Abstract
The accumulation of unfolded proteins in the endoplasmic reticulum (ER) triggers a stress response program that protects cells early in the response and can lead to apoptosis during prolonged stress. The basic leucine zipper transcription factor, CCAAT/enhancer-binding protein β (C/EBPβ), is one of the genes with increased expression during ER stress. Translation of the C/EBPβ mRNA from different initiation codons leads to the synthesis of two transcriptional activators (LAP-1 and -2) and a transcriptional repressor (LIP). The LIP/LAP ratio is a critical factor in C/EBPβ-mediated gene transcription. It is shown here that the LIP/LAP ratio decreased by 5-fold during the early phase of ER stress and increased by 20-fold during the late phase, mostly because of changes in LIP levels. The early decrease in LIP required degradation via the proteasome pathway and phosphorylation of the translation initiation factor, eIF2α. The increased LIP levels during the late phase were due to increased synthesis and increased stability of the protein. It is proposed that regulation of synthesis and degradation rates during ER stress controls the LIP/LAP ratio. The importance of C/EBPβ in the ER-stress response program was demonstrated using C/EBPβ-deficient mouse embryonic fibroblasts. It is shown that C/EBPβ attenuates expression of pro-survival ATF4 target genes in late ER stress and enhances expression of cell death-associated genes downstream of CHOP. The inhibitory effect of LIP on ATF4-induced transcription was demonstrated for the cat-1 amino acid transporter gene. We conclude that regulation of LIP/LAP ratios during ER stress is a novel mechanism for modulating the cellular stress response.
The accumulation of unfolded proteins in the endoplasmic reticulum triggers a complex regulatory program that involves regulation of both transcription and translation (1). In the early part of the unfolded protein response (UPR),2 there is a decrease in protein synthesis and increased expression of proteins that protect cells from stress (1). In contrast, during prolonged stress, the UPR can change to a proapoptotic program, leading to the expression of proteins that promote cell death (2–4).
The regulatory events in the UPR are linked to a key sensor within the ER lumen, the chaperone GRP78/BiP (5). In stressed cells, the binding of BiP to unfolded proteins lowers the level of free BiP, promoting the activation of three signaling pathways. (i) PERK protein kinase phosphorylates the α subunit of the translation initiation factor, eIF2 (eIF2α), to attenuate global translation initiation. Interestingly, the translation of some mRNAs increases (6, 7); increased translation of the bZIP transcription factor ATF4 mRNA leads to transcriptional activation of important stress-response genes (8, 9). (ii) The transmembrane protein ATF6, which is normally retained in the ER, transits to the Golgi, where it is cleaved to produce an active transcription factor. Its target genes include ER chaperones that assist with protein folding (BiP, GRP94, and calreticulin) (10). (iii) The endonuclease IRE1 is activated, initiating the cytoplasmic splicing of XBP1 mRNA to a form that encodes a potent transcription factor (11). XBP1 forms heterodimers with ATF6 or directly induces gene transcription (10). The level of downstream gene expression in these signaling pathways determines cell fate during the ER stress response (12), as recently shown for IRE1 signaling (13).
Expression of the bZIP transcription factor C/EBPβ is induced by stress conditions, such as ER stress and amino acid starvation (14, 15). C/EBPβ heterodimerizes with other bZIP factors, including ATF4 and CHOP, which are also induced during the UPR (16). CHOP is a C/EBP family member that has been associated with induction of apoptosis, as well as many diseases (4). During translation of the C/EBPβ mRNA, initiation at alternate in-frame AUG codons gives rise to at least three isoforms, LAP-1, LAP-2, and LIP (see Fig. 1C) (17). LAP-1 and -2 contain an N-terminal transcription transactivation domain, a central DNA binding domain, and a C-terminal leucine zipper domain that homo- or heterodimerizes with other bZIP proteins (18). LIP is missing the transactivation domain but can dimerize and bind DNA. As a consequence, LAP-1 and -2 are mostly transcriptional activators, whereas LIP acts as a repressor (19). C/EBPβ is expressed in many tissues (18), and high LIP levels are associated with aggressive tumors, suggesting that LIP is oncogenic (19). In addition, characterization of C/EBPβ–/– mice provides clues to its functions. These mice have two phenotypes, A and B (20). Mice with the B phenotype die in the perinatal period because of hypoglycemia and inability to mobilize glycogen. Mice with the A phenotype survive to adulthood but have defective glucose and lipid homeostasis. They have fasting hypoglycemia, lower hepatic glucose production, as well as reduced plasma insulin and free fatty acids. The importance of the individual C/EBPβ isoforms in vivo has not been demonstrated. However, in a recent report, mice containing a knock-in mutation of the LAP-2 translation initiation codon did not have the metabolic defects of the C/EBPβ–/– mice, suggesting important roles for LAP-1 and LIP in the cellular metabolism (21).
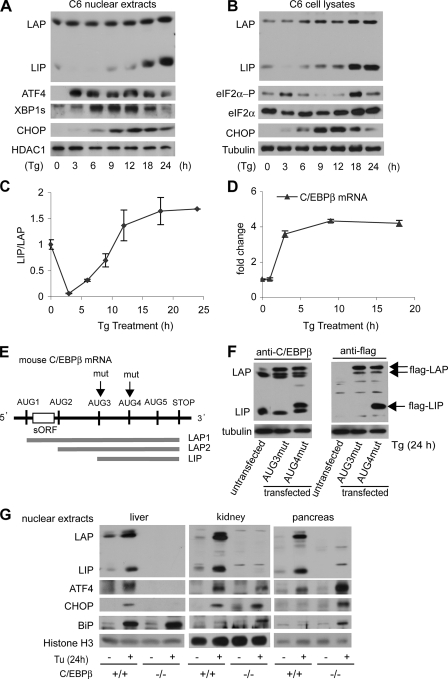
FIGURE 1.
Differential regulation of LIP and LAP levels during ER stress. A, Western blot analysis of nuclear extracts (A) or whole cell extracts (B) from C6 cells treated with Tg for the indicated times. Blots were probed with antibodies for C/EBPβ or the indicated proteins. C, Western blots in three independent experiments were quantified and used to calculate the LIP/LAP ratio. Means ± S.E. are shown. D, C/EBPβ mRNA levels were monitored by qRT-PCR using total RNA samples from C6 cells treated with Tg for the indicated times. Data were normalized to the 18 S ribosomal RNA signal. E, schematic of the translation products of C/EBPβ. The arrows show the mutations of the AUG codons in the AUG3mut and AUG4mut constructs. F, Western blot analysis of cell extracts from Tg-treated C6 cells stably expressing mutant FLAG-tagged CEBPβ proteins. Blots were probed with antibodies for C/EBPβ and the FLAG epitope. G, Western blot analysis of nuclear extracts from the indicated tissues of WT and C/EBPβ–/– mice following 24 h treatment with Tu. Blots were probed for antibodies for C/EBPβ and the indicated proteins.
The importance of C/EBPβ in the ER stress response has been shown by the reduced apoptosis of C/EBPβ–/– MEFs (22), which suggested that this protein might play a role in the regulation of gene expression during the response. In fact, C/EBPβ has been associated with regulation of the asparagine synthase (AS) gene (23) and genes downstream of CHOP (DOCs) during ER stress (9, 24). Because there are no clear consensus DNA-binding sites for the bZIP heterodimers induced during ER stress, regulation of individual genes needs to be determined experimentally. The opposing roles of LIP and LAP in the regulation of gene expression (23), the induction of C/EBPβ expression during ER stress (15), and the potential of LAP and LIP to form heterodimers with other bZIP transcription factors led us to undertake the studies in this report.
We explored the regulation of LIP and LAP levels during ER stress and the physiological significance of C/EBPβ in the stress-response program. We found that LIP levels decrease during the first few hours of ER stress, followed by a large increase, whereas LAP levels showed a steady increase. The result was a decline and then a large increase in the LIP/LAP ratio. Both LIP and LAP were found to be short lived proteins that are degraded by the proteasome. Our results suggest that changes in the rates of synthesis and degradation are important for regulating LIP and LAP levels. We also show that LIP can inhibit the transcription of stress-response genes that are activated by the binding of ATF4 to an amino acid-response element (AARE). In support of this idea, the expression of these genes was altered in C/EBPβ–/– MEFs. In wild-type cells and in C/EBPβ–/– MEFs expressing LIP (but not LAP), expression increased early in the UPR and then declined, whereas the expression remained elevated in C/EBPβ–/– MEFs. In contrast, a subset of genes that also require AAREs for induction of transcription during ER stress were attenuated in the absence of C/EBPβ. The transcription factor CHOP has been implicated in the regulation of these genes (9, 12). Our data suggest differential regulation of AARE-mediated transcription during ER stress by C/EBPβ. The findings in this study are consistent with the notion that the LIP/LAP ratio is an important regulator of the stress response, and that the high levels of LIP during the late response correlate with attenuated expression of ATF4-regulated genes.
EXPERIMENTAL PROCEDURES
Plasmid Constructs—pGL3-3xUPRE and pGL3-2xERSE luciferase reporter vectors were gifts from Dr. R. Kaufman. PA1.4N and pGL3-3xAARE luciferase reporter vectors were described previously (25, 26). FLAG-tagged mouse C/EBPβ expression vectors carrying mutations in AUG codons were generated from the mouse C/EBPβ expression vector pcDNA3.1(–)mC/EBPβ-5′-UTR (27) by inserting a DNA fragment encoding a FLAG tag into the mouse C/EBPβ open reading frame (ORF) just before the stop codon and mutating the third or fourth ATG codons to ATC using PCR methods. To construct the LIP expression vector, a fragment containing the LIP ORF was amplified by PCR methods and inserted into pcDNA3.1(–).
Cell Culture and DNA Transfections—C6 rat glioma cells were maintained in high glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal bovine serum (FBS) and 5% calf serum. WT and C/EBPβ–/– mouse embryonic fibroblasts (MEFs) (28) were maintained in high glucose DMEM supplemented with 10% FBS. Plasmid DNAs were transfected into C6 cells using FuGENE 6 (Roche Applied Science). Stably transfected mass cultures were generated by selection in 0.1% G418. Transfected cells were grown in medium containing 0.04% G418 and shifted to drug-free medium 1 day before experiments. To control for transfection efficiency in transient transfection experiments, cells were cotransfected with plasmids expressing either β-galactosidase or Renilla luciferase (29). To induce ER stress, cells were incubated with medium containing 400 nm thapsigargin (Tg) or 2 μg/ml tunicamycin (Tu) for the indicated times. Amino acid starvation was induced by incubation of cells in Met-, Cys-free high glucose DMEM supplemented with 10% dialyzed FBS as described previously (26). To inhibit protein synthesis or the proteasome, cells were incubated with medium containing 10 μg/ml cycloheximide (CHX), 1 μm hippuristanol (Hipp), 10 μm MG132, or 10 μm epoxomycin, for the indicated times. Luciferase and β-galactosidase activities were measured as described previously (29).
Animal Studies—Breeding of C/EBPβ–/– and wild-type mice was described previously (30). To induce ER stress, 8–10-week-old mice were injected intraperitoneally with Tu (1 μg/g body weight). After 24 h, livers, kidneys, and pancreata were harvested and homogenized in lysis buffer (20 mm Tris, pH 7.6, 0.1 mm EDTA, 0.5 mm EGTA, 1% Triton X-100, 250 mm sucrose) as described previously (31). The homogenates were centrifuged at 17,000 × g for 30 min at 4 °C, and the supernatants were discarded. The pellets were sonicated in 0.4 ml of lysis buffer/g of tissue and centrifuged at 17,000 × g for 10 min at 4 °C, and the supernatants were collected as nuclear extracts.
Western Blot Analysis—Cells were scraped in phosphate-buffered saline, collected by centrifugation, and suspended in 20 mm HEPES, pH 8.0, 380 mm NaCl, 6 mm MgCl2, 1% deoxycholic acid, incubated on ice for 15 min with occasional mixing, and centrifuged at 17,000 × g for 10 min at 4 °C. The supernatants were collected as total cell lysates. Nuclear extracts were isolated as described previously (31). Total cell lysates (20 μg of protein) or nuclear extracts (10 μg of protein) were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and probed with antibodies using standard procedures. Antibodies for the following proteins were purchased from Santa Cruz Biotechnology: ATF4 (C-20, sc-200), BiP (H-129, sc-13968), C/EBPβ C terminus (C-19, sc-150), CHOP (B-3, sc-7351), HDAC1 (H-11, sc-8410), and XBP1 (M-186, sc-7160). Anti-FLAG antibodies (PRB-132P) were from Covance. Antihistone H3 antibodies (sc-9715) were from Cell Signaling. Anti-eIF2α-P antibodies (44–728G) were from BIOSOURCE. Monoclonal anti-α-tubulin antibodies (T9026) were from Sigma. Anti-eIF2α antibodies were prepared by Quality Controlled Biochemicals.
Polysome Profiles and RNA Isolation—Control or Tg-treated C6 cells were incubated for the last 5 min of treatment with 100 μg/ml CHX. Cells were then washed twice with phosphate-buffered saline, scraped in phosphate-buffered saline, and centrifuged at 1,200 × g for 5 min. The cell pellets were suspended and homogenized by 15 passages through a 25-gauge needle in 500 μl of 10 mm HEPES-KOH, pH 7.4, 2.5 mm MgCl2, 100 mm KCl, 1 mm dithiothreitol, 0.25% Nonidet P-40, 200 units/ml RNase inhibitor, 100 μg/ml CHX, and 1 tablet of EDTA-free protease inhibitor mixture/10 ml (Roche Applied Science). Lysates were centrifuged at 17,000 × g for 15 min; the supernatants were collected, and absorbances were measured. About 7 A260 units of the cytosolic extracts were layered over 10–50% sucrose gradients and centrifuged at 16,000 rpm in a Beckman SW28 rotor for 19 h at 4 °C. After centrifugation, fractions (∼1.2 ml each) were collected. RNA was extracted with TRIzol reagent (Invitrogen) and used to determine the distribution of ribosomal RNAs (by agarose gel electrophoresis) and mRNAs (by qRT-PCR).
Metabolic Labeling and Immunoprecipitation—C6 cells were incubated for 1 h in l-Met-, l-Cys-deficient DMEM (Invitrogen catalog number 21013) containing 10% dialyzed FBS with or without 400 nm Tg. Cells were then incubated in Met-, Cys-deficient DMEM with 35S-Met and -Cys (100 μCi/ml Expre35S35S Protein labeling mix, PerkinElmer Life Sciences) with or without Tg for an additional hour. Cells were scraped in cold phosphate-buffered saline and lysed in RIPA buffer (50 mm Tris-HCl, pH 8, 150 mm NaCl, 1% Nonidet P-40, 0.1% SDS, 1 mm NaF, 1 mm Na3VO4, 2 mm imidazole) supplemented with protease inhibitor mixture (Roche Applied Science). Lysates (500 μg of protein) were incubated overnight with anti-C/EBPβ (2 μg, Santa Cruz Biotechnology, sc-150). Samples were separated on 12% SDS-polyacrylamide gels, dried, and analyzed by autoradiography.
DNA Affinity Pulldown—A biotinylated double-stranded DNA oligonucleotide containing two copies of the cat-1 AARE (5′-CGCGCGGCTGATGAAACCGGCTCGCGCGCGGCTGATGAAACCGGCTCG-3′) was used to isolate proteins from C6 cell nuclear extracts as described previously (32) with minor modifications. Extracts (500 μg of protein) were incubated streptavidin magnetic particles (Roche Applied Science) for 30 min at 4 °C; the beads were removed, and 100 μg/ml poly(dI-dC) was added to the supernatants for 30 min, followed by 100 pmol of the biotinylated oligonucleotide in binding buffer (12 mm HEPES-NaOH, pH 7.9, 4 mm Tris-HCl, pH 7.9, 12% glycerol, 60 mm KCl, 1 mm EDTA, 1 mm dithiothreitol) for 1 h at 4 °C. Streptavidin magnetic particles were added, incubated for 30 min at 4 °C, and the particles isolated. The particles were washed twice in binding buffer and once with binding buffer supplemented with 40 mm KCl at room temperature. The isolated proteins were separated on 12% SDS gels and analyzed by Western blotting.
Quantitative Real Time RT-PCR (qRT-PCR)—First-strand cDNA samples were synthesized from RNA samples using SuperScript III (Invitrogen). The PCR primers used are listed in supplemental Table 1.
RESULTS
Differential Regulation of the Transcriptional Activator LAP and Transcriptional Repressor LIP during ER Stress—Although C/EBPβ has been implicated in the transcriptional control of stress-response genes (14, 15), its role in the adaptive response of cells to ER stress is not clear. Because translation of the C/EBPβ mRNA produces two transcriptional activators, LAP1 and -2, and a transcriptional repressor, LIP (Fig. 1E), we first determined the levels of LAP and LIP in C6 cells treated with Tg for 0–24 h, which acts by depleting ER Ca2+ stores (6). As expected, Tg caused ER stress, as shown by the induction of key modulators of the stress response (1). These include the transcription factors ATF4, CHOP, and the spliced form of XBP1 in nuclear fractions (Fig. 1A), as well as the phosphorylated form of the translation initiation factor eIF2α in whole cell lysates (Fig. 1B). In contrast, the levels of histone deacetylase 1 and tubulin, which were used as loading controls for nuclear and whole cell fractions, respectively, were constant.
Tg caused a small increase in nuclear LAP2 levels (Fig. 1A). In contrast, LIP levels decreased in nuclear extracts from 0 to 3 h, followed by an increase from 9 to 24 h, with the final level greatly exceeding that in untreated cells. The levels of LIP and LAP were also examined in whole cell lysates (Fig. 1B). Similar to nuclear fractions, LIP declined and then showed a large increase; in contrast, LAP levels increased continuously throughout the experiment.
The LIP/LAP ratio has been proposed to be a critical modulator in the expression of C/EBPβ-regulated genes (33–35). During ER stress, the LIP/LAP ratio in total cell lysates decreased by 90% in the early response (3–6 h) and then increased by 30-fold from the minimum (Fig. 1C). This finding suggests that modulation of LAP and LIP levels may be required for the adaptive response to ER stress. Similar results were obtained in cells treated with Tu, which causes ER stress by blocking N-glycosylation (not shown). We also examined the levels of C/EBPβ mRNA during Tg treatment by qRT-PCR (Fig. 1D). The levels rose by ∼4-fold after 3 h of treatment and remained elevated. This finding can explain the increased levels of both LIP and LAP in whole cell extracts but not the differential accumulation of these proteins during late ER stress (Fig. 1B). Because LIP and LAP are synthesized from the same mRNA, the changes in the relative levels of these proteins must be due to independent regulation of their synthesis and degradation.
LIP is expressed by translation initiation at an in-frame AUG codon (AUG3) in the C/EBPβ ORF (Fig. 1E). To confirm that the LIP protein expressed during ER stress is the authentic p20 C/EBPβ isoform, we mutagenized AUG codons in an expression vector that encodes mouse C/EBPβ with a C-terminal FLAG epitope. These mutations preserve the C/EBPβ ORF. LAP, but not LIP, was expressed from the AUG3mut construct as seen in blots probed with anti-C/EBPβ and anti-FLAG antibodies (Fig. 1F), whereas both forms were expressed from the AUG4mut construct, demonstrating that C6 cells express authentic LIP that is translated from AUG3. These data exclude the possibility that the LIP isoform is a product of proteolysis in our cells, as suggested previously (27).
We next determined if nuclear C/EBPβ levels are regulated in mice treated with Tu, which is a rapid inducer of ER stress in vivo. Tuhas been shown to cause an acute ER stress response in the kidney (22). Among the other tissues, pancreas and to a lesser extent liver are most susceptible to ER stress (36). To evaluate the role of C/EBPβ in the ER stress response, we compared the induction of stress-response genes in wild-type (WT) and C/EBPβ–/– mice at 24 h of treatment. This time of treatment has been associated with kidney damage (22). In WT mice, Tu caused an induction of LAP and LIP levels in liver, kidney, and pancreas (Fig. 1G). The LIP/LAP ratio increased by 2.5-fold in the liver and decreased 3- and 7-fold in kidney and pancreas, suggesting differential responses of the three tissues to ER stress. As expected, LAP and LIP were not expressed in C/EBPβ–/– mice.
To correlate the LIP/LAP ratio with the stress response, we examined induction of three stress-response proteins, ATF4, CHOP, and BiP, in nuclear extracts. ATF4 is a transcription factor that has been implicated in supporting either survival or death of stressed cells (9, 37). CHOP is a transcription factor that promotes cell death (12), whereas BiP is a chaperone within the ER and nuclear envelope that has pro-survival functions (38). Liver showed the highest induction of BiP in WT and mutant mice (Fig. 1E), in agreement with previous reports that BiP induction depends on the ATF6 signaling pathway (39). In contrast, ATF4 and CHOP were expressed in the liver of wild-type but not C/EBPβ–/– mice, although these proteins were induced in kidney and pancreas of mutant animals. These data suggest that the ER stress gene expression program varies in different tissues. The absence of ATF4 and CHOP and the presence of BiP in C/EBPβ–/– livers may suggest that the liver uses the XBP1/ATF6 rather than the PERK/eIF2α pathway for response to late ER stress, as shown for pancreatic beta cells that lack PERK signaling (40). Interestingly, the decreased LIP/LAP ratios in pancreas and kidney are correlated with higher tissue injury. Given the importance of ATF4, CHOP, and C/EBPβ in gene regulation by forming heterodimers among them (9, 16, 24), our data also suggest that LIP/LAP ratios may play a role in modulating the ER stress-response program.
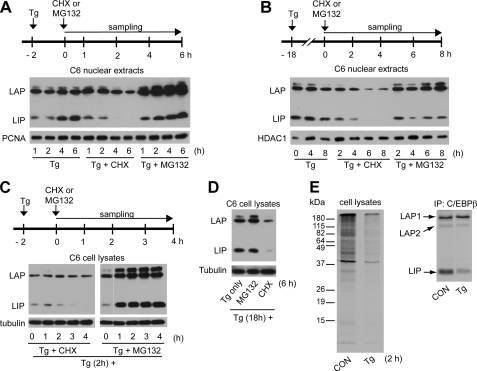
LAP and LIP Are Degraded by the Proteasome during ER Stress—We have shown that LIP levels increased by 30-fold between 3 and 18 h of Tg treatment (Fig. 1C). In contrast, LAP levels showed only a 1.5-fold increase. These differences may be due to differences in the rates of synthesis or degradation. We therefore determined the half-lives (t½) of LAP and LIP by measuring protein levels in the presence of the protein synthesis inhibitor, CHX, in cells after either 2 or 18 h of Tg treatment (Fig. 2). CHX caused decreases in these proteins in both nuclear extracts (Fig. 2, A and B) and whole cell extracts (Fig. 2, C and D). LAP in nuclear extracts decreased with t½ of 6 and 3 h at 2 and 18 h, respectively, whereas the t½ for LIP was 2 h at both times. Similar results were obtained from the analysis of total cell extracts. The levels of the long lived proteins proliferating cell nuclear antigen and HDAC1 did not show large changes during CHX treatment (Fig. 2, A and B). These data suggest that the increases in LIP levels between 2 and 18 h of Tg treatment are because of increased synthesis, rather than a decrease in degradation. In contrast, the decline in the t½ of LAP during late ER stress may contribute to the fact that LAP shows smaller increases than LIP during prolonged stress.
FIGURE 2.
Differential regulation of LIP and LAP levels during ER stress by inhibitors of the proteasome and protein synthesis. Western blot analysis of nuclear extracts (A and B) and total extracts (C and D) from C6 cells. Cells were treated with Tg for 2 h (A and C) or 18 h (B and D) before addition of CHX or MG132 as indicated. Blots were probed with antibodies for C/EBPβ, HDAC1, proliferating cell nuclear antigen (PCNA), and tubulin. E, cells treated with Tg for 1 h or control (CON) cells were labeled with 35S-Met/Cys for 1 h and immunoprecipitated (IP) with anti-C/EBPβ antibody. Samples of cell lysates and the immunoprecipitates were analyzed on SDS gels, and radioactivity was detected by autoradiography.
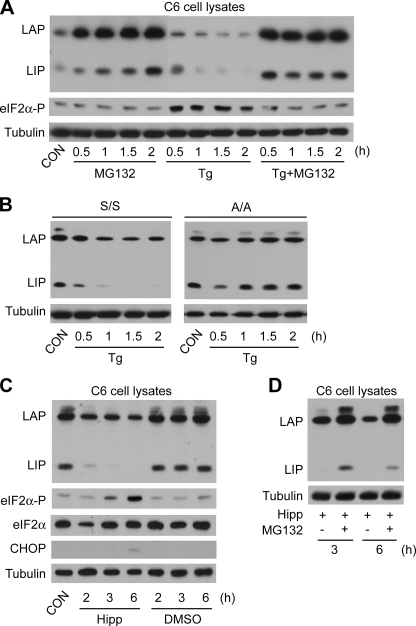
We next determined if the proteasome is involved in LAP and LIP degradation during ER stress by treating cells with Tg and the proteasome inhibitor MG132. Inhibition of the proteasome resulted in increased levels of LAP in early and late stress in both whole cell and nuclear extracts (Fig. 2, A–D), indicating that proteasomal degradation of this protein is an important route of turnover. MG132 caused increased LIP accumulation during the early phase (Fig. 2, A and C) but not in the late phase (Fig. 2, B and D). These data suggest that the rate of LIP synthesis exceeds its degradation in late ER stress, consistent with the large accumulation of LIP between 18 and 24 h (Fig. 1A). Interestingly, MG132 resulted in a dramatic increase of total LIP levels but a smaller increase of nuclear levels (compare Fig. 2, A and C), consistent with cytoplasmic degradation of the protein. The mechanism of degradation of LIP is under investigation in our laboratory. We also used MG132 to show that the proteasome degrades LAP and LIP in unstressed cells (Fig. 3A). There is a striking accumulation of LIP and LAP in cells treated with both Tg and MG132 for 2 h (Fig. 3A). It is worth noting that MG132 abolished accumulation of phospho-eIF2α during ER stress (Fig. 3A), in agreement with some reports (41) but in disagreement with others (42–44), possibly because of the short treatment and low MG132 concentration. Longer times and higher concentrations increased eIF2α phosphorylation (data not shown). We therefore conclude that LAP and LIP are degraded by the proteasome in untreated and ER-stressed cells and that the proteasome is involved in the decline in LIP during the first 2 h of stress (Fig. 3A). Similar results were obtained with epoxomycin, another proteasome inhibitor (not shown).
FIGURE 3.
Decreased LIP levels during early ER stress require eIF2α phosphorylation and an active proteasome. Western blot analysis of cell extracts from C6 cells (A) or S/S and A/A MEFs (B) treated with Tg and MG132 for the indicated times. Blots were probed with antibodies for C/EBPβ, eIF2α-P (Ser-51), and tubulin. C and D, C6 cells were treated with MG132 and/or the translation inhibitor hippuristanol (Hipp) for the indicated times. Proteins in whole cell lysates were analyzed on Western blots. CON, control.
An early event in the cellular response to ER stress is a decrease in global protein synthesis, which is caused by the phosphorylation of eIF2α (1). Consequently, we assessed the contribution of this change to the decreased LIP levels in early stress. Cells were treated with Tg for 1 h and then labeled with 35S-Met and -Cys for an additional hour. SDS gel analysis of whole cell lysates revealed the expected ∼50% decrease in protein synthesis (Fig. 2E). Immunoprecipitation of LIP and LAP revealed that similar amounts of LAP1 and -2 were synthesized in Tg-treated and control cells, whereas the amount of LIP declined by ∼50%. Therefore, decreased global protein synthesis may contribute to the lower LIP levels during early stress. However, the constant levels of LAP suggest that synthesis of these proteins may involve translation regulation.
We next tested directly the role of eIF2α phosphorylation in the regulation of LIP levels early in the stress response. This was accomplished by comparing WT MEFs (S/S) and MEFs with a S51A mutation in eIF2α (A/A) that cannot be phosphorylated. LIP levels decreased in WT MEFs treated with Tg for 2 h (Fig. 3B, S/S). In contrast, the decrease was not observed in mutant MEFs (Fig. 3B, A/A). These data demonstrate that eIF2α phosphorylation is required for the decreased LIP levels during early ER stress.
As an additional test of the relative half-lives of LIP and LAP, we tested the effects of Hipp, which blocks initiation by inhibiting eIF4A (45), in contrast to CHX, which inhibits elongation. Hipp treatment of C6 cells caused a large decrease in LIP levels, with a much smaller change in LAP (Fig. 3C). This decrease was prevented by MG132 (Fig. 3D), further supporting the idea that LIP is a short lived protein that is degraded by the proteasome in both stressed and unstressed cells.
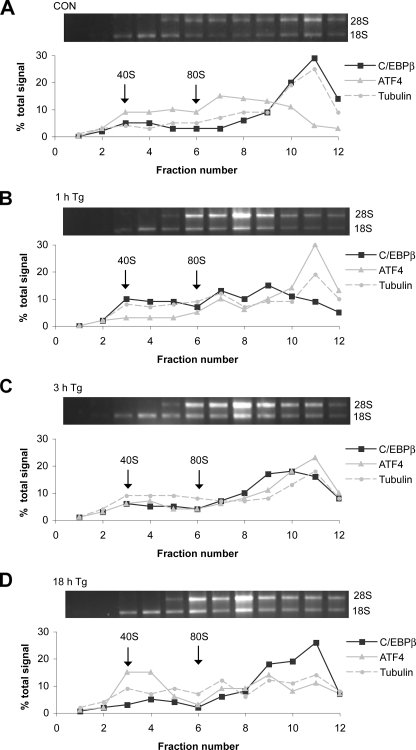
The results in Figs. 2 and 3 suggest that changes in synthesis rates contribute to the changes in LIP and LAP levels during ER stress. To evaluate this hypothesis, we examined the association of C/EBPβ mRNA with heavy polysomes as a measurement of translational efficiency. Samples were analyzed by gradient centrifugation, and mRNA levels were measured by quantitative RT-PCR. Control mRNAs included ATF4, because ER stress is known to increase the translation efficiency of this mRNA (7, 46), as well as tubulin mRNA.
The effect of ER stress on translation is indicated by the distribution of rRNAs (Fig. 4). In control cells, most of the rRNAs are in heavy polysomes (fractions 10–12); ER stress causes a large decrease in the heavy polysome fraction and a corresponding increase in lighter polysomes and free subunits. In unstressed C6 cells, 65% of the C/EBPβ mRNA was in the fractions (10–12) containing heavy polysomes (Fig. 4A), and the distribution was similar to that of tubulin mRNA. Tg treatment for 1 h caused a shift of the C/EBPβ mRNA to lighter fractions (Fig. 4B), consistent with less efficient translation. Starting at 3 h, the C/EBPβ mRNA returned to heavier polysomes, and by 18 h, 50% of the mRNA was in these fractions (Fig. 4D). In contrast, tubulin mRNA was translated efficiently in untreated cells and less efficiently in stressed cells. The distribution of ATF4 mRNA also showed characteristic changes. In unstressed cells, only 20% of the ATF4 mRNA was in heavy polysomes (Fig. 4A). In contrast, at 1 h of stress, 60% of the ATF4 mRNA was associated with heavy polysomes (Fig. 4B) and then declined at later times, consistent with its efficient translation in early stress (6, 47).
FIGURE 4.
The C/EBPβ mRNA associates with heavy polysomes during prolonged ER stress. C6 cells were treated with Tg for the indicated times, and cytoplasmic extracts were analyzed by sucrose gradient centrifugation as described under “Experimental Procedures.” Total RNA samples were run on agarose gels to analyze ribosomal RNAs (upper panels). qRT-PCR was used to monitor the distribution of C/EBPβ, tubulin, and ATF mRNAs (lower panels). CON, control.
These results suggest that changes in translation efficiency of C/EBPβ during ER stress play a role in regulating the levels of LIP and LAP. However, because both are translated from the same mRNA, this experiment does not reveal which protein is synthesized from the polysome-associated mRNA. Nevertheless, the large sustained increase in LIP levels during late ER stress is consistent with a selective increase in LIP synthesis at this time.
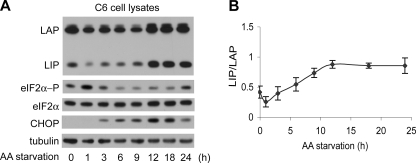
LIP Levels Increase during Amino Acid Starvation but the LIP/LAP Ratio Shows a Small Change—C/EBPβ has been implicated in the integrated stress response of cells to both ER stress (15) and nutrient limitation (amino acids or glucose, see Ref. 14). We therefore tested the levels of LIP and LAP in C6 cells during amino acid starvation (Fig. 5A). First, we showed that the response in C6 cells followed the previously reported gene expression pattern (15). This response involves phosphorylation of eIF2α by GCN2 kinase, which leads to the induction of the stress-response mediator ATF4. Fig. 5 shows that eIF2α phosphorylation increases after 1 h and then declines, followed by a small increase during prolonged starvation, as reported previously (48). In addition, CHOP expression, which is regulated by ATF4 (49), is induced during prolonged starvation. Similar to ER stress, LIP and LAP levels decreased during early starvation and increased during prolonged starvation (Fig. 5A). However, in contrast to ER stress, the changes of the LIP/LAP ratio during amino acid starvation were less than 2-fold (Fig. 5B) (50). Similar changes of LIP and LAP levels in nuclear extracts were also observed (not shown). These data suggest that involvement of LIP and LAP in stress response programs may vary with the type of stress (14, 15).
FIGURE 5.
Amino acid starvation induces LIP and LAP levels but has a small effect on the LIP/LAP ratio. A, Western blot analysis of extracts from C6 cells incubated in amino acid-deficient medium for the indicated times. Blots were probed with antibodies for the indicated proteins. B, quantification of the LIP/LAP ratio from three independent Western blot analyses shown in A.
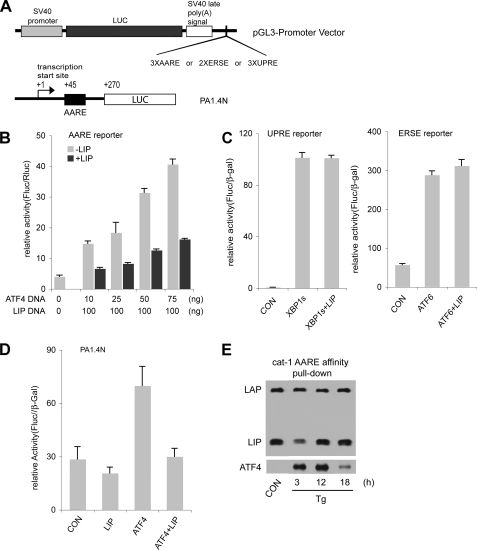
LIP Attenuates Transcription Mediated by ATF4 but Not XBP1 or ATF6—The induced transcription of stress-response genes during ER stress involves either ATF4 or XBP1 and ATF6 (8, 51–53). Consensus DNA sequences specific for ATF4 (AAREs), ATF6 (ERSEs), and XBP1 (UPREs) have been described (1). Because LIP associates with CHOP (22) and because CHOP expression is stimulated by ATF4 (49), the increased LIP levels during ER stress might attenuate transcription mediated by these pathways. There are no studies demonstrating heterodimers of LIP with ATF4, ATF6, or XBP1 in vivo. We therefore compared the effect of LIP on ATF4- and XBP1/ATF6-mediated transcription. We used luciferase expression vectors with the SV40 early promoter and 2–3 copies of the stress-response elements downstream of the luciferase ORF (Fig. 6A). The effect of cotransfecting LIP with ATF4, ATF6, or XBP1 was examined. LIP cotransfection with ATF4 inhibited luciferase expression from the construct containing the cat-1 AARE (Fig. 6B). In contrast, neither ATF6- nor XBP1-mediated luciferase expression from the constructs containing ERSEs or UPREs was affected by coexpression of LIP (Fig. 6C). As an additional test of the role of LIP in attenuating ATF-stimulated transcription, we examined its effect on the promoter of the gene encoding cat-1, which contains the AARE in the first exon (25). We used a construct with 1.4 kb of the promoter region and the 5′-UTR of the cat-1 mRNA linked to a luciferase reporter (Fig. 6A). Luciferase expression from this construct was not induced by LIP, but ATF4-stimulated expression was inhibited by the coexpression of LIP (Fig. 6D). These results demonstrate that LIP attenuates ATF4-mediated gene transcription and suggests that C/EBPβ regulates expression of genes that are directly or indirectly induced by the ATF4-mediated stress response program.
FIGURE 6.
LIP attenuates induction of transcription by ATF4 via the cat-1 AARE enhancer element. A, schematic of luciferase (LUC) expression vectors containing AAREs, ERSEs, or UPREs (top) and the cat-1 promoter-containing expression vector (bottom). B–D, C6 cells were transfected with the expression vectors in A along with expression vectors for the indicated transcription factors. Expression plasmids for β-galactosidase (β-gal) or Renilla luciferase were included to normalize for transfection efficiency. Enzymatic activities were measured in cell extracts 48 h after transfection and normalized as indicated. E, DNA affinity pulldown using nuclear extracts from control and Tg-treated C6 cells was conducted as described under “Experimental Procedures.” The biotin-labeled DNA probe contained two copies of the cat-1 AARE. Samples were analyzed for C/EBPβ and ATF4 on Western blots. CON, control.
To test the DNA binding ability of active transcription factors in stressed cells, DNA pulldown experiments were performed. A biotinylated double-stranded oligonucleotide containing two copies of the cat-1 AARE was incubated with nuclear extracts from control and stressed cells. The oligonucleotide-protein complexes were isolated, and LIP, LAP, and ATF4 were analyzed by Western blotting (Fig. 6E). LAP binding was high in all the extracts, consistent with the constant levels of this protein in nuclear extracts (Fig. 1A). LIP showed strong binding in controls, in contrast to the low levels of this protein in nuclear extracts. This suggests that LIP can bind preferentially under these conditions, consistent with its proposed role as a transcriptional repressor of the gene encoding cat-1. The level of binding then declined and increased, mirroring the changes in LIP protein levels in nuclear extracts. ATF4 binding was low in controls, consistent with the low level of this protein in the extracts. The level increased strongly during stress, but then declined at 18 h, even though levels in the extracts remained high. This is probably because of competition with LIP, although the involvement of another protein cannot be excluded.
C/EBPβ Attenuates the Induction of Gene Expression Mediated by the PERK-ATF4 Pathway but Not the XBP1/ATF6 Pathway during ER Stress—To test the hypothesis that C/EBPβ regulates expression of genes that are induced by the ATF4 (9, 12, 15, 54, 55) and not the ATF6/XBP1 (53, 56, 57) pathways, we compared the induction of stress-response genes in wild-type and C/EBPβ–/– MEF cells during ER stress. In addition, to examine the role of LIP expression, we analyzed C/EBPβ–/– (LIP) cells that constitutively expressed LIP from a transfected plasmid. This plasmid contained the LIP ORF without any 5′- or 3′-flanking sequences.
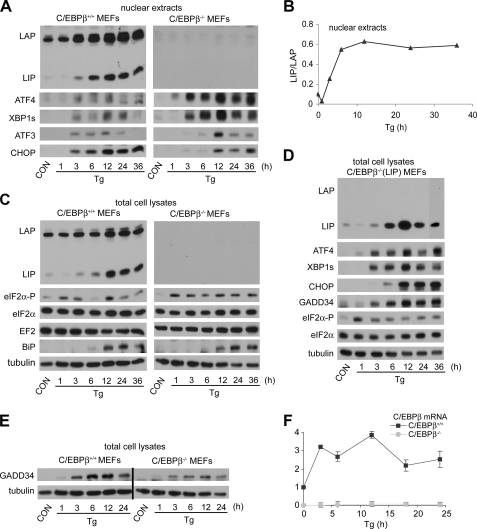
First, we measured the induction of LIP and LAP by ER stress. The results in WT MEFs (Fig. 7, A and C) were similar to those observed in C6 cells, with large increases in LIP and LAP levels following prolonged stress, and with the LIP/LAP ratio showing a transient decrease followed by a large increase (Fig. 7C). The changes in the LIP/LAP ratio were similar in nuclear and total extracts of Tg-treated cells. C/EBPβ mRNA levels showed a smaller increase (2–3-fold) than protein levels during ER stress (Fig. 7F), as reported previously (15). The C/EBPβ–/– (LIP) cells expressed LIP at all times, although there was a decrease in early stress followed by a large transient increase in late stress (Fig. 7D). Because the LIP expression construct only contains the LIP ORF (Fig. 1E), these changes are probably not because of translational control; changes in LIP stability are a more likely explanation. Furthermore, the sustained LIP levels in WT MEFs in late ER stress suggest efficient translation of the LIP ORF in the endogenous C/EBPβ mRNA but not in the transfected LIP expression plasmid.
FIGURE 7.
Induction of stress-response proteins in C/EBPβ+/+, C/EBPβ–/–, and C/EBPβ–/– (LIP) MEFs. WT, mutant, and C/EBPβ–/– cells stably expressing LIP (C/EBPβ–/– (LIP)) were treated with Tg for the indicated times. A and C–E, Western blot analysis of nuclear (A) or total (C–E) cell extracts probed with antibodies for the indicated proteins. B, quantification of the LIP/LAP ratio from A. F, quantification of C/EBPβ mRNA levels in MEFs treated with Tg for the indicated times. Data from qRT/PCR analysis of total RNA using C/EBPβ specific primers were normalized to the signal of 18 S ribosomal RNA. CON, control.
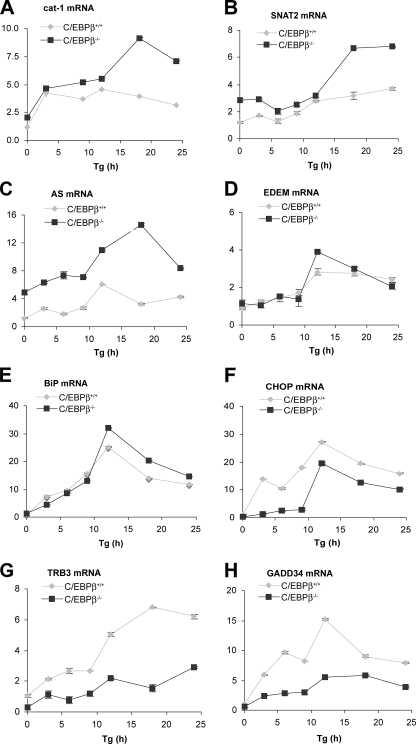
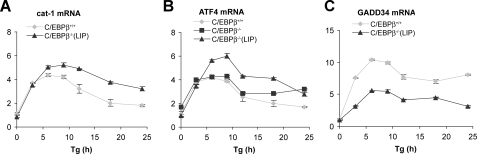
Next, mRNA levels of stress-response genes were analyzed by quantitative RT-PCR analysis on cells stressed with Tg (Fig. 8). Three patterns of expression were observed. 1) Some genes (AS and the amino acid transporters cat-1 and SNAT2) showed higher expression in C/EBPβ–/– cells than in wild-type cells, particularly after prolonged stress. These genes are known to be induced by ATF4 (15, 26, 54, 58). The attenuation of transcription was also seen in C/EBPβ–/– (LIP) cells (Fig. 9A). Similar regulation was seen for the AS gene (not shown). These observations are consistent with the inhibition of ATF4-induced transcription by elevated LIP during prolonged stress. 2) Some genes had lower expression levels in C/EBPβ–/– cells than in WT cells, suggesting that C/EBPβ is required for the induction of transcription. These genes encode CHOP (59), GADD34 (12), and TRB3 (9), which are either downstream of ATF2/ATF4 (CHOP) or CHOP (GADD34 and TRB3). This is probably an effect of LAP, because C/EBPβ–/– and C/EBPβ–/– (LIP) cells were similar (data not shown). 3) BiP and ER degradation enhancing α-mannosidase-like protein (EDEM) were expressed at similar levels in mutant and WT cells. The lack of induction is consistent with the regulation of these genes by ATF6 and XBP1, rather than ATF4. These data support our hypothesis that the expression of genes downstream of ATF4 is modulated by C/EBPβ. 4) The accumulation of ATF4 mRNA was increased more in C/EBPβ–/– (LIP) cells than in WT and C/EBPβ–/– cells (Fig. 9B), suggesting that LIP has stimulatory effects on Atf4 gene expression in the absence of LAP. The mechanism of this regulation needs further investigation.
FIGURE 8.
C/EBPβ differentially regulates expression of genes in the PERK/ATF4 pathway during ER stress. Quantification of mRNA levels for the indicated genes in C/EBPβ–/– and C/EBPβ+/+ MEFs treated with Tg for the indicated times. Data from qRT/PCR analysis of total RNA using gene-specific primers were normalized to the 18 S ribosomal RNA signal.
FIGURE 9.
Attenuation of cat-1 gene transcriptional activation in C/EBPβ–/– (LIP) cells. MEF cells (C/EBPβ+/+, C/EBPβ–/–, and C/EBPβ–/– (LIP)) were treated with Tg for the indicated times, and cat-1, ATF4, and GADD34 mRNA levels were analyzed by qRT-PCR as in Fig. 8. B shows results from all three cell lines, whereas A and C show only the C/EBPβ+/+ and C/EBPβ–/– (LIP) cells.
C/EBPβ–/– MEF Cells Have Lower Levels of GADD34 and CHOP Proteins and Reduced eIF2α Dephosphorylation—We have shown that the induction of CHOP and GADD34 mRNAs during ER stress is lower in C/EBPβ–/– than in WT cells (Figs. 8 and 9). To demonstrate that the changes in mRNA levels cause changes in protein levels, we measured these proteins by Western blotting during ER stress in WT and mutant cells. As expected, CHOP was induced more slowly in mutant cells and accumulated to lower levels (Fig. 7A). In contrast, BiP levels were induced to a similar extent in both cells, consistent with the similar induction in mRNA levels (Fig. 8). GADD34 levels were also 2-fold lower in mutant cells (Fig. 7E). In agreement with the decreased levels of GADD34, eIF2α phosphorylation was sustained in C/EBPβ–/– and C/EBPβ–/– (LIP) cells. In contrast, WT MEFs showed a transient decrease in eIF2α phosphorylation (Fig. 7C), which is a hallmark of the stress response in many cell types, including C6 cells (48). ATF4 levels were higher in the mutant and C/EBPβ–/– (LIP) cells, consistent with the sustained eIF2α phosphorylation. Furthermore, the levels of the transcriptional repressor ATF3, which is downstream of ATF4, were also higher in the mutant cells. These data are consistent with the hypothesis that LAP modulates the ER stress response by enhancing expression of genes encoding proteins that promote cell death (CHOP, GADD34, and TRB3) and attenuating expression of genes encoding proteins involved in amino acid synthesis and transport (cat-1, AS, and SNAT2).
DISCUSSION
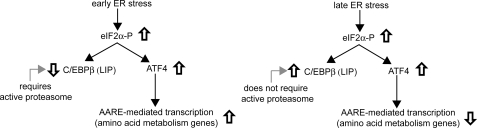
C/EBP transcription factors are involved in a variety of physiological processes, such as metabolic regulation, cellular differentiation, and stress responses (14, 15, 60, 61). In the case of C/EBPβ, translational control leads to synthesis of the transcriptional activators LAP-1 and LAP-2 and the transcriptional repressor LIP from a single mRNA (34, 62). The cellular LIP/LAP ratio has been suggested to be an important determinant of regulation of gene expression (35, 63, 64). It is shown in this study that LAP and LIP levels are regulated during ER stress. The findings can be summarized as follows. (i) Total LIP levels decrease during the early response (0–3 h), resulting in a 10-fold decrease in the LIP/LAP ratio. This decrease requires proteasome activity and the phosphorylation of eIF2α; it is paralleled by a decrease in C/EBPβ mRNA translation. (ii) Total LIP levels increase during the late ER stress response (>9 h), leading to a 30-fold increase of the nuclear LIP/LAP ratio because of a more efficient translation of the C/EBPβ mRNA and increased LIP protein. (iii) Expression of C/EBPβ proteins during ER stress has two opposite roles; it attenuates expression of some genes involved in amino acid metabolism (via LIP) and it enhances expression of proapoptotic genes, such as CHOP and its downstream targets (via LAP).
Differential Regulation of LAP and LIP Levels during ER Stress—A novel finding of this study is the complex changes in LIP and LAP levels during ER stress. This observation raises questions about the mechanisms that change the synthesis and degradation rates of these proteins and their physiological significance. C/EBPβ mRNA translation decreases during early stress and recovers during late stress. This is explained at least in part by the effects of eIF2α phosphorylation, which causes a decline of global protein synthesis in early stress (1). Moreover, because LIP has a shorter half-life than LAP (2 h for LIP and 6 h for LAP), decreased translation of C/EBPβ mRNA can account for the more pronounced reduction in LIP during the early period. During late stress, the efficient translation of C/EBPβ mRNA contributes to the increases in both LIP and LAP. However the increase in the LIP/LAP ratio suggests that there is preferential accumulation of LIP. Because these proteins are synthesized from a single mRNA, the stress response may regulate translation. This novel mode of regulation is an avenue that we will pursue in the future. A previous report suggested that CUGBP1 preferentially increases LIP translation (65). However, we found that depletion of CUGBP1 did not affect the accumulation of LIP during late ER stress (not shown).
Degradation also plays a role in the regulation of both LIP and LAP during ER stress. Our experiments with MG132 suggest that these proteins are degraded by the proteasome. The shorter half-life of LIPs could be due to differences in the N termini of the proteins or to differences in their molecular interactions, as has been shown for the homologue proteins of C/EBPβ and TRB3 in Drosophila (9, 66). Cyclin D1 is also degraded by proteasomes during ER stress (67). Although eIF2α phosphorylation is required for the decreased levels of both proteins, the decline is accompanied by decreased mRNA translation of LIP but sustained translation of cyclin D1.
Our data on LIP levels during ER stress present a paradox. Measurements of LIP stability with CHX showed that half-lives were similar in early and late stress. In contrast, MG132 increased LIP levels during early but not late stress, suggesting that LIP is more stable in late stress. Because the LIP level increases in late stress, our data could be explained by the fact that the rate of synthesis exceeds the rate of degradation, making the effects of MG132 difficult to detect. It is also possible that the drugs have indirect effects and alter LIP stability by their actions on other proteins. This could be the case in early stress if CHX indirectly inhibits LIP degradation. Although metabolic labeling to examine the LIP half-life would address this question, the labeling conditions will alter amino acid pools and induce stress by amino acid starvation. A striking finding was the accumulation of LIP protein during stress in C/EBPβ–/– (LIP) cells. The changes in LIP levels during ER stress in these cells paralleled those seen for protein expressed from the C/EBPβ mRNA in wild-type cells with one difference; accumulation of LIP in late ER stress was transient in C/EBPβ–/– (LIP) and continuous in C/EBPβ+/+ cells. These data suggest that LIP accumulation during ER stress is controlled in part by degradation of the protein, with rapid degradation during early stress and slower degradation in late stress. This conclusion appears to contradict the half-lives measured using inhibitors of protein synthesis, which gave similar values in early and late stress. The most likely explanation for this discrepancy is that a short-lived protein is involved in the control of LIP degradation during ER stress and that protein synthesis inhibitors interfere with normal regulation. We are currently investigating this hypothesis.
The proteasome plays a key role in the ER stress response. It has a pro-survival role in the early phase by degrading accumulated unfolded proteins and a pro-apoptotic role in the late phase by degrading pro-survival proteins such as Bcl-2 (68, 69). It is therefore possible that changes in LIP levels involve proteasome-mediated degradation of LIP in the pro-survival phase and protection of degradation by the proteasome in the proapoptotic phase of stress.
LIP Attenuates Transcriptional Induction of the PERK/eIF2α/ATF4 Arm of the Cellular Response to ER Stress—What is the physiological significance of the regulation of LIP levels during ER stress? We show that amino acid starvation, another stress that involves eIF2α phosphorylation, only caused small changes in the LIP/LAP ratio during prolonged stress (Fig. 5), suggesting that stress-specific mechanisms are involved in the regulation of C/EBPβ gene expression. Because LIP levels decrease during the pro-survival phase of stress and increase during the pro-apoptotic phase, we can speculate on the significance of its function as a mediator of pro-survival or pro-apoptotic processes. In at least one report, it was shown that LIP can bind DNA with higher affinity than LAP (34) and may form heterodimers with other members from the bZIP transcription factor family (70). Heterodimers in cell extracts have been suggested with LAP or CHOP (24). Because LIP is missing the transactivation domain, it is believed to act as a transcriptional repressor by heterodimerizing with transcriptional activators and either sequestering them away from DNA target sites or binding the DNA targets and conferring reduced transcriptional activation. Because of these properties, the ratio of LIP to potential partner activators can be critical for gene regulation (19, 34, 35). This is clearly demonstrated by the decreased ability of ATF4 in cell extracts with increased LIP to bind the AARE (Fig. 6E).
The early response of cells to ER stress involves the activation of the PERK/eIF2α signaling, which attenuates global protein synthesis and increases translation of specific mRNAs, among them the mRNA for the transcription factor ATF4 (7). The PERK/eIF2α/ATF4 pathway has been implicated in the induction of genes involved in amino acid transport (cat-1 and SNAT2) and biosynthesis (asparagine synthase), glutathione biosynthesis, and protection against oxidative stress (1). We found that LIP attenuated ATF4-mediated transcription from the gene encoding cat-1, in agreement with its function as a transcriptional repressor. It is therefore likely that reduced LIP levels early in the ER stress response enable ATF4-mediated transcriptional induction of target genes.
In an attempt to determine the role of C/EBPβ in the ER stress response, we compared mRNA and protein levels of genes that are targets of the PERK/eIF2α/ATF4 pathway in wild-type and C/EBPβ–/– MEFs. This signaling pathway targets pro-survival genes early in ER stress (3) as mentioned above. However, it also induces pro-apoptotic genes during prolonged stress (71). Among the pro-apoptotic genes are the ones shown to require CHOP for their induction during stress, such as those encoding TRB3 and GADD34 (9, 12). We observed delayed and reduced accumulation of CHOP mRNA and protein during ER stress (Figs. 7 and 8). It has been documented that PERK/eIF2α signaling is an absolute requirement for induction of CHOP gene transcription during ER stress and that ATF4 may activate CHOP transcription during the early stress response by binding to an AARE (6, 49). Our data show that induction of CHOP expression did not occur in the early stress response in C/EBPβ–/– cells despite high levels of ATF4 and induction of other ATF4 target genes (Figs. 7 and 8). We can also speculate that eIF2α phosphorylation early in ER stress induces CHOP expression in two ways as follows: (i) phosphorylation increases ATF4 expression; (ii) phosphorylation decreases LIP levels, allowing ATF4 to form heterodimers with the activator LAP. In agreement with increased transcriptional activity of LAP during the early stress response is its increased phosphorylation at Thr-188 (rat sequence; data not shown) as shown previously in other systems (72). Regulation of CHOP gene expression via ATF4-C/EBPβ heterodimers binding to the CHOP AARE has been suggested (73). The CHOP gene promoter is also a target for the IRE-1 and ATF6 pathway, which has been implicated in induction of CHOP transcription in late ER stress (74). Our data are consistent with C/EBPβ inducing transcription of the CHOP gene early but not late stress response and are supported by a recent report that C/EBPβ binds to the CHOP gene promoter in vivo earlier than XBP-1 during ER stress (52).
The disruption of the C/EBPβ gene had opposite effects on CHOP, GADD34, and TRB3 expression as compared with AS, cat-1, and SNAT2 during ER stress. The induction of the latter mRNAs was significantly increased in C/EBPβ–/– cells during the late response, suggesting a loss of a transcriptional repressor during this phase of ER stress. We have shown in this study that LIP attenuates ATF4-mediated cat-1 transcriptional activation via the AARE. ATF3 is also a transcriptional repressor of these genes, and it inhibits ATF4-mediated transcription via their AAREs (75, 76). However, in C/EBPβ–/– cells, in contrast to the higher levels of these mRNAs, ATF3 levels were higher than the WT cells (Fig. 7). This can be explained by the overexpression of ATF4 and the fact that ATF3 is downstream of ATF4 (47). Therefore, we can speculate that LIP levels, which increase dramatically during late ER stress, attenuate transcription of ATF4-mediated transcription of genes via AAREs. C/EBPβ has been suggested to be a negative regulator of expression of the AS gene, at a time when AS mRNA levels were declining during ER stress (23). Our findings can explain this correlation by LIP being the attenuator of positive regulation of AARE/ATF4-mediated gene transcription during late ER stress. A model summarizing the key finding of this work is shown in Fig. 10. It was recently shown that LAP may also inhibit induction of AS gene expression during amino acid starvation (50). Our studies showed that the expression of LIP only in MEFs resulted in attenuation of ATF4-mediated cat-1 transcription. The regulation of AARE-containing genes by LAP and LIP during stress needs further study to clarify these issues.
FIGURE 10.
LIP modulates the PERK/ATF4-mediated gene expression program during ER stress. Diagrammatic representation of the regulation and functional significance of LIP during ER stress.
C/EBPβ Is Required for Expression of Genes Downstream of CHOP—Induction of CHOP has been associated with pro-apoptotic functions (22). These functions include down-regulation of Bcl-2 and up-regulation of genes that either compromise cell survival or promote apoptosis (71, 77). CHOP can form heterodimers with either ATF4 or C/EBPβ proteins and induce gene expression during ER stress (9, 16). Two of its target genes encode the activator of protein phosphatase 1, GADD34, which is induced by CHOP as a heterodimer with either ATF4 or C/EBPβ, and TRB3, which is induced by CHOP/ATF4 heterodimers (9, 16). We show here that GADD34 and TRB3 expression was significantly reduced in C/EBPβ–/– cells. This is in agreement with a recent report that C/EBPβ induces TRB3 expression in mitogen-activated lymphocytes (78). The reduction of GADD34 is consistent with the sustained eIF2α phosphorylation in C/EBPβ–/– cells during ER stress (Fig. 7). This extended phosphorylation can explain the observation that ATF4 levels are higher in C/EBPβ–/– MEFs than in wild-type cells. However, the increased ATF4 levels may also be due to decreased levels of TRB3, which has been shown to attenuate ATF4-mediated transcription (79).
Our data support the idea that C/EBPβ is required for maximum induction of CHOP and its downstream targets. However, these genes were induced in C/EBPβ–/– cells, although at lower levels and with delayed kinetics. This is in contrast to a previous report on a different group of downstream genes called DOCs (24); induction of DOCs during ER stress was abolished in C/EBPβ–/– cells. It is therefore likely that GADD34 and TRB3 transcription is activated by homo- or heterodimers of ATF4 and CHOP in C/EBPβ–/– cells. However, because of the higher expression of these proteins in wild-type cells, it is likely that heterodimers of C/EBPβ with other transcription factors are the most efficient activators of these genes. We prepared MEFs that express LIP in the absence of LAP, and we showed that LIP attenuates expression of genes involved in amino acid metabolism. We also plan to develop LAP-expressing cells in the absence of LIP. The successful development of these cell types will be a valuable tool to dissect the importance of LAP and LIP on pro-apoptotic gene expression during ER stress. It is therefore left in future studies to determine whether LIP has pro-survival functions during late ER stress by regulating the concentrations of transcription factors that compete for crucial DNA-binding sites.
Physiological Significance of LAP/LIP Ratios during ER Stress and Future Directions—Our studies have demonstrated a physiological role of LIP in attenuation of ATF4-mediated transcription, in agreement with the role of ATF4 in the induction of genes required for amino acid metabolism and transport. Our studies will also shed light on diseases of bone formation, where high protein synthesis rates supported by high ATF4 contribute to skeletal abnormalities (80).
Finally, we have shown that Tu induced ER stress in mice and caused accumulation of LAP and LIP after 24 h of treatment in liver, kidney, and pancreas. Although the significance of the different LIP/LAP ratios in the three ER-stressed organs is not known, it is clear that ER stress regulates C/EBPβ levels in animals and the stress-response gene expression program is tissue-specific. The most striking observation is that induction of CHOP is abolished in the liver but maintained in the kidneys of C/EBPβ–/– mice. In view of the recent reports on the importance of ER stress response pathways in cell fate (13), future studies in mice can address the differential response of tissues to ER stress and the importance of the C/EBPβ transcription factor.
Previous studies in mice have shown that Tu treatments, similar to the one used here, caused impairment in renal function and death of kidney cells (22). In agreement with the implication of CHOP and GADD34 in apoptosis during the ER stress response, mice lacking these proteins showed protection from kidney damage following treatment with Tu (12, 22). Because C/EBPβ is the major heterodimerization partner of CHOP (16, 81), its expression has also been implicated in promoting cell death (82). In fact, ER stress-induced apoptosis was lower in C/EBPβ–/– MEFs than WT MEFs (22). Based on these findings, we conclude that changes in LAP and LIP levels during ER stress may be important modulators of the integrated response to stress and may participate in decisions of cell fate during severe stress conditions.
Supplementary Material
Acknowledgments
We thank Dr. Randal Kaufman (University of Michigan) for UPRE and ERSE plasmids and for S/S and A/A MEF cells. We also thank Carrie Millward for assistance with the animal experiments.
This work was supported, in whole or in part, by National Institutes of Health Grants DK060596, DK053307 (to M. H.), and DK075040 (to C. M. C.) and by National Institutes of Health Intramural Research Program of the Center for Cancer Research, NCI (to P. J.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table I.
Footnotes
The abbreviations used are: UPR, unfolded protein response; ATF, activating transcription factor; AARE, amino acid-response element; AS, asparagine synthase; BiP, immunoglobulin heavy chain-binding protein; bZIP, basic leucine zipper transcription factor; C/EBPβ, CCAAT/enhancer-binding protein β; CHOP, C/EBP homologous protein; CHX, cycloheximide; eIF2α, eukaryotic translation initiation factor 2α; ER, endoplasmic reticulum; ERSE, endoplasmic reticulum stress-response element; HDAC1, histone deacetylase 1; Hipp, hippuristanol; LAP, liver-enriched transcriptional activating protein; LIP, liver-enriched transcriptional inhibitory protein; MEFs, mouse embryo fibroblasts; ORF, open reading frame; PERK, PKR-like ER kinase; UPRE, unfolded protein-response element; SNAT2, sodium-coupled neutral amino acid transporter 2; TRB3, pseudokinase tribble 3; WT, wild type; FBS, fetal bovine serum; Tg, thapsigargin; Tu, tunicamycin; RT, reverse transcription; qRT-PCR, quantitative real-time PCR; DMEM, Dulbecco's modified Eagle's medium.
References
- 1.Schroder, M., and Kaufman, R. J. (2005) Mutat. Res. 569 29–63 [DOI] [PubMed] [Google Scholar]
- 2.Eizirik, D. L., Cardozo, A. K., and Cnop, M. (2008) Endocr. Rev. 29 42–61 [DOI] [PubMed] [Google Scholar]
- 3.Yoshida, H. (2007) FEBS J. 274 630–658 [DOI] [PubMed] [Google Scholar]
- 4.Zhang, K., and Kaufman, R. J. (2006) Neurology 66 S102–S109 [DOI] [PubMed] [Google Scholar]
- 5.Bertolotti, A., Zhang, Y., Hendershot, L. M., Harding, H. P., and Ron, D. (2000) Nat. Cell Biol. 2 326–332 [DOI] [PubMed] [Google Scholar]
- 6.Harding, H. P., Novoa, I., Zhang, Y., Zeng, H., Wek, R., Schapira, M., and Ron, D. (2000) Mol. Cell 6 1099–1108 [DOI] [PubMed] [Google Scholar]
- 7.Vattem, K. M., and Wek, R. C. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siu, F., Bain, P. J., LeBlanc-Chaffin, R., Chen, H., and Kilberg, M. S. (2002) J. Biol. Chem. 277 24120–24127 [DOI] [PubMed] [Google Scholar]
- 9.Ohoka, N., Yoshii, S., Hattori, T., Onozaki, K., and Hayashi, H. (2005) EMBO J. 24 1243–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamamoto, K., Sato, T., Matsui, T., Sato, M., Okada, T., Yoshida, H., Harada, A., and Mori, K. (2007) Dev. Cell 13 365–376 [DOI] [PubMed] [Google Scholar]
- 11.Calfon, M., Zeng, H., Urano, F., Till, J. H., Hubbard, S. R., Harding, H. P., Clark, S. G., and Ron, D. (2002) Nature 415 92–96 [DOI] [PubMed] [Google Scholar]
- 12.Marciniak, S. J., Yun, C. Y., Oyadomari, S., Novoa, I., Zhang, Y., Jungreis, R., Nagata, K., Harding, H. P., and Ron, D. (2004) Genes Dev. 18 3066–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin, J. H., Li, H., Yasumura, D., Cohen, H. R., Zhang, C., Panning, B., Shokat, K. M., Lavail, M. M., and Walter, P. (2007) Science 318 944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, C., Dudenhausen, E., Chen, H., Pan, Y. X., Gjymishka, A., and Kilberg, M. S. (2005) Biochem. J. 391 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, C., Dudenhausen, E. E., Pan, Y. X., Zhong, C., and Kilberg, M. S. (2004) J. Biol. Chem. 279 27948–27956 [DOI] [PubMed] [Google Scholar]
- 16.Sok, J., Wang, X. Z., Batchvarova, N., Kuroda, M., Harding, H., and Ron, D. (1999) Mol. Cell. Biol. 19 495–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calkhoven, C. F., Muller, C., and Leutz, A. (2000) Genes Dev. 14 1920–1932 [PMC free article] [PubMed] [Google Scholar]
- 18.Nerlov, C. (2007) Trends Cell Biol. 17 318–324 [DOI] [PubMed] [Google Scholar]
- 19.Zahnow, C. A., Younes, P., Laucirica, R., and Rosen, J. M. (1997) J. Natl. Cancer Inst. 89 1887–1891 [DOI] [PubMed] [Google Scholar]
- 20.Croniger, C. M., Millward, C., Yang, J., Kawai, Y., Arinze, I. J., Liu, S., Harada-Shiba, M., Chakravarty, K., Friedman, J. E., Poli, V., and Hanson, R. W. (2001) J. Biol. Chem. 276 629–638 [DOI] [PubMed] [Google Scholar]
- 21.Uematsu, S., Kaisho, T., Tanaka, T., Matsumoto, M., Yamakami, M., Omori, H., Yamamoto, M., Yoshimori, T., and Akira, S. (2007) J. Immunol. 179 5378–5386 [DOI] [PubMed] [Google Scholar]
- 22.Zinszner, H., Kuroda, M., Wang, X., Batchvarova, N., Lightfoot, R. T., Remotti, H., Stevens, J. L., and Ron, D. (1998) Genes Dev. 12 982–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siu, F., Chen, C., Zhong, C., and Kilberg, M. S. (2001) J. Biol. Chem. 276 48100–48107 [DOI] [PubMed] [Google Scholar]
- 24.Wang, X. Z., Kuroda, M., Sok, J., Batchvarova, N., Kimmel, R., Chung, P., Zinszner, H., and Ron, D. (1998) EMBO J. 17 3619–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez, J., Lopez, A. B., Wang, C., Mishra, R., Zhou, L., Yaman, I., Snider, M. D., Hatzoglou, M., and Hatzolgou, M. (2003) J. Biol. Chem. 278 50000–50009 [DOI] [PubMed] [Google Scholar]
- 26.Lopez, A. B., Wang, C., Huang, C. C., Yaman, I., Li, Y., Chakravarty, K., Johnson, P. F., Chiang, C. M., Snider, M. D., Wek, R. C., and Hatzoglou, M. (2007) Biochem. J. 402 163–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baer, M., and Johnson, P. F. (2000) J. Biol. Chem. 275 26582–26590 [DOI] [PubMed] [Google Scholar]
- 28.Sebastian, T., Malik, R., Thomas, S., Sage, J., and Johnson, P. F. (2005) EMBO J. 24 3301–3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandez, J., Yaman, I., Mishra, R., Merrick, W. C., Snider, M. D., Lamers, W. H., and Hatzoglou, M. (2001) J. Biol. Chem. 276 12285–12291 [DOI] [PubMed] [Google Scholar]
- 30.Millward, C. A., Heaney, J. D., Sinasac, D. S., Chu, E. C., Bederman, I. R., Gilge, D. A., Previs, S. F., and Croniger, C. M. (2007) Diabetes 56 161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yaman, I., Fernandez, J., Sarkar, B., Schneider, R. J., Snider, M. D., Nagy, L. E., and Hatzoglou, M. (2002) J. Biol. Chem. 277 41539–41546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chodosh, L. A. (2001) in Current Protocols in Molecular Biology (Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K., eds) pp. 12.6.1–12.6.9, John Wiley and Sons, Hoboken
- 33.Zahnow, C. A. (2002) Breast Cancer Res. 4 113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Descombes, P., and Schibler, U. (1991) Cell 67 569–579 [DOI] [PubMed] [Google Scholar]
- 35.Raught, B., Liao, W. S., and Rosen, J. M. (1995) Mol. Endocrinol. 9 1223–1232 [DOI] [PubMed] [Google Scholar]
- 36.Scheuner, D., Mierde, D. V., Song, B., Flamez, D., Creemers, J. W., Tsukamoto, K., Ribick, M., Schuit, F. C., and Kaufman, R. J. (2005) Nat. Med. 11 757–764 [DOI] [PubMed] [Google Scholar]
- 37.Fels, D. R., and Koumenis, C. (2006) Cancer Biol. Ther. 5 723–728 [DOI] [PubMed] [Google Scholar]
- 38.Li, J., and Lee, A. S. (2006) Curr. Mol. Med. (Hilversum) 6 45–54 [DOI] [PubMed] [Google Scholar]
- 39.Li, M., Baumeister, P., Roy, B., Phan, T., Foti, D., Luo, S., and Lee, A. S. (2000) Mol. Cell. Biol. 20 5096–5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oyadomari, S., Araki, E., and Mori, M. (2002) Apoptosis 7 335–345 [DOI] [PubMed] [Google Scholar]
- 41.Wojcik, C., Rowicka, M., Kudlicki, A., Nowis, D., McConnell, E., Kujawa, M., and DeMartino, G. N. (2006) Mol. Biol. Cell 17 4606–4618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yerlikaya, A., Kimball, S. R., and Stanley, B. A. (2008) Biochem. J. 412 579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang, H. Y., and Wek, R. C. (2005) J. Biol. Chem. 280 14189–14202 [DOI] [PubMed] [Google Scholar]
- 44.Mazroui, R., Di Marco, S., Kaufman, R. J., and Gallouzi, I. E. (2007) Mol. Biol. Cell 18 2603–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindqvist, L., Oberer, M., Reibarkh, M., Cencic, R., Bordeleau, M. E., Vogt, E., Marintchev, A., Tanaka, J., Fagotto, F., Altmann, M., Wagner, G., and Pelletier, J. (2008) PLoS. ONE 3 e1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu, P. D., Harding, H. P., and Ron, D. (2004) J. Cell Biol. 167 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang, H. Y., Wek, S. A., McGrath, B. C., Lu, D., Hai, T., Harding, H. P., Wang, X., Ron, D., Cavener, D. R., and Wek, R. C. (2004) Mol. Cell. Biol. 24 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fernandez, J. M., Yaman, I., Sarnow, P., Snider, M. D., and Hatzoglou, M. (2002) J. Biol. Chem. 277 19198–19205 [DOI] [PubMed] [Google Scholar]
- 49.Averous, J., Bruhat, A., Jousse, C., Carraro, V., Thiel, G., and Fafournoux, P. (2004) J. Biol. Chem. 279 5288–5297 [DOI] [PubMed] [Google Scholar]
- 50.Thiaville, M. M., Dudenhausen, E. E., Zhong, C., Pan, Y. X., and Kilberg, M. S. (2007) Biochem. J. 410 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oyadomari, S., and Mori, M. (2004) Cell Death Differ. 11 381–389 [DOI] [PubMed] [Google Scholar]
- 52.Donati, G., Imbriano, C., and Mantovani, R. (2006) Nucleic Acids Res. 34 3116–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baumeister, P., Luo, S., Skarnes, W. C., Sui, G., Seto, E., Shi, Y., and Lee, A. S. (2005) Mol. Cell. Biol. 25 4529–4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palii, S. S., Chen, H., and Kilberg, M. S. (2004) J. Biol. Chem. 279 3463–3471 [DOI] [PubMed] [Google Scholar]
- 55.Ma, Y., and Hendershot, L. M. (2003) J. Biol. Chem. 278 34864–34873 [DOI] [PubMed] [Google Scholar]
- 56.Shang, J., and Lehrman, M. A. (2004) Biochem. Biophys. Res. Commun. 317 390–396 [DOI] [PubMed] [Google Scholar]
- 57.Lee, A. H., Iwakoshi, N. N., and Glimcher, L. H. (2003) Mol. Cell. Biol. 23 7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaccioli, F., Huang, C. C., Wang, C., Bevilacqua, E., Franchi-Gazzola, R., Gazzola, G. C., Bussolati, O., Snider, M. D., and Hatzoglou, M. (2006) J. Biol. Chem. 281 17929–17940 [DOI] [PubMed] [Google Scholar]
- 59.Bruhat, A., Averous, J., Carraro, V., Zhong, C., Reimold, A. M., Kilberg, M. S., and Fafournoux, P. (2002) J. Biol. Chem. 277 48107–48114 [DOI] [PubMed] [Google Scholar]
- 60.Bezy, O., Vernochet, C., Gesta, S., Farmer, S. R., and Kahn, C. R. (2007) Mol. Cell. Biol. 27 6818–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seagroves, T. N., Krnacik, S., Raught, B., Gay, J., Burgess-Beusse, B., Darlington, G. J., and Rosen, J. M. (1998) Genes Dev. 12 1917–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ossipow, V., Descombes, P., and Schibler, U. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 8219–8223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gingras, A. C., Raught, B., and Sonenberg, N. (1999) Annu. Rev. Biochem. 68 913–963 [DOI] [PubMed] [Google Scholar]
- 64.Hu, B., Wu, Z., Jin, H., Hashimoto, N., Liu, T., and Phan, S. H. (2004) J. Immunol. 173 4661–4668 [DOI] [PubMed] [Google Scholar]
- 65.Dudaronek, J. M., Barber, S. A., and Clements, J. E. (2007) J. Immunol. 179 7262–7269 [DOI] [PubMed] [Google Scholar]
- 66.Rorth, P., Szabo, K., and Texido, G. (2000) Mol. Cell 6 23–30 [DOI] [PubMed] [Google Scholar]
- 67.Raven, J. F., Baltzis, D., Wang, S., Mounir, Z., Papadakis, A. I., Gao, H. Q., and Koromilas, A. E. (2008) J. Biol. Chem. 283 3097–3108 [DOI] [PubMed] [Google Scholar]
- 68.Kondratyev, M., Avezov, E., Shenkman, M., Groisman, B., and Lederkremer, G. Z. (2007) Exp. Cell Res. 313 3395–3407 [DOI] [PubMed] [Google Scholar]
- 69.Basu, A., and Haldar, S. (2002) Int. J. Oncol. 21 597–601 [PubMed] [Google Scholar]
- 70.Vinson, C., Myakishev, M., Acharya, A., Mir, A. A., Moll, J. R., and Bonovich, M. (2002) Mol. Cell. Biol. 22 6321–6335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Puthalakath, H., O'Reilly, L. A., Gunn, P., Lee, L., Kelly, P. N., Huntington, N. D., Hughes, P. D., Michalak, E. M., McKimm-Breschkin, J., Motoyama, N., Gotoh, T., Akira, S., Bouillet, P., and Strasser, A. (2007) Cell 129 1337–1349 [DOI] [PubMed] [Google Scholar]
- 72.Kim, J. W., Tang, Q. Q., Li, X., and Lane, M. D. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 1800–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen, H., Pan, Y. X., Dudenhausen, E. E., and Kilberg, M. S. (2004) J. Biol. Chem. 279 50829–50839 [DOI] [PubMed] [Google Scholar]
- 74.Yoshida, H., Okada, T., Haze, K., Yanagi, H., Yura, T., Negishi, M., and Mori, K. (2000) Mol. Cell. Biol. 20 6755–6767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hai, T., Wolfgang, C. D., Marsee, D. K., Allen, A. E., and Sivaprasad, U. (1999) Gene Expr. 7 321–335 [PMC free article] [PubMed] [Google Scholar]
- 76.Wolfgang, C. D., Chen, B. P., Martindale, J. L., Holbrook, N. J., and Hai, T. (1997) Mol. Cell. Biol. 17 6700–6707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McCullough, K. D., Martindale, J. L., Klotz, L. O., Aw, T. Y., and Holbrook, N. J. (2001) Mol. Cell. Biol. 21 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Selim, E., Frkanec, J. T., and Cunard, R. (2007) Mol. Immunol. 44 1218–1229 [DOI] [PubMed] [Google Scholar]
- 79.Jousse, C., Deval, C., Maurin, A. C., Parry, L., Cherasse, Y., Chaveroux, C., Lefloch, R., Lenormand, P., Bruhat, A., and Fafournoux, P. (2007) J. Biol. Chem. 282 15851–15861 [DOI] [PubMed] [Google Scholar]
- 80.Elefteriou, F., Benson, M. D., Sowa, H., Starbuck, M., Liu, X., Ron, D., Parada, L. F., and Karsenty, G. (2006) Cell Metab. 4 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ron, D., and Habener, J. F. (1992) Genes Dev. 6 439–453 [DOI] [PubMed] [Google Scholar]
- 82.Kapadia, R., Tureyen, K., Bowen, K. K., Kalluri, H., Johnson, P. F., and Vemuganti, R. (2006) J. Neurochem. 98 1718–1731 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.