson, L. C., Weissbach, H., Brot, N., Lively, M. O., and Lowther, W. T. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 9597-9602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurley, J. H. (2003) Sci. STKE 2003, PE1. [DOI] [PubMed]
- 12.Martinez, S. E., Beavo, J. A., and Hol, W. G. (2002) Mol. Interv. 2 317-323 [DOI] [PubMed] [Google Scholar]
- 13.Francis, S. H., Bessay, E. P., Kotera, J., Grimes, K. A., Liu, L., Thompson, W. J., and Corbin, J. D. (2002) J. Biol. Chem. 277 47581-47587 [DOI] [PubMed] [Google Scholar]
- 14.Rybalkin, S. D., Rybalkina, I. G., Feil, R., Hofmann, F., and Beavo, J. A. (2002) J. Biol. Chem. 277 3310-3317 [DOI] [PubMed] [Google Scholar]
- 15.Rybalkin, S. D., Rybalkina, I. G., Shimizu-Albergine, M., Tang, X. B., and Beavo, J. A. (2003) EMBO J. 22 469-478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rybalkin, S. D., Yan, C., Bornfeldt, K. E., and Beavo, J. A. (2003) Circ. Res. 93 280-291 [DOI] [PubMed] [Google Scholar]
- 17.Zoraghi, R., Bessay, E. P., Corbin, J. D., and Francis, S. H. (2005) J. Biol. Chem. 280 12051-12063 [DOI] [PubMed] [Google Scholar]
- 18.Blount, M. A., Zoraghi, R., Ke, H., Bessay, E. P., Corbin, J. D., and Francis, S. H. (2006) Mol. Pharmacol. 70 1822-1831 [DOI] [PubMed] [Google Scholar]
- 19.Sung, B. J., Hwang, K. Y., Jeon, Y. H., Lee, J. I., Heo, Y. S., Kim, J. H., Moon, J., Yoon, J. M., Hyun, Y. L., Kim, E., Eum, S. J., Park, S. Y., Lee, J. O., Lee, T. G., Ro, S., and Cho, J. M. (2003) Nature 425 98-102 [DOI] [PubMed] [Google Scholar]
- 20.Wang, H., Liu, Y., Huai, Q., Cai, J., Zoraghi, R., Francis, S. H., Corbin, J. D., Robinson, H., Xin, Z., Lin, G., and Ke, H. (2006) J. Biol. Chem. 281 21469-21479 [DOI] [PubMed] [Google Scholar]
- 21.Card, G. L., England, B. P., Suzuki, Y., Fong, D., Powell, B., Lee, B., Luu, C., Tabrizizad, M., Gillette, S., Ibrahim, P. N., Artis, D. R., Bollag, G., Milburn, M. V., Kim, S. H., Schlessinger, J., and Zhang, K. Y. (2004) Structure 12 2233-2247 [DOI] [PubMed] [Google Scholar]
- 22.Zhang, K. Y., Card, G. L., Suzuki, Y., Artis, D. R., Fong, D., Gillette, S., Hsieh, D., Neiman, J., West, B. L., Zhang, C., Milburn, M. V., Kim, S. H., Schlessinger, J., and Bollag, G. (2004) Mol. Cell 15 279-286 [DOI] [PubMed] [Google Scholar]
- 23.Kameni Tcheudji, J. F., Lebeau, L., Virmaux, N., Maftei, C. G., Cote, R. H., Lugnier, C., and Schultz, P. (2001) J. Mol. Biol. 310 781-791 [DOI] [PubMed] [Google Scholar]
- 24.Martinez, S. E., Wu, A. Y., Glavas, N. A., Tang, X. B., Turley, S., Hol, W. G., and Beavo, J. A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13260-13265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, W., and Malcolm, B. A. (1999) BioTechniques 26 680-682 [DOI] [PubMed] [Google Scholar]
- 26.Sekharan, M. R., Rajagopal, P., and Klevit, R. E. (2005) J. Biomol. NMR 33 75. [DOI] [PubMed] [Google Scholar]
- 27.Schwede, T., Kopp, J., Guex, N., and Peitsch, M. C. (2003) Nucleic Acids Res. 31 3381-3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., and Bax, A. (1995) J. Biomol. NMR 6 277-293 [DOI] [PubMed] [Google Scholar]
- 29.Johnson, B. A., and Blevins, R. A. (1994) J. Biomol. NMR 4 603-614 [DOI] [PubMed] [Google Scholar]
- 30.Salzmann, M., Pervushin, K., Wider, G., Senn, H., and Wuthrich, K. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 13585-13590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pervushin, K., Riek, R., Wider, G., and Wuthrich, K. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 12366-12371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guntert, P., Mumenthaler, C., and Wuthrich, K. (1997) J. Mol. Biol. 273 283-298 [DOI] [PubMed] [Google Scholar]
- 33.Herrmann, T., Guntert, P., and Wuthrich, K. (2002) J. Mol. Biol. 319 209-227 [DOI] [PubMed] [Google Scholar]
- 34.Cornilescu, G., Delaglio, F., and Bax, A. (1999) J. Biomol. NMR 13 289-302 [DOI] [PubMed] [Google Scholar]
- 35.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Section D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 36.Linge, J. P., Williams, M. A., Spronk, C. A., Bonvin, A. M., and Nilges, M. (2003) Proteins 50 496-506 [DOI] [PubMed] [Google Scholar]
- 37.Schuttelkopf, A. W., and van Aalten, D. M. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60 1355-1363 [DOI] [PubMed] [Google Scholar]
- 38.Kleywegt, G. (1995) CCP4/ESF-EACBM Newsletter on Protein Crystallography 31 45-50 [Google Scholar]
- 39.Laskowski, R. A., Rullmann, J. A. C., MacArthur, M. W., Kaptein, R., and Thornton, J. M. (1996) J. Biomol. NMR 8 477-486 [DOI] [PubMed] [Google Scholar]
- 40.Thompson, W. J., Higgins D. G., and Gibson T. J. (1994) Nucleic Acids Res. 22 4673-4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., and Bairoch, A. (2005) in The Proteomics Protocols Handbook (Walker, J. M., ed) pp. 571-607, Humana Press, Totowa, NJ
- 42.Martinez, S. E., Bruder, S., Schultz, A., Zheng, N., Schultz, J. E., Beavo, J. A., and Linder, J. U. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 3082-3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Charbonneau, H., Prusti, R. K., LeTrong, H., Sonnenburg, W. K., Mullaney, P. J., Walsh, K. A., and Beavo, J. A. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 288-292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turko, I. V., Haik, T. L., McAllister-Lucas, L. M., Burns, F., Francis, S. H., and Corbin, J. D. (1996) J. Biol. Chem. 271 22240-22244 [DOI] [PubMed] [Google Scholar]
- 45.Bruder, S., Schultz, A., and Schultz, J. E. (2006) J. Biol. Chem. 281 19969-19976 [DOI] [PubMed] [Google Scholar]
- 46.Wu, A. Y., Tang, X. B., Martinez, S. E., Ikeda, K., and Beavo, J. A. (2004) J. Biol. Chem. 279 37928-37938 [DOI] [PubMed] [Google Scholar]
- 47.Hebert, M. C., Schwede, F., Jastorff, B., and Cote, R. H. (1998) J. Biol. Chem. 273 5557-5565 [DOI] [PubMed] [Google Scholar]
- 48.Ho, Y. S., Burden, L. M., and Hurley, J. H. (2000) EMBO J. 19 5288-5299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stroop, S. D., and Beavo, J. A. (1991) J. Biol. Chem. 266 23802-23809 [PubMed] [Google Scholar]
- 50.Fink, T. L., Francis, S. H., Beasley, A., Grimes, K. A., and Corbin, J. D. (1999) J. Biol. Chem. 274 34613-34620 [DOI] [PubMed] [Google Scholar]
- 51.Thomas, M. K., Francis, S. H., and Corbin, J. D. (1990) J. Biol. Chem. 265 14964-14970 [PubMed] [Google Scholar]
- 52.Handa, N., Mizohata, E., Kishishita, S., Toyama, M., Morita, S., Uchikubo-Kamo, T., Akasaka, R., Omori, K., Kotera, J., Terada, T., Shirouzu, M., and Yokoyama, S. (May 13, 2008) J. Biol. Chem. 10.1074/jbc.M800595200 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.